Abstract
Ron receptor activation induces numerous cellular responses in vitro, including proliferation, dissociation, and migration. Ron is thought to be involved in blood cell development in vivo, as well as in many aspects of the immune response including macrophage activation, antigen presentation, and nitric oxide regulation. In previous studies to determine the function of Ron in vivo, mice were generated with a targeted deletion of the extracellular and transmembrane regions of this gene. Mice homologous for this deletion appear to die early during embryonic development. To ascertain the in vivo function of Ron in more detail, we have generated mice with a germline ablation of the tyrosine kinase domain. Strikingly, our studies indicate that this domain of Ron, and therefore Ron cytoplasmic signaling, is not essential for embryonic development. While mice deficient in this domain are overtly normal, mice lacking Ron signaling have an altered ability to regulate nitric oxide levels and, in addition, have enhanced tissue damage following acute and cell-mediated inflammatory responses.
Introduction
Tyrosine kinase receptors have been shown to regulate cellular migration, growth, and differentiation and are also involved in the defense against infectious agents and wound repair. One crucial family of multifunctional tyrosine kinase receptors is composed of the proto-oncogene products encoded by c-Met (the hepatocyte growth factor receptor), c-sea, and Ron/STK (stem cell–derived tyrosine kinase). These tyrosine kinases are synthesized as a single polypeptide chain of approximately 190 kDa. Each receptor undergoes proteolytic cleavage and is presented at the cell surface as a disulfide-linked heterodimer consisting of a 45-kDa α chain and a 150-kDa β chain. The heterodimeric receptors contain an extracellular ligand-binding domain, a single transmembrane-spanning region, and a highly conserved intracellular catalytic tyrosine kinase domain (1–3). Signaling following activation by the appropriate growth factor induces complex pleiotropic effects known as “invasive growth,” including proliferation, tubular morphogenesis, angiogenesis, cellular motility, and invasiveness (4, 5).
In the adult mouse, Ron is expressed ubiquitously (3, 6–8). Developmental expression studies in the mouse indicate that Ron is expressed in extraembryonic trophoblasts as early as embryonic day 3.5 (E3.5) (9). Ron expression in the embryo proper begins in the liver around day E12.5 and is followed by expression in the central nervous system, developing bone, lung, and glandular epithelia along the digestive tract (8, 10, 11). It is thought that Ron expression in the liver during early development may correlate with its involvement in hematopoiesis. This is consistent with recent findings suggesting that a truncated version of the Ron protein, possibly resulting from a transcript derived from an internal promoter, confers susceptibility to Friend virus–induced erythroleukemia, indicating a functional link between Ron and blood cell development (12).
The ligand for Ron is a serum protein called hepatocyte growth factor–like protein/macrophage-stimulating protein (HGFL/MSP) (3, 13, 14). To date, several functions for HGFL and Ron have been elucidated, including the ability to stimulate chemotactic migration of murine resident peritoneal macrophages (15, 16) and to stimulate ingestion of complement-coated erythrocytes via the macrophage CR3 receptor (17). Recently, in vitro activation of Ron in macrophages has been shown to inhibit inducible nitric oxide synthase (iNOS) expression and nitric oxide (NO) production following LPS or INF-γ stimulation (9, 18, 19). This inhibition appears to occur during the latter stages of the inflammatory response and is associated with phosphatidylinositol-3 kinase activation (18).
To determine in vivo functions for Ron and HGFL, mice carrying null alleles for either of the respective genes have been generated. Mice containing the targeted disruption of HGFL develop to term and have no obvious phenotypic abnormalities (20). These mice produce and sustain offspring. Two laboratories have produced mice deficient in portions of Ron with different results (9, 11). Our laboratory has reported that deletion of exons 1–14 of the Ron gene (the extracellular and transmembrane portion of Ron) leads to early embryonic death before the peri-implantation period (9). Conversely, mice with a targeted deletion of the first exon of Ron develop normally, are fertile, and display no obvious developmental defects (11). However, in support of previous in vitro studies, macrophages isolated from each of these mice exhibit increased levels of NO after exposure to LPS and INF-γ in vitro. In vivo, an increased sensitivity to endotoxic shock was exhibited, strengthening the link of Ron’s involvement in modulating the inflammatory response (9, 11).
Given the fact that the loss of the extracellular and transmembrane domain of Ron results in early embryonic lethality, and a limited deletion of the extracellular domain of Ron is compatible with life, we set out to determine the biological role of a specific domain of this pivotal receptor. Remarkably, our studies indicate that the cytoplasmic tyrosine kinase-containing (TK-containing) domain of Ron is dispensable for mouse development. Adult mice devoid of the intracellular domain of Ron are phenotypically normal with inflammatory defects apparent only upon stress.
Methods
Gene targeting of the mouse Ron TK domain.
The mouse Ron gene isolated from a 129/Ola genomic library was used to construct a replacement-type targeting vector (8). A 938-bp PCR fragment encompassing exons 10–12 (Ron gene nucleotides 10,235 to 11,173 (8), was cloned into pKOV901 (Lexicon Genetics Inc., The Woodlands, Texas, USA). The long arm of homology consisted of a 2.3-kb PCR fragment encompassing nucleotides 13,628 to 15,942 of the Ron gene (see Figure 1). This arm contained a loxP site. The internal portion of the targeting vector was a PCR product containing 5′ and 3′ loxP sites surrounding nucleotides 11,235 to 13,620 of the Ron gene. For positive selection, a hypoxanthine phosphoribosyltransferase (HPRT) cassette was subcloned from pKOSelect HPRT into the unique AcsI site of pKOV901. For negative selection, a thymidine kinase expression cassette was subcloned from pKOSelect TK (Lexicon Genetics Inc.) and placed into the RsrII site of pKOV901. The targeting vector was linearized with NotI and introduced into mouse embryonic stem (ES) cells as described previously (9).
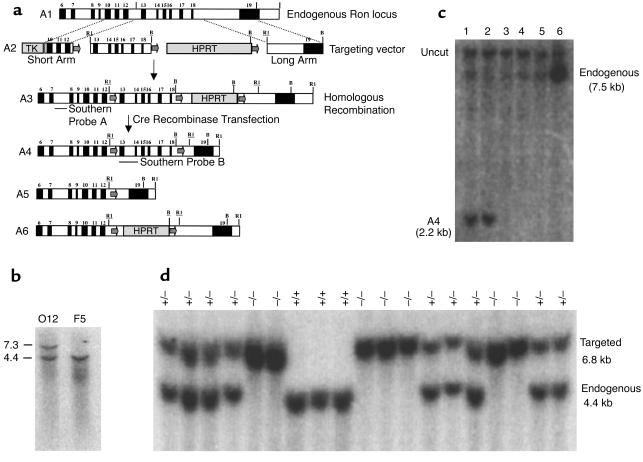
Figure 1.
