Abstract
Low genetic diversity is predicted to negatively impact species viability and has been a central concern for conservation. In contrast, the possibility that some species may thrive in spite of a relatively poor diversity has received little attention. The wandering and Amsterdam albatrosses (Diomedea exulans and Diomedea amsterdamensis) are long-lived seabirds standing at an extreme along the gradient of life strategies, having traits that may favour inbreeding and low genetic diversity. Divergence time of the two species is estimated at 0.84 Myr ago from cytochrome b data. We tested the hypothesis that both albatrosses inherited poor genetic diversity from their common ancestor. Within the wandering albatross, per cent polymorphic loci and expected heterozygosity at amplified fragment length polymorphisms were approximately one-third of the minimal values reported in other vertebrates. Genetic diversity in the Amsterdam albatross, which is recovering from a severe bottleneck, was about twice as low as in the wandering albatross. Simulations supported the hypothesis that genetic diversity in albatrosses was already depleted prior to their divergence. Given the generally high breeding success of these species, it is likely that they are not suffering much from their impoverished diversity. Whether albatrosses are unique in this regard is unknown, but they appear to challenge the classical view about the negative consequences of genetic depletion on species survival.
Keywords: genetic diversity, life-history traits, albatrosses, AFLP
1. Introduction
Theory predicts that species with higher genetic diversity may enjoy a lower risk of inbreeding depression, increased fitness through heterozygote benefits and better evolutionary potential than species with more limited diversity (Sherwin & Moritz 2000; Frankham et al. 2002). While these predictions have received empirical or experimental support (e.g. Saccheri et al. 1998; England et al. 2003; Spielman et al. 2004), they have also contributed to the spread of the idea that having low genetic diversity is necessarily bad. Such a perception is widely acknowledged, explicitly or implicitly, in both scientific and popular publications, and perhaps strengthened by the frequent attribution (rightly or wrongly) of low levels of genetic diversity to population bottlenecks (O'Brien 1994; Hoelzel et al. 2002; Russello et al. 2004; Zhang et al. 2004). Since a bottleneck is a period of severe demographic contraction where a species or a population may approach extinction, this might reinforce the idea that exhibiting low genetic diversity cannot be a normal state in a ‘healthy’ species. Hence, while conservation concerns regarding genetically depleted populations have been widely discussed, the possibility that some species may thrive in spite of relatively poor genetic diversity has received little attention from empirical investigations.
A number of studies that examined both historical (prior to known bottleneck) and contemporary samples, or that compared closely related species or populations, showed that bottlenecks can reduce genetic diversity to different extents in different species (e.g. Groombridge et al. 2000; Wisely et al. 2002; Bellinger et al. 2003). However, claims about the role of bottlenecks in depleting genetic diversity are not always realistic with regards to theoretical expectations (Amos & Balmford 2001). For example, to actually have its heterozygosity decrease substantially, a population must usually experience an extreme contraction over a large number of generations. Even species notoriously depleted, such as the African Cheetah (Acinonyx jubatus), might not have experienced such conditions (Merola 1994; Amos & Harwood 1998; Amos & Balmford 2001).
Alternatively, life-history traits might cause some species to retain little genetic variability. Demographic patterns associated with life history (e.g. low fecundity or metapopulation dynamics) may result in a small effective population size (Ne) and associated loss of genetic diversity over long evolutionary times (Frankham et al. 2002). Likewise, recovery of genetic diversity following a bottleneck or a founder event will depend upon the specific demography of each species. For instance, the dating of the cheetah's last bottleneck as 6000–20 000 years ago (Menotti-Raymond & O'Brien 1993) is evocative of the species' slowness to re-establish its genetic diversity.
