Abstract
Quantitative assessment of post-translational modifications in proteins by mass spectrometry often requires the consideration of the alteration in ionization efficiency of peptides induced by the modification. Herein, we introduced a method to measure the relative ionization efficiencies of peptides using specifically designed unlabeled peptides. In our design, the peptide under study, in either the unmodified or modified form, is linked with an internal standard peptide via an enzyme cleavage site; thus, after enzymatic digestion, we could obtain readily a 1:1 ratio between the peptide under investigation and the internal standard peptide. The relative ionization efficiencies of the modified and unmodified peptides can then be calculated from the modification-induced change in the ratio of relative abundances of the ion of the peptide of interest over that of the internal standard peptide. We demonstrated the usefulness of the method by assessing the change in ionization efficiencies of four peptides introduced by phosphorylation.
Introduction
Post-translational modification (PTM) of a peptide often induces a change in the ionization efficiency of the peptide, which draws great concerns while mass spectrometric techniques are employed for the quantitative measurement of PTMs in proteins [1]. Methods have been developed to achieve the unbiased PTM analysis by using mass spectrometry [2]. In this respect, isotope-labeled internal standard was used to achieve absolute quantification of PTMs in proteins. For instance, isotope-coded affinity tag (ICAT) [3], stable isotope labeling with amino acids in cell culture (SILAC) [4], introducing 18O labeling by enzymatic digestion in H218O [5] have been employed. In addition, an isotope-dilution method, where the absolute or relative quantification of PTM levels of peptides are achieved by doping known amount of the synthetic isotope-labeled peptides into the peptide samples as the internal standard, was reported [6].
In recent years, an isotope-free method for the quantification of protein phosphorylation was also developed [7]. In this method, after normalizing against the signal of suitable proteolytic peptides, the extent of phosphorylation could be determined. In addition, methods for direct measurement of the ionization efficiency changes caused by PTM on peptides were reported [1, 8, 9]. In these prior studies, the synthetic peptides were first quantified by weight [8] or amino acid analysis [1] and, after comparing the LC-MS results with known quantification data, the variation in ionization efficiency caused by certain PTMs could be measured.
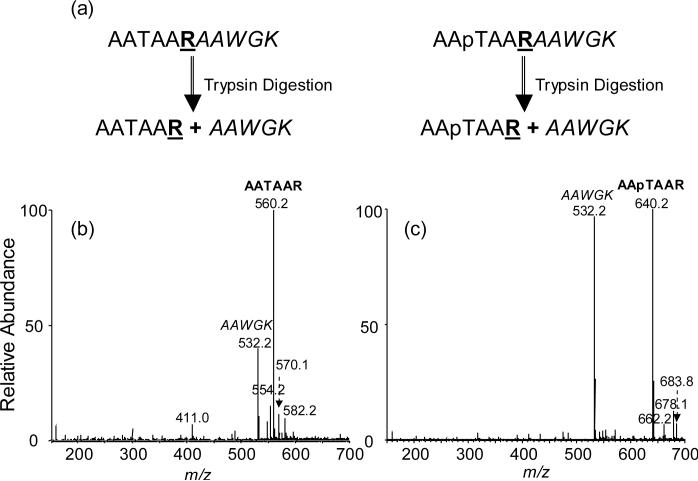
Herein we developed a simple isotope-free method to measure the change in ionization efficiency introduced by PTMs and we demonstrated that the method allowed for the measurement of the alteration in ionization efficiency introduced by phosphorylation of threonine residues in peptides. In our method, the target peptide, in the unmodified or phosphorylated form, is fused with a standard peptide through the recognition site of a proteolytic enzyme (Figure 1). After digestion with the protease, the standard peptide and the peptide under study can be released at a molar ratio of 1:1. The change in ionization efficiency can then be gauged from the alteration in the relative ion abundances of the peptide of interest over the standard peptide.
Figure 1.
Experimental Design: (a) Design of starting peptide; Positive-ion ESI-MS of the tryptic digestion mixtures of AATAARAAWGK (b) and AApTAARAAWGK (c).
Experimental
Synthetic peptides (Table 1) were obtained from Quality Controlled Biochemicals, Inc. (Hopkinton, MA) and purified by HPLC prior to use. The purified peptides were subjected directly to digestion by trypsin (Roche Applied Science, Indianapolis, IN) in a 50-mM NH4HCO3 buffer (pH 8.0) at 37°C for 18 hrs.
Table 1.