Shown are the Ron genomic locus (A1), the targeting vector (A2), the resulting locus obtained after homologous recombination (A3), and the resulting products obtained after Cre recombinase–mediated excision (A4, A5, A6). LoxP recombinase recognition sites are indicated by arrows. Homologous arms are indicated by text and by dotted lines. Exons are indicated by black boxes. Probes for Southern blot analyses are indicated. Restriction enzyme notations are as follows, with underlining representing sites specific for the targeted locus: B, BamH1; RI, EcoR1. (b) Southern blot analysis of a positively selected ES clone (12) following electroporation of the targeting vector. (c) Transient transfection of targeted ES cell clone O12 with Cre recombinase will result in three types of Cre-mediated excision (A4, A5, A6). Transient transfected ES cell lysates were digested with EcoR and were probed with probe B. Lanes 1 and 2 have DNA from ES cell clones that have undergone the A4 recombination event, while loss of the TK domain in lanes 3–6 suggests ES cells clones were obtained from the A5 recombination event. (d) Southern blot analysis of BamHI-digested DNA isolated from mice of Ron TK+/– crosses containing the A5 recombination event, probed with probe A.
ES cell clones that had undergone homologous recombination were identified by Southern analyses on BamHI-digested genomic DNA using a probe outside the targeting vector. Targeted ES cell clone O12 (Figure 1b) was used further to recombine regions encompassed by the loxP sites. For this recombination, 107 cells were transfected with 10 μg of the supercoiled plasmid pML78, which contains a Cre recombinase cDNA with a nuclear localization signal driven by the human β-actin promoter. Four days after electroporation, the ES cells were split 1:2, and the next day the cells were placed into complete media containing 6-thioguanine (6-TG; Sigma Chemical Co., St. Louis, Missouri, USA). Approximately 5 to 7 days later, individual resistant clones were isolated, expanded, and analyzed. Specific Cre-driven recombination events were analyzed by Southern blot analyses using multiple probe sets.
Generation of mouse colonies and genotype analysis.
Given that it was not known if viable mice would be obtained following a germline deletion of the TK domain of Ron, two targeted ES cell clones from each of the A4 and A5 lines (Figure 1a) were used to generate chimeric founders. Chimeric founders generated from each ES cell line were crossed to NIH Black Swiss mice to generate F1 offspring. Hemizygous F1 offspring were crossed to NIH Black Swiss females to generate F2 offspring. F2 hemizygous mice were mated to produce mice homozygous for the given Ron mutation. In the case of the A4 line, the mutation would generate loxP sites flanking the TK domain (exons 13 through 18 of the Ron gene). To conditionally delete the floxed allele, F3 offspring were then crossed to HNF3αCre-expressing mice (R. Behr and K.H. Kaestner, unpublished observations). The HNF3αCre mice were in a CD-1 background. Mice from this latter mating that were hemizygous for both Cre and the floxed Ron TK allele were mated to mice that were homozygous for the floxed Ron TK allele. Mice derived from the A4 and A5 lines were found to have similar phenotypes and thus mice containing a homozygous deletion of Ron TK and cytoplasmic domains are collectively referred to as Ron TK–/– mice in this report. Wild-type mice (for the A5 line) or mice with loxP sites surrounding the TK domain of Ron are collectively referred to as Ron TK+/+.
Derivation of HNF3αCre mice.
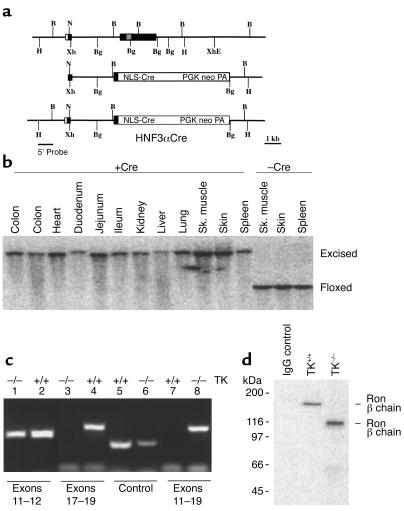
A gene-targeting vector was constructed to replace exon 2 of the HNF3α gene with a nuclear localization sequence–Cre (NLS-Cre) cassette. The vector design was similar to that used previously to replace HNF3α with the lacZ gene (21). The targeting vector containing 9.6 kb of HNF3α homology was electroporated into E14/1 ES cells and targeted clones identified as described previously (21). Of 300 ES cell clones analyzed, five had undergone targeted gene replacement (Figure 2a). Chimeric mice were derived from the targeted ES cells as described above and used to establish the HNF3αCre line.
Figure 2.
HNF3αCre targeting strategy and characterization of the mutant mice. (a) The top line depicts the gene structure of the HNF3α gene. Exons are indicated as boxes; the gray box represents the winged helix domain. The targeting vector used for homologous recombination is shown in the middle line, while the structure of the targeted allele (HNF3αCre) is shown on the bottom. PGK neo PA was used for selection. The 5′ probe was used for the Southern blot analyses to identify targeted clones. B, BamHI; Bg, BglII; E, EcoRI; H, HindIII; N, NotI; Xh, XhoI. (b) Southern hybridization analysis was performed on DNA from the indicated organs of an experimental and a control mouse. Complete Cre-mediated recombination was seen in every tissue analyzed in the experimental animals (+Cre, Excised) compared with the control mice (–Cre, Floxed). (c) Ron expression was evaluated by RT-PCR from RNA from control (TK+/+; lanes 2, 4, 5, and 7) and experimental (TK–/–; lanes 1, 3, 6, and 8) mice. Expression of a RT-PCR product specific for exons 11 and 12 is shown in lanes 1 and 2. Primers specific for the tyrosine kinase domain of Ron is shown in lanes 3 and 4. Primers specific for a truncated Ron gene product (with primers in exons 11 and 19) are shown in lanes 7 and 8. GAPDH expression is shown in lanes 5 and 6. (d) Western blot analysis of Ron from resident peritoneal macrophages. A truncated form of Ron is expressed in the experimental animals.
Ron expression and protein analysis.
Total RNA was isolated from tissues with Trizol (Life Technologies Inc., Rockville, Maryland, USA) according to the manufacturer’s instructions. Total RNA was subjected to DNaseI treatment followed by first-strand cDNA synthesis using Superscript II (Life Technologies Inc.), followed by PCR analysis using the following primer sets. The first primer set was specific for the extracellular/transmembrane domain of Ron and included a 5′ primer in exon 11 (5′-TGGGTGGTGAGGTCTGCCAACATGAGCTCC-3′) and a 3′ primer in exon 12 (5′-CCGTCTTCGGGAGTTAAAGATCA-GGGCAAC-3′). These primers would produce a 251-bp product from reverse-transcribed RNA and a 398-bp genomic band. The second primer set was specific for the TK domain of Ron with a 5′ primer in exon 17 (5′-ATCGCCATGCTCGCCTGCCAGTCAAATGGA-3′) and a 3′ primer in exon 19 (5′-CATAGTGGTCCCCAAGCAGTGAGGCCACTA-3′). This combination would produce an RNA-specific band of 330 bp with a genomic band being too large to be detected. The third primer set analyzed the deletion in the Ron TK–/– mice. The 5′ primer was in exon 11 (as indicated above) and the 3′ primer was in exon 19 (5′-TGGCTCTGAGAGAGGCCGGGGCTTTGACGT-3′). This primer set would produce an RNA-specific product of 508 bp only if recombination occurred with deletion of exons 13 through 18 of the Ron gene. All other combinations from this primer set would be much larger (ranging from 1.4 kb to over 5 kb). This primer pair was used to sequence the junction from exon 12 to exon 19. Primer sequences for the control GAPDH reactions were 5′-TGGTGCAGGATGCATTGCTG-3′ and 5′-AGTGGAGATTGTTGCCATCA-3′.