As the build up of genetic variability is time-bound by the lifespan of each species, it may be that some are unlikely to ever reach the level of genetic diversity observed in others. This might apply to the wandering albatross (Diomedea exulans). This long-lived seabird is positioned at the ‘slow’ end of the gradient of life strategies, characterized by a very low reproductive output (rearing one chick every other year), extensive overlapping generations (lifespan may exceed 50 years), natal and breeding philopatry and small colony size (from less than 10 to less than 2000 pairs for most colonies; Weimerskirch & Jouventin 1987; Tickell 2000; Inchausti & Weimerskirch 2002). Inbreeding and genetic drift within colonies may be more prevalent under the influence of these traits. Actually, a moderately low level of genetic variation was uncovered at microsatellite loci in that species, as well as a rather shallow mitochondrial DNA (mtDNA) phylogeny (Burg & Croxall 2004; Alderman et al. 2005). In addition, in a pilot study conducted with amplified fragment length polymorphism (AFLP) markers, we found an extremely low genetic diversity.
In this study, we compared AFLP variation in two sister species, the wandering and the Amsterdam (Diomedea amsterdamensis) albatrosses, to test the hypothesis that they have had impoverished genetic diversity since their divergence, which began some 0.8 million years ago according to cytochrome b sequence data and molecular clock calibration (Nunn & Stanley 1998; Penhallurick & Wink 2004). While the wandering albatross is widespread across the Southern Ocean, the Amsterdam albatross is recovering from an extreme bottleneck (Weimerskirch et al. 1997). Support in favour of our hypothesis would suggest that, despite quite different population histories, these albatrosses never reached levels of diversity observed in other vertebrates.
We chose AFLPs because they generate a larger number of loci (100s, compared to typically 5–20 loci for microsatellites or 1 locus for mtDNA) and do not specifically target sequences evolving neutrally (Campbell & Bernatchez 2004). Thus, they integrate variation in genetic diversity within a species' whole genome, unlike microsatellites markers which target a specific type of DNA (tandem repeats) and are generally selected on the basis of their level of polymorphism. Moreover, AFLPs will always exhibit a maximum of two alleles, thus ensuring that interspecific comparisons of diversity are not flawed by large variation in the number of alleles between loci and between species, as such variation not only reflects intraspecific diversity, but also contingencies (chance and effort invested) during marker development. Thus, AFLPs allow for more straightforward interspecific comparisons of genetic parameters.
So far, empirical studies of the relationship between life-history traits and genetic diversity have provided an ambiguous picture (Amos & Harwood 1998). Here, comparison between wandering and Amsterdam albatrosses helped disentangle the two main factors (bottleneck, life-history traits reducing Ne) potentially responsible for the genetic diversity observed today in albatrosses.
2. Material and methods
According to a recent, albeit still debated, taxonomical revision, the wandering albatross (D. exulans) breeds on four archipelagos and one isolated island throughout the Southern Ocean (Robertson & Nunn 1998; Burg & Croxall 2004; Alderman et al. 2005; see electronic supplementary material). Formerly, the species also included two subspecies from islands nearby New Zealand (now referred as Gibson's albatross, D. Gibsoni and Antipodean albatross, D. antipodensis) and one from the Tristan da Cuhna archipelago (now the Tristan albatross, D. dabbenena). In this study, we examined genetic variation in D. exulans as defined according to this new taxonomy. This species numbers approximately 8500 annual breeding pairs (Gales 1997). In contrast, Amsterdam albatrosses were represented by only 38 pairs in 2004–2005 (H. Weimerskirch, unpublished), all breeding on Amsterdam Island in the Indian Ocean, but were down to five pairs in 1984 (Weimerskirch et al. 1997).
(a) Sampling and AFLP amplification
Samples of 400 wandering albatrosses were collected at nine breeding colonies from four archipelagos across the species' range (see electronic supplementary material for colony locations and sizes). Altogether, these archipelagos are home to more than 99% of the world population of D. exulans, while more than 60% of all couples breed in the colonies sampled for this study. Thirty-four Amsterdam albatross samples were collected in the sole existing colony. Since this species is critically endangered, this study has been approved by the ethics committee of Institut Polaire Français Paul-Émile-Victor and received permission from the French Ministry of Environment for handling protected species. Samples consisted of blood except for Marion Island birds and Amsterdam albatrosses for which feathers were plucked off chicks. DNA was extracted using either the QIAamp DNA blood mini kit (blood samples) or by isoamyl alcohol extraction and ethanol precipitation (feathers; Bello et al. 2001).