Sequences of synthetic peptides examined in this paper
| Peptide Number | Peptide Sequence |
|---|---|
| 1 | NH2-AAWGKAAAATR-COOH |
| 2 | NH2-AAWGKAAAA(pT)R-COOH |
| 3 | NH2-AATAARAAWGK-COOH |
| 4 | NH2-AA(pT)AARAAWGK-COOH |
| 5 | NH2-TAAAARAAWGK-COOH |
| 6 | NH2-(pT)AAAARAAWGK-COOH |
| 7 | NH2-AAAAKTTTTPGR-COOH |
| 8 | NH2-AAAAKTTT(pT)PGR-COOH |
* pT designates a phosphothreonine
MS experiments were carried out on an LCQ Deca XP ion-trap mass spectrometer (ThermoFinnigan, San Jose, CA). An equal-volume solvent mixture of acetonitrile and water with 0.6% acetic acid was used as the electrospray solvent. The MS experiments were carried out in either positive- or negative-ion mode, in which the spray voltages were 4.5 and 3.5 kV, respectively. Each spectrum reported here was averaged from approximately 75 scans, and the time for each scan was 0.1 s.
Results
We chose to develop a label-free mass spectrometry-based method for the measurement of the change in ionization efficiencies introduced by PTMs. As illustrated in Figure 1, our method begins with the enzymatic digestion of two synthetic peptides in which the peptide of interest, in either phosphorylated or unmodified form, is fused with an internal standard peptide via a trypsin recognition site. In this respect, the standard peptide can be conjugated to either the C- or N-terminus of the peptide of interest. After trypsin cleavage, the peptide under investigation and the internal standard peptide are released in equal quantities. The change in ionization efficiency introduced by phosphorylation can be determined from the variation in relative ion abundances of the peptide of interest (i.e., AATAAR or AApTAAR) over the internal standard peptide (AAWGK, Figure 1). In this context, it is important to emphasize that the synthetic fusion peptides need to be purified by HPLC to remove the truncated peptides, or failure sequences because their presence may result in the formation of the standard peptide or the peptide of interest, but not both, thereby introducing errors for the ionization efficiency measurement. In addition, it is worth noting that, other than the singly protonated species, we also found alkali metal ion-associated species in positive-ion ESI-MS (Figure 1b&c). The sum of intensities of peaks for the protonated and metal ion-associated species for the same peptide was employed for the calculation of the relative ion abundances.
By using this method, we determined the relative ionization efficiencies of three pairs of alanine-rich peptides which carry either an unmodified or phosphorylated threonine. The results showed that threonine phosphorylation apparently affects the ionization efficiencies of these peptides (Figure 1b&c, Table 2). In addition, the different location of phosphothreonine bears different effects on the ionization efficiencies (Table 2). For peptide 1, AAAATR, where threonine is neighboring to the C-terminal arginine, the ionization efficiency dropped by more than 60% after the threonine was phosphorylated (Table 2). However, while the threonine resides in the middle of the peptide sequence, i.e., AATAAR, the ionization efficiency was reduced by less than 30% upon phosphorylation (Table 2). In stark contrast, when threonine is placed on the N-terminus, that is, TAAAAR, the ionization efficiency of the peptide increased by 20% upon phosphorylation (Table 2). The reason for the different effects induced by the location of the phosphorylated residue is not clear, though we speculate that the intramolecular interactions, i.e., salt bridges, may change with the location of the phosphothreonine.
Table 2.
Relative ionization efficiency for phosphorylated over unphosphorylated peptides measured by ESI-MS via direct infusion. The data represent the mean and standard deviation of three independent measurements.
| Relative Ionization Efficiency | ESI Positive Mode | ESI Negative Mode |
|---|---|---|
| AAAApTR/AAAATR | 0.36 ± 0.03 | 1.0 ± 0.2 |
| AApTAAR/AATAAR | 0.76 ± 0.02 | 1.15 ± 0.09 |
| pTAAAAR/TAAAAR | 1.21 ± 0.09 | 1.1 ± 0.2 |
| TTTpTPGR/TTTTPGR | 1.1 ± 0.1 | 1.8 ± 0.3 |
Unlike in the positive-ion mode, the ionization efficiencies of peptides in the negative-ion mode did not vary with the location of the phosphorylated residue (Table 2). In addition, threonine phosphorylation leads to increase in the ionization efficiency. This result is consistent with the previous MALDI study [9] and is in line with the fact that a phosphorylated side chain can deprotonate readily to render the negatively charged peptide, thereby increasing the ionization efficiency. In this respect, the relative ionization efficiencies were measured following the same procedures as described above for the positive-ion mode.