Immunoprecipitations were performed on resident peritoneal macrophages from Ron TK–/– and Ron TK+/+ mice. Approximately 108 macrophages were lysed as described previously (14). Cellular proteins were immunoprecipitated with a rat anti-mouse Ron monoclonal Ab (clone 2B) (7) and separated on an 8% SDS-PAGE gel under reducing conditions. Wild-type mouse Ron and truncated Ron proteins were detected by Western analysis using a rabbit IgG Ab to the extracellular domain of Ron (22), followed by a goat anti-rabbit IgG Ab conjugated with horseradish peroxidase. The reaction was developed with enhanced chemiluminescence (ECL; Amersham Pharmacia Biotech, Piscataway, New Jersey, USA) reagents and exposed to film.
Tissue histology and immunohistochemistry.
For ovarian analyses, mice were hyperstimulated by hormonal priming as described previously (23). For histology, mice were sacrificed and organs of interest were harvested, fixed in 10% neutral-buffered formalin, and paraffin embedded. Tissues were sectioned, mounted on slides, and stained with hematoxylin and eosin. Immunohistochemistry was performed on paraffin-embedded tissue sections using an anti-nitrotyrosine Ab (Upstate Biotechnology Inc., Lake Placid, New York, USA) or a monoclonal Ab to the Ron α chain (Transduction Laboratories, Lexington, Kentucky, USA). The detection system used secondary Ab’s conjugated to biotin (Vector Laboratories, Burlingame, California, USA), the Vectastain ABC Kit (Vector Laboratories), and a DAB peroxidase substrate (3,3′-diaminobenzidine tablet sets; Sigma Chemical Co.). The slides were counterstained with hematoxylin.
Production of HGFL and activation of peritoneal macrophage.
HGFL was produced following transfection of a wild-type human HGFL cDNA clone into Chinese hamster ovary (CHO) cells as described previously (24). Adult mouse peritoneal macrophages were isolated by peritoneal lavage as described previously (24). Macrophages were treated with conditioned media from CHO cells or CHO cells expressing human HGFL as described previously (24). Cell morphology was analyzed at indicated time intervals under a phase-contrast microscope. In each assay, the number of activated and nonactivated macrophages were determined by blind counting of multiple microscopic fields containing approximately 100 macrophages. Macrophages were scored as activated if the cells displayed a compact, spindle-shaped morphology with elongated cytoplasmic processes. Statistical analyses were performed using StatView and the unpaired t test. For nitric oxide (NO) production, the macrophages were stimulated with 1 μg/ml of LPS from Escherichia coli (Sigma Chemical Co.) and 100 U/ml recombinant mouse INF-γ in the presence or absence of recombinant HGFL (200 ng/ml). NO levels from in vitro–cultured macrophages were determined by measuring the levels of nitrite, a stable by-product of NO, in the serum-free conditioned culture media of plated peritoneal macrophages as described (9). Values were normalized to control levels, and fold induction of nitrite levels were plotted.
LPS challenge.
Mice were injected in the peritoneum with LPS in sterile PBS at 8.9 μg/g body weight. Survival curves were generated by monitoring the mice at daily intervals for 10 days. Eighteen mice were used in the control group (Ron TK+/+), and 23 mice were used in the experimental group (Ron TK–/–).
Contact dermatitis.
To induce an acute irritation, a 15% phenol solution (in absolute ethanol, vehicle) was applied to the dorsal surface of the right ear of each mouse, while vehicle was applied to the left ear. Four hours later, the mice were sacrificed and the ears were taken for histological analyses. Antigen-dependent contact dermatitis (CD) was induced with dinitrofluorobenzene (DNFB) (25). Mice were placed under metophane anesthesia, and a 0.5% DNFB (Sigma Chemical Co.) solution in 4:1 acetone/olive oil (vehicle) was applied to a 2-cm2 area of the shaved abdomen. This treatment was repeated after 24 hours. Five days later, DNFB was applied on the dorsal surface of the right ear, with the left ear being treated with vehicle alone. Twenty-four hours later, the mice were sacrificed and the ears were harvested. The extent of ear swelling was measured microscopically from histological sections using a graticule. Ear thickness was used to quantify CD and was measured by four blinded measurements taken for each ear. The percentage of increase in ear thickness was calculated based on measuring the size of the exposed ear versus the unexposed (vehicle treated) ear for each animal. Results are indicated as averages taken from 10 to 14 mice per group between 8 and 12 weeks of age.
Results
Generation of mice containing a deletion of the TK domain of Ron.
A targeting vector was constructed to delete the TK domain of the Ron gene (Figure 1a). Due to the fact that this mutation may lead to embryonic lethality, the targeting vector was designed to allow the generation of conditional Ron deletions, which would allow the dissection of Ron-specific functions in vivo. The targeting vector was designed such that loxP recombinase recognition sites were incorporated 5′ to exon 13 and 3′ to exon 18, thereby floxing exons encoding the TK domain of Ron. Deletion of the TK domain would render the Ron protein catalytically inactive. Following transfection of the ES cells with the targeting vector, homologous recombination was confirmed by Southern hybridization analyses using probes from outside the targeted region (probe A in Figure 1a). Homologous recombination occurred in 3 out of 340 ES cell clones that were analyzed (Figure 1b).
Given the possibility that the HPRT selection cassette could interfere with the endogenous expression pattern of the mutated Ron gene, a strategy was designed to eliminate HPRT before generation of the mice. The targeted ES cell clone, O12, was transiently transfected with a Cre recombinase expression vector. After transfection with Cre, recombination events between any two of the three loxP sites can be obtained (Figure 1a: A4, A5, A6). To enrich for recombinant forms A4 and A5, the Cre-transfected ES cells were selected against HPRT. After selection, 150 ES cell clones were isolated, and Southern blot analyses were performed to determine the extent of recombination (Figure 1c). Of the six clones analyzed in Figure 1c, two had the HPRT cassette deleted (Figure 1c, lanes 1 and 2), while the other clones have had the entire floxed region excised (Figure 1c, lanes 3–6, and data not shown). Analysis of all 150 ES cell clones indicated that 20% contained the A4 recombination and 80% contained the A5 recombination. The only modification to the floxed Ron allele (A4) is the inclusion of portions of bacterial polylinkers used in cloning the targeting vector and the addition of two 34-bp loxP sites surrounding the Ron TK domain in the intervening sequences. The A5 line contains a germline deletion of the TK domain of Ron and an additional loxP site incorporated into an intronic region.