The AFLP procedure followed the method of Vos et al. (1995). Restriction enzymes EcoRI and MseI were used for digestion of whole genomic DNA. Six EcoRI/MseI primer pairs were used in the selective PCR, with each primer ending with three selective nucleotides: AAC/CAT, AAG/CCG, AAG/CGT, ACA/CAC, ACC/CGT and ACT/CCG. PCR products were loaded into an ABI Prism 3100 capillary sequencer and DNA fragments were analysed with GeneMarker v. 1.4 (Softgenetics). Only clear peaks in the range of 50–500 bp were retained as markers. Negative controls were included at each step to make sure that no contamination occurred. To test the repeatability of AFLP fragments, DNA was re-extracted for 47 individuals and the whole AFLP procedure was repeated for these individuals.
(b) Polymorphism and genetic diversity
The proportion of polymorphic loci (expressed in per cent) in the two albatross species was calculated as the proportion of loci for which the frequency of the most common allele was less than 0.95 (herein P5%). P5% is the parameter reported for all organisms to which we compared albatross data, except one (table 2). Markers with a frequency of more than 0% and less than 5% are considered monomorphic. This 5% threshold is intended to buffer for unequal sample sizes among studies. Fragments present in a single individual were excluded from the dataset, because a single genotyping error (e.g. due a slightly incomplete digestion) would create a false marker and therefore artificially increase the number of monomorphic loci under the 5% criterion.
Table 2.
Genetic diversity at AFLPs in albatrosses and other vertebrates in the literature. Data is representative of the range found in vertebrates.
| species | region/area covered | extensive coverage of species' range? | sample size | number of loci | % polymorphic locia | expected heterozygosity | source |
|---|---|---|---|---|---|---|---|
| wandering Albatross | Indian and Atlantic Oceans | yes | 400 | 234 | 5.1 | 0.028 | this study |
| Amsterdam Albatross | Amsterdam island (Indian Ocean) | yes | 34 | 234 | 2.1 | 0.010 | this study |
| snow goose | Bylot Island | no | 745 | 191 | 17 | 0.10 | Lecomte et al. (in preparation) |
| willow flycatcher (1 subspecies) | Southwestern United States | no | 290 | 708 | 27.8 | 0.221–0.348b | Busch et al. (2000) |
| horned grebe | Yukon, Québec, Iceland | yes | 90 | 96 | 29.2 | n.a. | M. Boulet (2006, personal communication) |
| house finch | North America and Hawaii | yes | 163 | 269 | 29.9c | 0.10c | Wang et al. (2003) |
| bluethroat (1 population) | Northwestern France | no | 162 | 232 | 34.9 | n.a. | Questiau et al. (1999) |
| warblers (2 subspecies) | Pyrenees | no | 30 | 251 | 56.2 | n.a. | Bensch et al. (2002) |
| wild turkey (5 subspecies) | North America and Mexico | yes | 379 | n.a. | 37–58, 54–73d | n.a. | Mock et al. (2002) |
| American eel | US Atlantic coast | yes | 193 | 373 | 51.7 | n.a. | V. Albert, (2006, personal communication) |
| European eel | Northeast Atlantic and Mediterranean Sea | no | 186 | 186 | 50.7 | n.a. | V. Albert, (2006, personal communication) |
| mole-rats | Israel/<1 ha | no | 20 | 729 | 59.9 | 0.097–0.159 | Polyakov et al. (2004) |
| Anolis lizards (two species) | Caribbean's/transects less than 7 km | no | 48–90 | 310–313 | 61.9–62.9 | n.a. | Ogden & Thorpe (2002) |
| giant salamander | Southern British Columbia (Canada) | no | 20–28 | n.a. | 53.7–97.6c | 0.192–0.386c | Curtis & Taylor (2004) |
5% criterion except for giant salamander for which it was close or equal to 5% as sample sizes within populations ranged 20–28.
Range of values for 20 sampling sites
Average of values within sampling localities.
Range of confidence intervals within defined subspecies.