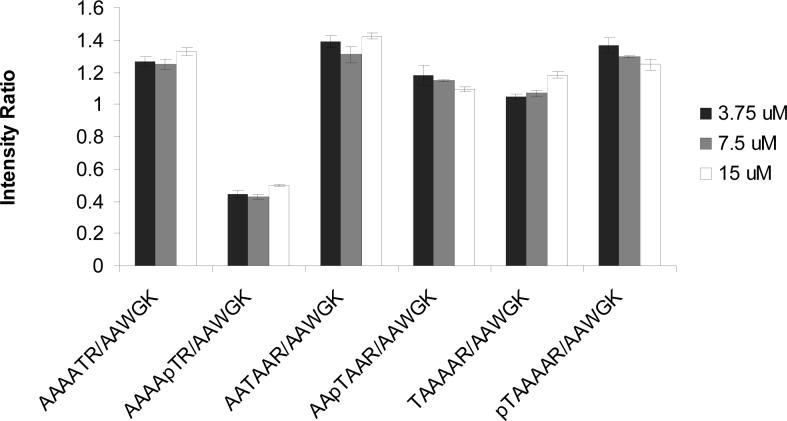
Next, we examined whether the concentration of the peptide sample would affect the measurement. For different peptide samples, we chose three different concentrations, that is, 15, 7.5 and 3.75 μM. It turned out that the peak intensity ratio between the target peptide and internal standard peptide did not vary appreciably with the concentrations of the peptides (Figure 2). We also carried out the measurements on different days, and it turned out that the variation among measurements on different days is within a few percent.
Figure 2.
The effects of concentration effects on the the intensity ratios, measured by positive-ion ESI-MS, between the target peptide and internal standard peptide. The data represent the mean and standard deviation of three independent measurements.
To further demonstrate the general applicability of our methods, we chose a peptide from the HMGA1a protein, T74TTTPGR80 , as the target peptide. In this regard, Thr77 along with other threonine and serine residues in this protein could be phosphorylated in PC-3 human prostate cancer cells [10]. Here, we designed a starting peptide sequence to explore the effects of phosphorylation on the ionization efficiency of the peptide containing Thr77 (Table 1, peptides 7 & 8, AAAAK served as the internal standard peptide).
We carried out the experiment by direct infusion method. Different from peptides 1−6, peptide 7, TTTTPGR, gave abundant doubly protonated ion, i.e., the ion of m/z 367.2. We also detected ions corresponding to sodium (+22 Da) and potassium (+38 Da) adducts. However, phosphorylation did not cause significant change to the ionization efficiency of the peptide, and the relative ionization efficiency of TTTpTPGR/TTTTPGR was measured to be 1.1±0.1 (Table 2).
Discussion and Conclusions
Measuring the relative ionization efficiencies of a post-translationally modified peptide and its unmodified counterpart is important for quantifying the level of PTMs by using mass spectrometry. Here we introduced an isotope-free method that can facilitate the determination of the change in ionization efficiency introduced by the PTMs. By using three alanine-rich peptides, we showed that threonine phosphorylation can lead to change in ionization efficiency of the peptides. In addition, the location of the phosphorylated threonine affects both the direction and the magnitude of the change in ionization efficiency. We believe that the method introduced in this paper will be very useful for the systematic studies of the effect of PTMs on the ionization efficiencies of peptides, which can be correlated with theoretical approaches to understand, in general, how a specific PTM affects the ionization process.
There are several advantages associated with our method. First, the method does not require isotope-labeled peptides. In addition, our experimental data supported that the difference in peptide concentration does not introduce obvious difference in the relative ionization efficiencies of the phosphorylated peptides and their unmodified counterparts (Figure 2). Therefore, the measured ionization efficiency difference between the unmodified and the corresponding modified peptide is not restricted to a certain concentration range. This is different from the isotope-dilution method [6] where the quantification of the isotope-labeled standard peptide is necessary. In this context, absolute quantifications of standard peptides are sometimes challenging. For instance, it is often difficult to measure peptides by weight because lyophilized peptides may contain bound water or salt. In addition, many peptides do not carry tryptophan or tyrosine residue, which prevent the quantification of these peptides by using spectrophotometric methods. Moreover, enzyme miss-cleavage does not interfere with the quantitative assessment of ionization efficiency because the miscleavage does not give rise to the peptide under study or the internal standard peptide. It is worth noting that one could also design starting peptides by linking directly the phosphorylated peptide with its unphosphorylated analog [11]. In this design, the introduction of internal standard peptides becomes unnecessary, which may reduce variations among measurements. The use of an internal standard peptide, however, facilitates the simultaneous quantification of multiple PTMs by using LC-MS/MS with a single reference peptide.