Due to the fact that a germline mutation of the TK domain of Ron may result in embryonic lethality, two of each of the A4- and A5-targeted ES cell clones were injected into mouse blastocysts using standard technologies. Germline transmission was obtained from each of the ES cell lines.
Germline deletion of the TK domain of Ron.
To determine the effect of the homozygous loss of the TK domain of Ron in mice, hemizygous Ron TK+/– siblings (from the A5 line) were intercrossed, and genotypes of the offspring were determined by Southern blot analysis (Figure 1d). The expected mendelian ratio of inheritance was observed from heterozygous matings. The null mice (TK–/–) are devoid of the TK domain of Ron. These studies indicate that this mutation is compatible with mouse viability.
Cre-mediated excision of the TK domain of Ron.
To conditionally target the floxed TK Ron allele derived from the A4 ES cell line, homozygous floxed TK Ron mice were crossed to mice expressing Cre recombinase. Cre recombinase expression was driven by the endogenous sequences of the HNF3α locus (21, 26, 27). The derivation of the HNF3αCre allele by gene targeting is outlined in Figure 2a. Due to the fine mapping of HNF3α expression during development and through adulthood, it was anticipated that Cre expression would initiate at E8–8.5 and continue through adulthood in endoderm-derived tissues, namely, the lung, liver, and gastrointestinal tract. Heterozygosity at the HNF3α locus does not result in any known phenotypic abnormalities (21). Mice homozygous for the floxed TK Ron allele were bred to mice hemizygous for both the floxed TK Ron allele and Cre. The generation of these animals followed a Mendelian pattern of inheritance. Mice homozygous for the floxed TK allele and heterozygous for HNF3α-driven Cre are born with no embryonic loss. To map the extent and location of the Cre-mediated recombination event, Southern hybridization and PCR analyses on DNA isolated from various organs from adult experimental mice were performed (Figure 2b). Surprisingly, this conditional deletion strategy resulted in mice with a systemic deletion of the floxed TK Ron sequences in every tissue analyzed (Figure 2b).
To determine the timing and extent of the excision, embryos and extraembryonic tissue were isolated and analyzed at E9. Southern analyses indicated that complete excision of the kinase domain of Ron had occurred in the whole embryo and extraembryonic tissues (data not shown). Therefore, our conditional mutation appeared to result in a very early embryonic systemic deletion of the TK domain of Ron. These studies further support data generated from the A5 mutation in that deletion of the TK domain of Ron is compatible for life. While the A4 line containing HNF3aCre confirms our germline deletion, we intend to use mice containing the floxed Ron alleles in future experiments to determine the effect of cell type–specific deletions of Ron following stressful insults.
RNA and protein analysis of the mutated allele.
RT-PCR analysis was used to determine the expression levels of Ron from the mutant allele (Figure 2c). Primers specific for the extracellular and transmembrane domains of Ron were used to analyze reverse-transcribed liver RNA from experimental (Ron TK–/–, lane 1) and control (Ron TK+/+, lane 2) mice. PCR products in both lanes indicate that this region of Ron is expressed in both animals. When primers were designed to specifically analyze the expression of the TK domain of Ron, a PCR product was seen only in the control mice (lane 4 compared with lane 3). Conversely, when primers were designed to specifically analyze the TK-deletion product, a PCR product was seen only in the experimental mice (lane 8 compared with lane 7). Furthermore, this product corresponds in size to what is expected if Ron exons 13–18 were deleted and the resultant splicing of exon 12 to 19 occurred. In addition to the liver, identical RT-PCR results were obtained from RNA isolated from the lungs, kidney, intestine, mammary gland, and skin (data not shown). By bringing exons 12 and 19 together, a stop codon is generated three codons into exon 19. This junction has been sequenced from the RT-PCR product, and our data suggest that this sequence would encode the amino acids lysine-lysine-glutamine-leucine-glycine-isoleucine-threonine, followed by the TGA stop codon. The glycine is produced by a fusion between exons 12 and 19. These data indicate that the experimental mice express the predicted truncated form of Ron mRNA, which, when translated, would produce a protein with only the extracellular and transmembrane domains of Ron.
Immunoprecipitation analyses were performed on resident peritoneal macrophages isolated from control and experimental mice. Using Ab’s specific for the extracellular domain of Ron, the full-length Ron β chain was detected in macrophages from the control mice (Ron TK+/+, Figure 2d), while a truncated protein product was detected in macrophages from the experimental animals (Ron TK–/–; Figure 2d). The truncated product is approximately 100 kDa and corresponds to the expected size of the mutant protein. Therefore, Southern blot, RT-PCR, and Western blot analyses all indicate a complete deletion of the TK domain in our experimental animals. Furthermore, this deletion encompasses all but five amino acids of the cytoplasmic domain of Ron. Similar results were obtained from mice harboring the Cre-mediated or germline (A5) Ron deletions (data not shown).
Pathological analysis of mice deficient in the TK domain of Ron.
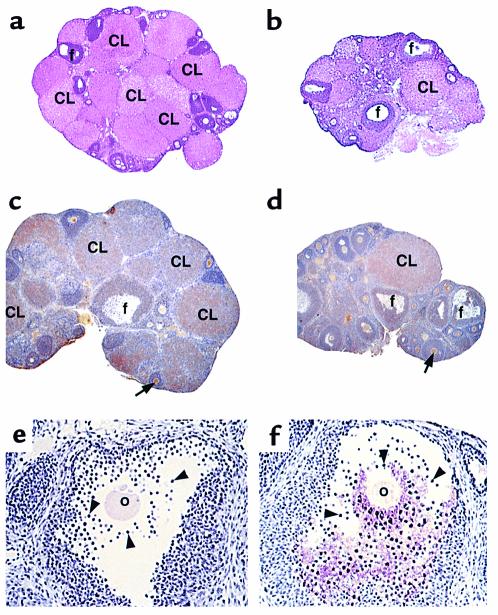
An extensive histological and clinical analysis was performed in parallel on Ron TK–/– and Ron TK+/+ mice. Histological analyses demonstrated that, in general, most tissues and organs were comparable, including sections of the eye, conjunctiva, harderian gland, masseter muscle, mandibular lymph node, heart, aorta, renal arteries, lung, trachea, diaphragm, liver, stomach, duodenum, pancreas, jejunum, ileum, cecum, colon, adrenal gland, quadriceps muscle, cervix, uterus, pituitary, cerebrum, and cerebellum. Interestingly, although folliculogenesis appears normal, alterations were seen in the ovaries of the Ron TK–/– animals (Figure 3). These alterations include reduced ovarian size, mild epithelial hyperplasia, a reduced number of corpora lutea (CL), and a change in density of the CL (Figure 3, a and b).
Figure 3.