We estimated allele frequencies with two methods accounting for the dominant nature of AFLPs. The first method simply uses the square root of the recessive genotype frequency as a maximum-likelihood estimator of the frequency q of the non-amplifying allele (assuming random mating), but is statistically biased particularly for small values of q (Lynch & Milligan 1994). The Bayesian method proposed by Zhivotovsky (1999) provides better estimates of q when the number of null-homozygotes is small or zero, as found in several of our markers. Parameters a and b for the prior beta distribution of genotype frequencies were computed from eqn (13) in Zhivotovsky (1999) using data from the 400 wandering and 34 Amsterdam albatrosses for the estimates of q within each species, respectively. Expected heterozygosity was obtained from He=2q(1−q). Within-colonies values of P5% and He were calculated for six out of the nine colonies for which sample size was greater than or equal to 20 (the minimal size for detecting polymorphism at the 5% threshold). Sampling locations with N<20 were used to increase geographical coverage when measuring global genetic diversity.
(c) Simulations to estimate ancestral polymorphism
To test the hypothesis that both albatross species have retained a set of AFLP markers which were already fixed in their most recent ancestor, we ran simulations in Maple v. 9.0 (Maplesoft) to estimate the total number of ancestral loci (ALtotal) from which the loci presently fixed in both species are derived, and the number of these ancestral loci which were already fixed for the dominant genotype (ALfixed) at time of speciation (see Appendix A for details). We define LP5% as the number of polymorphic loci in the current sample of 400 wandering +34 Amsterdam albatrosses. An estimate for the number of the ancestral polymorphic loci among those loci that gave rise to the currently observed fixed loci is ALtotal−ALfixed and so an estimate for the total number of ancestral polymorphic loci from which all observed loci were derived (both fixed and polymorphic) is (ALtotal−ALfixed+LP5%). Similarly, (ALtotal+LP5%) is an estimate for the total number of ancestral loci (fixed and polymorphic) from which all observed loci were derived. This leads to the following ancestral polymorphism estimate:
Hence, currently polymorphic loci were assumed to have been polymorphic at the time of speciation. This means that state transformations from fixed (ancestral) to polymorphic (current) due to mutation are not accounted for. Therefore, the above estimate is conservative, given our perspective which is to test whether polymorphism was already low in the ancestral albatross.
3. Results
(a) Polymorphism and heterozygosity in the wandering albatross
The six primer pairs used to generate AFLPs led to the amplification of 234 loci that could be unambiguously scored as present (peak) or absent (no peak). Genotyping errors and homoplasy were unlikely to affect our conclusions (see electronic supplementary material for details). Per cent polymorphic loci (P5%) averaged for six colonies of wandering albatrosses was 5.4±1.7% (s.d.) and global polymorphism across the species was similar (P5%=5.1%; table 1). Such a low level of polymorphism is roughly one-third of the minimum value reported in vertebrate populations for which AFLP data are available (table 2).
Table 1.
Within-colony and global genetic diversity at AFLPs in wandering and Amsterdam albatrosses.
| % polymorphic loci | expected heterozygosity | |||||||
|---|---|---|---|---|---|---|---|---|
| species | archipelago | colony | sample size | all loci | CG loci only | non-CG loci | square root estimator | Bayesian estimator |
| wandering | Kerguelen | Pointe Morne | 44 | 4.7 | 8.1 | 0.9 | 0.017 | 0.073 |
| albatross | Howe & islets | 28 | 6.0 | 10.5 | 0.9 | 0.015 | 0.066 | |
| Crozet | Baie du Marin | 83 | 4.3 | 4.0 | 1.8 | 0.018 | 0.075 | |
| Baie Américaine | 23 | 3.0 | 6.4 | 1.8 | 0.015 | 0.075 | ||
| Pointe Basse | 152 | 6.8 | 11.3 | 1.8 | 0.026 | 0.080 | ||
| Ile aux Cochons | 20 | 7.7 | 12.1 | 2.7 | 0.016 | 0.024 | ||
| alla | all colonies | 400 | 5.1 | 8.1 | 1.8 | 0.028 | 0.028 | |
| Amsterdam albatross | St-Paul & Amsterdam | Amsterdam island | 34 | 2.1 | 2.4 | 1.8 | 0.010 | 0.011 |
| two species pooled | 434 | 5.1 | 7.3 | 2.7 | 0.030 | 0.031 | ||
Global genetic variation also includes samples from Baie Larose (n=9), Courbet (n=9), Marion island (n=17) and Bird island (n=16; see electronic supplementary material).