Our method also bears several shortcomings. First, the measurement can be difficult if the peptide of interest carries a methionine or cysteine because these two amino acids are susceptible to oxidative modification. Second, when attempting to measure the relative ionization efficiencies by LC-MS, we found that none of the alanine-rich peptides retained well on the LC column, which resulted in poor precision for the measurements (data not shown). Thus, the retention issue must be taken into consideration when designing the starting peptides for LC-MS measurements. We believe that the application of our design to LC-MS measurement of ionization efficiency is achievable if both internal reference peptide and the peptide under investigation have good retention on the LC column. In addition, one pair of peptides could only allow for the measurement of the effect of a specific modification on the ionization efficiency of one peptide. Therefore, the method might be costly for large-scale proteomic study where many different types of PTMs need to be quantified simultaneously. Moreover, sometimes the sequence of the standard peptide may need to be optimized so that its ionization efficiency is not drastically different from that of the peptide under investigation. Marked difference of this nature may introduce errors in the ionization efficiency determination because mass analyzers have a certain dynamic range for quantitative measurements. If these limitations are taken into consideration, the method constitutes a simple approach for assessing the change in peptide ionization efficiency caused by its PTMs.
Acknowledgment
The authors want to thank the National Institutes of Health for supporting this research (Grant No. R01 CA 116522).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Steen H, Jebanathirajah JA, Rush J, Morrice N, Kirschner MW. Phosphorylation analysis by mass spectrometry: myths, facts, and the consequences for qualitative and quantitative measurements. Mol. Cell. Proteomics. 2006;5:172–181. doi: 10.1074/mcp.M500135-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.McLachlin DT, Chait BT. Analysis of phosphorylated proteins and peptides by mass spectrometry. Curr. Opin. Chem. Biol. 2001;5:591–602. doi: 10.1016/s1367-5931(00)00250-7. [DOI] [PubMed] [Google Scholar]
- 3.Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- 4.Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–130. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- 5.Bonenfant D, Schmelzle T, Jacinto E, Crespo JL, Mini T, Hall MN, Jenoe P. Quantitation of changes in protein phosphorylation: a simple method based on stable isotope labeling and mass spectrometry. Proc. Natl. Acad. Sci. USA. 2003;100:880–885. doi: 10.1073/pnas.232735599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bronstrup M. Absolute quantification strategies in proteomics based on mass spectrometry. Expert Rev. Proteomics. 2004;1:503–512. doi: 10.1586/14789450.1.4.503. [DOI] [PubMed] [Google Scholar]
- 7.Steen H, Jebanathirajah JA, Springer M, Kirschner MW. Stable isotope-free relative and absolute quantitation of protein phosphorylation stoichiometry by MS. Proc. Natl. Acad. Sci. USA. 2005;102:3948–3953. doi: 10.1073/pnas.0409536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakanishi T, Shimizu A. Determination of ionization efficiency of glycated and non-glycated peptides from the N-terminal of hemoglobin beta-chain by electrospray ionization mass spectrometry. J. Chromatogr. B, Biomed. Sci. Appl. 2000;746:83–89. doi: 10.1016/s0378-4347(00)00115-8. [DOI] [PubMed] [Google Scholar]
- 9.Janek K, Wenschuh H, Bienert M, Krause E. Phosphopeptide analysis by positive and negative ion matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2001;15:1593–1599. doi: 10.1002/rcm.417. [DOI] [PubMed] [Google Scholar]
- 10.Jiang X, Wang Y. Acetylation and phosphorylation of high-mobility group A1 proteins in PC-3 human tumor cells. Biochemistry. 2006;45:7194–7201. doi: 10.1021/bi060504v. [DOI] [PubMed] [Google Scholar]
- 11.Lehmann WD, Ripkorn R, Hung C-W. Enzyme-cleavable tandem peptides as tools for determination of relative ionization efficiencies. MP402. 55th ASMS Conference on Mass Spectrometry; Indianapolis, IN. June 3 − 7, 2007. [Google Scholar]