Ovarian alterations. Histological analysis of ovaries from Ron TK+/+ (a) and Ron TK–/– (b) mice. The Ron TK–/– mice possess developing follicles (f) but fewer corpora lutea (CL), compared with controls. Immunohistochemistry demonstrating expression of the extracellular domain of Ron in control (c) and Ron TK–/– mice (d). Staining is detected throughout both ovaries, but appears more pronounced in the oocytes (arrows) and in the CL. Mature graafian follicles of experimental ovaries (f) demonstrate an increase in the amount of nitrosylated proteins within the extracellular matrix between the cumulus cells (arrowheads) surrounding the developing oocyte (o) compared with controls (e). a–d, ×40; e and f, ×200.
Immunohistological analysis, with an Ab specific for the extracellular domain of Ron, demonstrated that ovaries from both the control (Figure 3c) and Ron TK–/– animals (Figure 3d) express approximately equal amounts of Ron. Furthermore, expression appears throughout the ovaries but is more intense in the CL and oocytes (CL and arrows in Figure 3, c and d). Given the involvement of Ron in NO regulation, the importance of NO in reproduction, and that NO exerts many of its functions through nitrosylation of proteins (28), the ovaries were analyzed for the amount of nitrosylated tyrosines by immunohistochemistry. Following immunohistochemistry with an anti-nitrotyrosine Ab, the cumulus extracellular matrix of experimental animals was found to exhibit a significant increase in the amount of nitrosylated proteins (Figure 3f) compared with control ovaries (Figure 3e). Similar results were obtained from mice harboring the Cre-mediated or germline (A5) Ron deletions (data not shown).
Clinical analyses indicate that the experimental animals had normal hematological values (including erythrocyte count, hemoglobin level, hematocrit, mean corpuscular volume, leukocyte count with differential, including morphology of neutrophils, eosinophils, monocytes, and lymphocytes), serum creatinine, serum electrolytes (sodium, chloride), alkaline phosphatase, and total protein (data not shown).
In vitro response of macrophage.
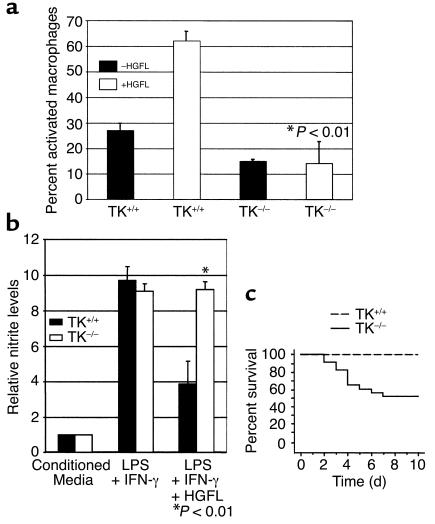
Hematological analyses suggest that the circulating numbers of monocytes are similar in both control and experimental animals. Furthermore, the number of resident peritoneal macrophages did not differ between groups, indicating that the TK domain of Ron is nonessential for the development of blood cells or in the recruitment of monocytes to the peritoneum. We and others have shown that macrophages isolated from the peritoneal cavity show dramatic morphological changes in response to HGFL in vitro (9, 15, 17, 24, 29–31) including the ability to assume elongated shapes. To assay for in vitro morphogenic responses to HGFL, peritoneal macrophages were isolated from control and experimental mice. In response to HGFL, macrophages from control mice undergo well-established characteristic morphological shape changes indicative of activation. The control mice had similar baseline levels of activated macrophages, as reported previously, and their response to HGFL is consistent with published analyses (Figure 4a) (9, 20, 24). In contrast, macrophages from experimental mice do not respond to HGFL, indicating that the TK domain of Ron is essential for the ability to activate macrophages in response to HGFL (Figure 4a). Similar results were obtained from mice harboring the Cre-mediated (A4) or germline (A5) Ron mutations (data not shown).
Figure 4.
Evaluation of macrophage activation, NO production, and LPS-induced endotoxemia. Peritoneal macrophages were isolated from control (Ron TK+/+) and experimental (Ron TK–/–) mice. (a) The macrophages were incubated with control conditioned media (–HGFL, filled bars) or conditioned media containing recombinant HGFL (+HGFL, open bars). After 3 hours, the average percentage of activated macrophages were scored. Values represent the averages of three independent experiments performed in triplicate. (b) Peritoneal macrophages were isolated from control (filled bars) and experimental (open bars) mice. The macrophages were plated in the presence of conditioned media, conditioned media plus LPS and INF-γ, or conditioned media plus LPS, INF-γ, and HGFL. Nitrite levels in the supernatants of cell cultures were measured and normalized to levels found in macrophages cultured in conditioned media. Experiments were performed in triplicate with SE values indicated. (c) Loss of the TK domain of Ron results in an increased susceptibility to LPS-induced endotoxic shock. Cumulative survival data are shown for control (dotted lines) and experimental (solid lines) mice.
Abrogated regulation of NO in mice deficient in the TK domain of Ron.
Stimulation of Ron by HGFL has been shown to inhibit LPS- and/or INF-γ–induced production of NO (9, 11, 19). This regulation is thought to be an important factor involved in attenuating the inflammatory activities of macrophages. To test the ability of HGFL to repress NO activity, peritoneal macrophages from experimental and control mice were isolated, and the amount of nitrite released by the macrophages was then determined (Figure 4b). Baseline levels of nitrite were similar in macrophages isolated from each group following addition of control HGFL-conditioned media. In the presence of LPS and INF-γ, the amount of nitrite was dramatically upregulated in both populations, indicating the functionality of macrophages in the mutant mice. In striking contrast, however, in the presence of HGFL the amount of nitrite production in the control macrophages was significantly reduced compared with the mutant macrophages in response to LPS and INF-γ (Figure 4b; P < 0.01 by ANOVA). Thus, macrophages from mutant mice are unable to attenuate the level of NO production in response to LPS or INF-γ.
Mice lacking the TK domain of Ron are susceptible to endotoxic shock.
Regulation of inducible NO is critical in the pathogenesis of septic shock, a systemic inflammatory response to bacterial-derived endotoxin, most importantly LPS from gram-negative bacteria (32, 33). Death can ensue due to shock and multiorgan failure. Elevated serum levels of NO are thought to play a central role in inducing the cardiovascular dysfunction (vasodilatation) and tissue damage observed during sepsis, and NO is thought to be the final common mediator in the inflammatory cascade leading to this condition (34). Due to the aberrant regulation of NO in peritoneal macrophages of our mutant mice, control and mutant mice were challenged with sublethal doses of LPS and monitored for survival (Figure 4c). While none of the control animals died, only 52% of the mutant mice survived beyond 10 days. The survival curves are significantly different—P values less than 0.001 determined by the log-rank (Mantel-Cox) analysis. Thus, the TK domain of Ron is a critical factor regulating physiologic responses to endotoxemia.
Altered inflammatory responses in Ron TK-deficient mice.