Since methylation tends to reduce the relative frequency of CGs in the genome (Beutler et al. 1989), AFLP primers containing this dinucleotide at their selective 3′-end were found to amplify a lower number of bands but a higher proportion of polymorphic fragments than other primers (Bensch & Akesson 2005). This variation has the potential to confound interspecific comparisons when primers differ among studies. Our four primer pairs containing a ‘CG’ dinucleotide amplified a higher proportion of polymorphic fragments than the two ‘non-CG’ pairs (one-tailed pair-sample test on arcsin transformed data: t=4.82, d.f.=6, n=7 colonies (six wandering +one Amsterdam albatrosses), p<0.003; table 1). The difference between ‘CG’ and ‘non-CG’ primers varied with colony and reached a maximum of ten times at Howe and islets. However, P5% values always remained well below those reported for other vertebrates.
Heterozygosity (He) in albatrosses was also much smaller than that found in other species (table 2). However, this comparison is limited to five vertebrate species for which He was estimated in a comparable way from AFLP data. Global square root and Bayesian estimates were identical (0.028; table 1), but the second method gave consistently higher estimates of intra-colony heterozygosity (one-tailed pair-sample test on arcsin transformed data: t=−4.53, d.f.=6, n=7 colonies, p<0.003), likely reflecting the bias of the square root estimator when sample size is smaller (Zhivotovsky 1999). However, the magnitude of difference between the two methods was not significantly correlated to within-colony sample size (Spearman's rs =0.214, n=7, p>0.5). Likewise, correlation between He and sample size (rs=0.639, n=7 colonies, 0.2>p>0.1) or colony size (rs=−0.473, n=7 colonies, p>0.2) was not significant (results here are for He from Bayesian estimates but are alike for square root estimates). Thus, heterozygosity was very low for all colonies regardless of sample sizes or the number of breeding couples they count.
(b) Comparison between wandering and Amsterdam albatrosses
Genetic diversity was more than twice as low in Amsterdam than in wandering albatrosses (table 1). When genotypes from the two species were grouped, P5% remained identical to that measured in the wandering albatross alone, while heterozygosity slightly increased. Therefore, the pooled genetic diversity of albatrosses is much lower than that uncovered in any other, single, vertebrate population. This last observation did not follow from the unequal sample sizes between the two species. In fact, the Amsterdam albatross has only five polymorphic loci (5% threshold), two of them being also polymorphic in the wandering albatross. The pooled genetic diversity thus shows that both species almost share the same set of fixed markers. Indeed, out of 234 markers, 184 were fixed for the dominant phenotype in both species while only one was differentially fixed (i.e. present in one species and absent in the other species). Among the remaining markers, 15 had a frequency between 0.05 and 0.95 in at least one species, and 34 markers were considered as fixed for null-homozygotes because their frequency was less than 0.05 in both albatrosses.
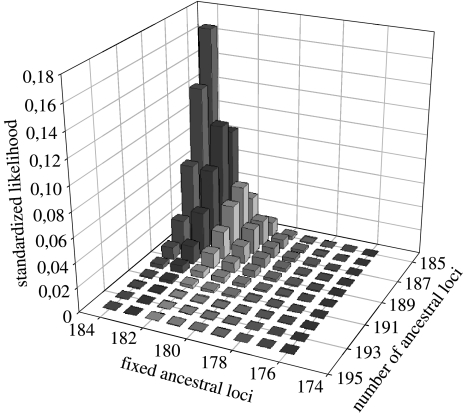
Simulations supported the hypothesis that the large proportion of fixed markers shared by the two albatross species was already fixed in their most recent ancestor. Under a pure drift model, the likelihood that ALtotal ancestral loci, among which ALfixed were fixed prior to speciation, led to the current pattern of fixation within the two species was maximal for values of ALtotal=185 and ALfixed=184 (figure 1). Moreover, the likelihood value decreases very quickly as one is moving away from these two values (99% CI for ALfixed: 179–184 loci). Ancestral polymorphism based on lower and upper bounds of this confidence interval was 8 and 12.8%, respectively. Since this estimate does not account for monomorphic loci with frequencies between 0 and 5%, it likely represents an overestimate of ancestral polymorphism.