Given the involvement of NO as a mediator of inflammation, we hypothesized that the mutant animals would have differences in their ability to regulate various types of immune and inflammatory responses. To test this hypothesis, mutant and control mice were challenged in two models of CD, a model of nonimmune-mediated cutaneous (irritant) inflammation and in a model of antigen-dependent contact (allergic) hypersensitivity. Although allergic and irritant CD share many clinical and histopathological features, they are thought to be initiated by separate pathways (35–39). Allergic CD requires antigen presentation by antigen-presenting cells to T cells, while irritant dermatitis is believed to activate the inflammatory cascade independent of antigen presentation. Both pathways, however, involve the release of cytokines that attract other inflammatory cells to the site, causing dilation of cutaneous blood vessels and resulting in dermal edema.
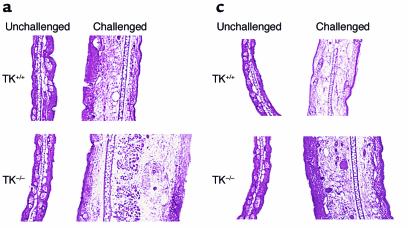
To determine whether the TK domain of Ron is required for acute (nonimmune-mediated) irritant dermatitis, challenged and unchallenged ears of mutant and control mice were isolated after acute exposure to phenol and analyzed. Histological analyses indicate a similar influx of inflammatory cells in the control and mutant animals (Figure 5a). However, the extent of edema was significantly greater in the experimental mice compared with controls (Figure 5, a and b). Ear thickness increased an average of twofold in the control animals compared with 3.1-fold in the mutant animals over that of the unchallenged ear.
Figure 5.
Enhanced irritant and allergic CD in mice lacking the TK domain of Ron. For acute irritant CD, experimental (Ron TK–/–) and control (Ron TK+/+) mice were challenged with a phenol solution as described in Methods (a and b). (a) Representative histological sections of ears following vehicle (unchallenged) and phenol application (challenged). ×100 magnification. (b) The average thickness of the ears were calculated and expressed as a percentage of increase over the control ear for each animal. The scatter diagram shows the values for each animal along with mean values (line) for each group. Mice lacking the TK domain of Ron displayed a significantly greater amount of ear swelling (P < 0.01). (c and d) To induce allergic contact hypersensitivity, mice were sensitized and challenged with DNFB as described in Methods. (c) Representative histological sections of ears. (d) Ear measurements were taken and expressed as a percentage of increase over the control ear. The scatter diagram depicts values determined for each animal as well as the population mean (line). Deficiencies in the TK domain of Ron result in a significantly exaggerated contact hypersensitivity response (P < 0.01).
Mice were also studied in a DNFB-induced model of contact hypersensitivity (Figure 5, c and d). Following DNFB sensitization, ears of mutant and control mice were challenged and evaluated histologically (Figure 5c). As in irritant-induced dermatitis, histological evaluation disclosed a comparable inflammatory cell infiltrate in both groups analyzed, suggesting that the TK domain of Ron was not important for cellular migration to this site. However the extent of edema in mutant mice was dramatically increased compared with control mice (Figure 5, c and d). Ear thickness measurements indicate that ear swelling was increased 3.4-fold in the mutant mice compared with 2.4-fold in the control animals relative to the unchallenged ear. Thus, the CD results suggest that the TK domain is important in regulating the extent of delayed type hypersensitivity responses and acute inflammatory responses, possibly as a result of altering NO levels, resulting in increased vascular permeability and edema.
Discussion
In an attempt to determine the in vivo functional significance of a specific domain of the Ron receptor, a targeting strategy was designed to delete the TK domain of this protein. Mice with a germline deletion of this domain were derived. Strikingly, our results demonstrate that complete ablation of the cytoplasmic domain, resulting in loss of Ron intracellular signaling, is compatible with life. There does not appear to be overt defects in embryonic development, hematopoiesis, or organ development or function in the nonstressed animal.
Previous studies from our laboratory have indicated that deletion of exons 1 through 14, as well as several hundred base pairs of 5′ flanking sequences of the Ron gene, causes early embryonic death in mice (9). This is intriguing, because deficiency of HGFL is compatible with normal development, growth, and propagation (20). In addition, disruption of the first exon of Ron, potentially resulting in a hypomorphic allele, produces animals with a phenotype similar to mice lacking the TK domain of Ron described in this report (11). The simplest explanation for these seemingly paradoxical results may be that our previous deletion resulted in the disruption or alteration of another gene near the Ron locus. This possibility cannot be entirely excluded. However, an early embryonic lethal phenotype with our previous Ron mutation was documented with multiple independent ES cell lines, and even after extensive inbreeding there is no evidence of segregation of the Ron-targeted allele from the lethal phenotype. We have also shown that Ron is expressed during the time frame of this early lethality and that mice hemizygous for this mutation display macrophage-related phenotypes similar to those displayed by mice characterized in this report (9). These data argue strongly that deletion of the extracellular and transmembrane domains of Ron results in embryonic loss. In contrast to our previous Ron mutation, there are no data to indicate that the disruption of the first exon of Ron generates a null allele (11). The targeting strategy for this latter Ron mutation was to simply insert an alteration into the first exon of this gene. This insertion is not expected to eliminate possible alternative Ron transcripts that have been reported subsequently (12). Given the fact that no data are presented at the mRNA or protein levels for Ron in the mutant animals containing a targeted alteration of the first exon, mice generated from this mutation may likely produce alternative Ron transcripts.
These combined data lead to an intriguing hypothesis that Ron signaling in vivo may occur through receptor heterodimerization. Recent reports have indicated that Ron functionally interacts with a number of receptors, namely the erythropoietin receptor, Met (the hepatocyte growth factor receptor), and with the common β signal transducer that associates with Il-3, Il-5, and GM-CSF (12, 40–42). During embryonic development, mutant mice generated in this report, as well as mutant mice containing an alteration of exon 1, may be capable of signaling through heterodimerization with alternative receptors (9, 11). Furthermore, potential activation of Ron-through heterodimerization is also consistent with the lack of phenotype observed in HGFL-deficient mice. In contrast, deletion of the extracellular and transmembrane domains of Ron may result in the complete loss of Ron function and/or the ability to heterodimerize, leading to embryonic lethality. Interestingly, based on regions deleted in the three mutant mice, Ron receptor dimerization possibly may occur through the transmembrane domain.
The studies in this report clearly indicate that deletion of all but five amino acids of the intracellular domain of Ron produces viable mice. These studies suggest a potential novel mechanism associated with Ron signaling in that the extracellular and transmembrane domains of Ron and not TK activity are required for developmental success. Furthermore, even though the TK domain is not essential, it is apparent that one, nonredundant function of Ron signaling is to attenuate the inflammatory response. Our present findings, as well as results of the previous Ron mutant animals, are in complete agreement that Ron expression is critical in the regulation of macrophage-mediated inflammatory responses (9, 11). One mechanism that Ron uses to limit the extent of inflammation may be by limiting the production of NO. NO is an important mediator of inflammation and microbicidal and tumoricidal activity of macrophages, and Ron may thus protect tissues from the potential harmful effects of NO and inflammation.
NO regulation has been found to be necessary for optimal mammalian reproduction, especially during the events of ovulation (43). In rodents, improper regulation of NO affects luteal function, steroidogenesis, and oocyte maturation that leads to a decrease in the number of ovulated cumulus oocyte complexes (44–46). Thus, the reduced number of CL in the ovaries from mice devoid of the TK domain of Ron suggests that these animals ovulated fewer oocytes compared with controls, possibly due to altered NO levels and increased amounts of nitrosylated proteins within the cumulus extracellular matrix (Figure 3). In addition to ovarian defects, the hyper-responsiveness to allergic and irritant CD imply that mice deficient in the TK domain of Ron do not have a defect in antigen presentation, but instead have a general inability to limit the extent of tissue damage following an inflammatory response. Ongoing studies are being performed to elucidate the molecular mechanism regulating these reproductive and immunological responses.
In summation, our current work provides an analysis of the potential roles of Ron signaling in vivo. We have shown that the absence of Ron signaling is compatible with life, but results in potentiation of the inflammatory response. A pivotal in vivo role of the TK domain of Ron in the mouse is to attenuate the harmful effects of NO, which may include increased vascular permeability and tissue edema in various models of inflammation of the skin and lungs (data not shown) and increased mortality resulting from LPS-induced endotoxemia. The mice generated in this report should be beneficial in elucidating the effect of the Ron TK domain on NO generation and regulation in inflammation as well as in ultimately determining the cell type–specific roles for this protein in vivo using conditional gene targeting.
Acknowledgments
The authors would like to acknowledge Angela Drew, Mike Leonis, Ali Mallakin, William Sun, and Cindy Wetzel for their assistance and reading of this manuscript. This work was supported in part by the Marion Merrel Dow Foundation (S.E. Waltz), by Public Health Service grants HD-36888 (S.E. Waltz), DK-47003 and DK-58182 (S.J.F. Degen), DK-53839 (K.H. Kaestner), training grant T-32-HL0775 (B.E. Pierce) and postdoctoral fellowship grant HD08659 (K.A. Hess) from the NIH, and grants from the March of Dimes Organization (S.E. Waltz), American Heart Association (S.E. Waltz), and a Board of Trustees grant from the Children’s Hospital Research Foundation (S.E. Waltz).
References
- 1.Bottaro DP, et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science. 1991;251:802–804. doi: 10.1126/science.1846706. [DOI] [PubMed] [Google Scholar]
- 2.Naldini L, et al. Hepatocyte growth factor (HGF) stimulates the tyrosine kinase activity of the receptor encoded by the proto-oncogene c-MET. Oncogene. 1991;6:501–504. [PubMed] [Google Scholar]
- 3.Gaudino G, et al. RON is a heterodimeric tyrosine kinase receptor activated by the HGF homologue MSP. EMBO J. 1994;13:3524–3532. doi: 10.1002/j.1460-2075.1994.tb06659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zarnegar R. Regulation of HGF and HGFR gene expression. EXS. 1995;74:33–49. doi: 10.1007/978-3-0348-9070-0_3. [DOI] [PubMed] [Google Scholar]
- 5.Medico E, et al. The tyrosine kinase receptors Ron and Sea control “scattering” and morphogenesis of liver progenitor cells in vitro. Mol Biol Cell. 1996;7:495–504. doi: 10.1091/mbc.7.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwama A, Okano K, Sudo T, Matsuda Y, Suda T. Molecular cloning of a novel receptor tyrosine kinase gene, STK, derived from enriched hematopoietic stem cells. Blood. 1994;83:3160–3169. [PubMed] [Google Scholar]
- 7.Iwama A, et al. Terminal differentiation of murine resident peritoneal macrophages is characterized by expression of the STK protein tyrosine kinase, a receptor for macrophage-stimulating protein. Blood. 1995;86:3394–3403. [PubMed] [Google Scholar]
- 8.Waltz SE, et al. Characterization of the mouse Ron/Stk receptor tyrosine kinase gene. Oncogene. 1998;16:27–42. doi: 10.1038/sj.onc.1201508. [DOI] [PubMed] [Google Scholar]
- 9.Muraoka RS, et al. The Ron/STK receptor tyrosine kinase is essential for peri-implantation development in the mouse. J Clin Invest. 1999;103:1277–1285. doi: 10.1172/JCI6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaudino G, et al. The proto-oncogene RON is involved in development of epithelial, bone and neuro-endocrine tissues. Oncogene. 1995;11:2627–2637. [PubMed] [Google Scholar]
- 11.Correll PH, et al. Deregulated inflammatory response in mice lacking the STK/RON receptor tyrosine kinase. Genes Funct. 1997;1:69–83. doi: 10.1046/j.1365-4624.1997.00009.x. [DOI] [PubMed] [Google Scholar]
- 12.Persons DA, et al. Fv2 encodes a truncated form of the Stk receptor tyrosine kinase. Nat Genet. 1999;23:159–165. doi: 10.1038/13787. [DOI] [PubMed] [Google Scholar]
- 13.Han S, Stuart LA, Degen SJ. Characterization of the DNF15S2 locus on human chromosome 3: identification of a gene coding for four kringle domains with homology to hepatocyte growth factor. Biochemistry. 1991;30:9768–9780. doi: 10.1021/bi00104a029. [DOI] [PubMed] [Google Scholar]
- 14.Wang MH, Iwama A, Skeel A, Suda T, Leonard EJ. The murine stk gene product, a transmembrane protein tyrosine kinase, is a receptor for macrophage-stimulating protein. Proc Natl Acad Sci USA. 1995;92:3933–3937. doi: 10.1073/pnas.92.9.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leonard EJ, Skeel A. A serum protein that stimulates macrophage movement, chemotaxis and spreading. Exp Cell Res. 1976;102:434–438. doi: 10.1016/0014-4827(76)90065-3. [DOI] [PubMed] [Google Scholar]
- 16.Leonard EJ, Skeel AH. Isolation of macrophage stimulating protein (MSP) from human serum. Exp Cell Res. 1978;114:117–126. doi: 10.1016/0014-4827(78)90043-5. [DOI] [PubMed] [Google Scholar]
- 17.Skeel A, et al. Macrophage stimulating protein: purification, partial amino acid sequence, and cellular activity. J Exp Med. 1991;173:1227–1234. doi: 10.1084/jem.173.5.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YQ, Fisher JH, Wang MH. Activation of the RON receptor tyrosine kinase inhibits inducible nitric oxide synthase (iNOS) expression by murine peritoneal exudate macrophages: phosphatidylinositol-3 kinase is required for RON-mediated inhibition of iNOS expression. J Immunol. 1998;161:4950–4959. [PubMed] [Google Scholar]