Figure 1.
Likelihood that a set ALtotal of ancestral AFLP loci, among which a number ALfixed are fixed, leads to the observed pattern of fixation in the two albatross species after their divergence. The current pattern is 184 loci fixed in both species and one locus fixed differentially (i.e. present in the wandering Albatross and absent in the Amsterdam Albatross). Results are from 10 000 simulations for each starting values of ALtotal and ALfixed. Note that the maximum number of fixed ancestral loci cannot exceed its current number (184) since pure drift will maintain monomorphic loci as monomorphic. In addition, the number of ancestral loci has to be at least as large as the number of loci found in the dataset (185) since loci subjected to pure drift can only disappear through drift, i.e. after fixating for absence in both species (see the worked out example in Appendix A).
The interpretation of a shared impoverished genetic diversity that was inherited from an ancestral species implicitly assumes negligible gene flow between the two derived species. Despite their overall high similarity, these species nonetheless exhibited pronounced genetic differences at a few loci. One locus was diagnostic, being present in all wandering, but absent in all Amsterdam albatrosses. Among polymorphic loci, several exhibited sharp frequency differences among species. Assignment tests performed with Aflpop software (Duchesne & Bernatchez 2002) excluding the diagnostic locus supported a much greater differentiation between the two species than between any pair of wandering albatross colonies (see electronic supplementary material). Moreover, breeding phenology of the two species is distinct, making the occurrence of hybridization events unlikely.
4. Discussion
AFLP data revealed an extreme genetic uniformity in the wandering albatross, a species widespread in the Southern Ocean although with limited numbers. Among all vertebrates surveyed for AFLP to date, only the endangered Amsterdam albatross has a lower diversity. Admittedly, however, interspecific comparisons of AFLP variation may be complicated by several factors. First, among-studies differences exist in sample size and proportion of species' ranges covered. Nevertheless, we believe that the reduced genetic diversity in albatrosses is not a sampling artefact. Our sampling was extensive: all archipelagos where wandering albatrosses breed were sampled, except for the isolated Macquarie island which housed only 19 pairs in 2004 (Terauds et al. 2006). On the other hand, only a fraction of the range was sampled for several species included in table 2. Thus, any additional sampling in those species with limited coverage would be more likely to increase the difference observed with the wandering albatross, not the opposite. Moreover, when comparing within-colonies diversity—which represents much smaller geographical coverage and sample sizes similar to those for less extensively sampled species—wandering albatrosses are still well below other vertebrates. Second, scoring method may vary greatly among studies or investigators (Bonin et al. 2004). One way to ensure confidence in genotyping quality is to assess levels of error and their potential effects on estimates of genetic parameters (Pompanon et al. 2005). In our case, error rate was too low to have a significant effect on our genetic diversity estimates.
Several studies did not report levels of polymorphism and could not be included in our comparisons. In addition, AFLPs have rarely been used in population genetic studies of animals compared with plants (Bensch & Akesson 2005). These factors greatly limit the number of species to which albatrosses can be compared with. Nevertheless, given their present rank on the diversity scale, it appears very unlikely that albatrosses do not in fact lie at the lower bound of genetic diversity among vertebrates.
(a) Origin of impoverished genetic diversity in albatrosses
Our results suggested that two species of albatross with different population histories have inherited a poor genetic diversity from their common ancestor. Given the available estimate of time since divergence (which is ca 0.84 Myr ago based on cytochrome b), Ne must have been small over long evolutionary times so that low levels of genetic diversity have been maintained until the advent of contemporary populations. This would not rule out the possibility of bottleneck episodes as well. In particular, the twofold lower heterozygosity in the Amsterdam compared with the wandering albatross colonies may reflect the current bottleneck in that species.