- 19.Wang MH, et al. Macrophage-stimulating protein inhibits induction of nitric oxide production by endotoxin- or cytokine-stimulated mouse macrophages. J Biol Chem. 1994;269:14027–14031. [PubMed] [Google Scholar]
- 20.Bezerra JA, Carrick TL, Degen JL, Witte D, Degen SJF. Biological effects of targeted inactivation of hepatocyte growth factor-like protein in mice. J Clin Invest. 1998;101:1175–1183. doi: 10.1172/JCI1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaestner KH, Katz J, Liu Y, Drucker DJ, Schutz G. Inactivation of the winged helix transcription factor HNF3alpha affects glucose homeostasis and islet glucagon gene expression in vivo. Genes Dev. 1999;13:495–504. doi: 10.1101/gad.13.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang MH, et al. Identification of the ron gene product as the receptor for the human macrophage stimulating protein. Science. 1994;266:117–119. doi: 10.1126/science.7939629. [DOI] [PubMed] [Google Scholar]
- 23.Hess KA, Chen L, Larsen WJ. Inter-alpha-inhibitor binding to hyaluronan in the cumulus extracellular matrix is required for optimal ovulation and development of mouse oocytes. Biol Reprod. 1999;61:436–443. doi: 10.1095/biolreprod61.2.436. [DOI] [PubMed] [Google Scholar]
- 24.Waltz SE, et al. Functional characterization of domains contained in hepatocyte growth factor-like protein. J Biol Chem. 1997;272:30526–30537. doi: 10.1074/jbc.272.48.30526. [DOI] [PubMed] [Google Scholar]
- 25.Wang M, et al. Matrix metalloproteinase deficiencies affect contact hypersensitivity: stromelysin-1 deficiency prevents the response and gelatinase B deficiency prolongs the response. Proc Natl Acad Sci USA. 1999;96:6885–6889. doi: 10.1073/pnas.96.12.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaestner KH, Hiemisch H, Luckow B, Schutz G. The HNF-3 gene family of transcription factors in mice: gene structure, cDNA sequence, and mRNA distribution. Genomics. 1994;20:377–385. doi: 10.1006/geno.1994.1191. [DOI] [PubMed] [Google Scholar]
- 27.Monaghan AP, Kaestner KH, Grau E, Schutz G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 alpha, beta and gamma genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development. 1993;119:567–578. doi: 10.1242/dev.119.3.567. [DOI] [PubMed] [Google Scholar]
- 28.Stamler JS. Redox signaling: nitrosylation and related target interactions of nitric oxide. Cell. 1994;78:931–936. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 29.Wang MH, Montero-Julian FA, Dauny I, Leonard EJ. Requirement of phosphatidylinositol-3 kinase for epithelial cell migration activated by human macrophage stimulating protein. Oncogene. 1996;13:2167–2175. [PubMed] [Google Scholar]
- 30.Wang MH, Skeel A, Leonard EJ. Proteolytic cleavage and activation of pro-macrophage-stimulating protein by resident peritoneal macrophage membrane proteases. J Clin Invest. 1996;97:720–727. doi: 10.1172/JCI118470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang MH, Yoshimura T, Skeel A, Leonard EJ. Proteolytic conversion of single chain precursor macrophage-stimulating protein to a biologically active heterodimer by contact enzymes of the coagulation cascade. J Biol Chem. 1994;269:3436–3440. [PubMed] [Google Scholar]
- 32.Hoffman WD, Natanson C. Endotoxin in septic shock. Anesth Analg. 1993;77:613–624. doi: 10.1213/00000539-199309000-00032. [DOI] [PubMed] [Google Scholar]
- 33.Parrillo JE. Pathogenetic mechanisms of septic shock. N Engl J Med. 1993;328:1471–1477. doi: 10.1056/NEJM199305203282008. [DOI] [PubMed] [Google Scholar]
- 34.Cobb JP, Danner RL. Nitric oxide and septic shock. JAMA. 1996;275:1192–1196. [PubMed] [Google Scholar]
- 35.Brasch J, Sterry W. Immunophenotypical characterization of inflammatory cellular infiltrates in tinea. Acta Derm Venereol. 1992;72:345–347. [PubMed] [Google Scholar]
- 36.Brasch J, Burgard J, Sterry W. Common pathogenetic pathways in allergic and irritant contact dermatitis. J Invest Dermatol. 1992;98:166–170. doi: 10.1111/1523-1747.ep12555804. [DOI] [PubMed] [Google Scholar]
- 37.Willis CM, Young E, Brandon DR, Wilkinson JD. Immunopathological and ultrastructural findings in human allergic and irritant contact dermatitis. Br J Dermatol. 1986;115:305–316. doi: 10.1111/j.1365-2133.1986.tb05745.x. [DOI] [PubMed] [Google Scholar]
- 38.Nater JP, Hoedemaeker PJ. Histological differences between irritant and allergic patch test reactions in man. Contact Dermatitis. 1976;2:247–253. doi: 10.1111/j.1600-0536.1976.tb03042.x. [DOI] [PubMed] [Google Scholar]
- 39.Hartman A, Hoedemaeker PJ, Nater JP. Histological aspects of DNCB sensitization and challenge tests. Br J Dermatol. 1976;94:407–416. doi: 10.1111/j.1365-2133.1976.tb06118.x. [DOI] [PubMed] [Google Scholar]
- 40.Follenzi A, et al. Cross-talk between the proto-oncogenes Met and Ron. Oncogene. 2000;19:3041–3049. doi: 10.1038/sj.onc.1203620. [DOI] [PubMed] [Google Scholar]
- 41.Mera A, Suga M, Ando M, Suda T, Yamaguchi N. Induction of cell shape changes through activation of the interleukin-3 common beta chain receptor by the RON receptor-type tyrosine kinase. J Biol Chem. 1999;274:15766–15774. doi: 10.1074/jbc.274.22.15766. [DOI] [PubMed] [Google Scholar]
- 42.Ney PA, D’Andrea AD. Friend erythroleukemia revisited. Blood. 2000;96:3675–3680. [PubMed] [Google Scholar]
- 43.McCann SM, et al. The role of nitric oxide in reproduction. Braz J Med Biol Res. 1999;32:1367–1379. doi: 10.1590/S0100-879X1999001100007. [DOI] [PubMed] [Google Scholar]
- 44.Jablonka-Shariff A, Olson LM. Nitric oxide is essential for optimal meiotic maturation of murine cumulus-oocyte complexes in vitro. Mol Reprod Dev. 2000;55:412–421. doi: 10.1002/(SICI)1098-2795(200004)55:4<412::AID-MRD9>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 45.Motta AB, Gimeno MA. Nitric oxide participates in the corpus luteum regression in ovaries isolated from pseudopregnant rats. Can J Physiol Pharmacol. 1997;75:1335–1339. [PubMed] [Google Scholar]
- 46.Zackrisson U, et al. Cell-specific localization of nitric oxide synthases (NOS) in the rat ovary during follicular development, ovulation and luteal formation. Hum Reprod. 1996;11:2667–2673. doi: 10.1093/oxfordjournals.humrep.a019189. [DOI] [PubMed] [Google Scholar]