To understand how genetic diversity could remain at low levels, one needs to consider both gains of mutational variants and their eventual loss or fixation through genetic drift (in this case the fixation of either AFLP allele). Obviously, the low annual fecundity of wandering and Amsterdam albatrosses, along with their limited numbers, slow down the accumulation of mutations relative to species with higher fecundity and/or population sizes (for a given mutation rate). Moreover, Nunn & Stanley (1998) found evidence for a slower rate mtDNA cytochrome b evolution in larger Procellariformes. The mutation rate was estimated at 0.62% per million year for albatrosses, but 0.78 and 0.92% for intermediate-sized and smallest species of Procellariiformes, respectively. These results were attributed to metabolic rates differences, which are known to correlate with body size (Nunn & Stanley 1998). Therefore, the extreme reduction in genetic diversity we observed in these two albatrosses might be partly attributable to a similar reduction in mutation rate at nuclear loci relative to smaller species.
Small long-term population size thus appears as the basic limiting factor in the accumulation of new polymorphisms, a phenomenon potentially accentuated by low mutation rates within the albatross lineage. It is hard, however, to identify a single basic factor that might explain the rate of loss of diversity through drift, for it is influenced not only by population size but also by a number of specific life-history traits. Single-locus simulations showed that the combined effect of generation overlap, life expectancy and age at first reproduction of albatrosses, lead to a rate of decay of heterozygosity per generation between two and three times faster than that of an ideal population of equal size (see electronic supplementary material). This is consistent with the expectations from theoretical population models for overlapping generations and under random mating (Rogers & Prügel-Bennett 2000). On the other hand, annual drift rate as estimated from simulations was 13 times slower than that of an ideal population with a 1 year generation time (see electronic supplementary material).
Other factors might have a large effect on the extent of genetic drift. Namely, metapopulation dynamics involving recurrent population extinction–colonization is known to reduce Ne compared to a classical island model of population structure (Whitlock 2003). Indeed, such dynamics represent a plausible alternative hypothesis for the genetic depletion in cheetahs (Hedrick 1996). From an analysis of 40 years of mark–recapture data, Inchausti & Weimerskirch (2002) found evidence for metapopulation dynamics in the wandering albatross. In addition, past glaciations are likely to have provoked frequent colonizations and extinctions of albatross populations (Alderman et al. 2005). On the other hand, assortative mating according to age and partner fidelity (Jouventin et al. 1999), along with the small variance in family size (Weimerskirch et al. 2005), may result in a maximal number of effective breeders contributing to the next generation, for a given census size. Therefore, alleles might be retained for a longer period than expected under a random mating scheme. In addition, Amos et al. (2001) reported a slight but significant negative relationship between parental similarity and reproductive success in the wandering albatross. As stated by Amos & Balmford (2001), greater fitness of outbred pairs may provide populations with ‘extra resilience against inbreeding depression and genetic erosion’.
Another key factor influencing the rate of genetic drift is temporal fluctuations in Ne. Sæther et al. (2004) showed that bird species located towards the slow end of the ‘slow–fast’ gradient of life histories (albatrosses representing the most extreme slow case) are better buffered against the demographic stochasticity. In other words, the amplitude of fluctuations in effective population size should be less important. Thus, even if albatross populations remained small, they might have been able to limit further loss of genetic diversity that is expected to be caused by the Ne fluctuations.
One scenario suggested by the above considerations is that albatrosses may have a very low accumulation of mutations coupled with a low rate of heterozygosity loss per year as compared to most other vertebrates. However, the hypothesis that the peculiar life-history traits of albatrosses may result in the maintenance of a low genetic diversity for the entire species lifespan will need to be confirmed by means of thorough demographically and spatially realistic simulations.
(b) Genetic diversity in other species
Poor genetic diversity has been uncovered in several top predators and/or long-lived species. For some of them such as Mauritius kestrels (Falco punctatus; Groombridge et al. 2000), data unequivocally point out to the effect of recent bottlenecks. In contrast, bottlenecks have had little impact on genetic diversity in fur seals (Arctocephalus gazella and A. tropicalis; Wynen et al. 2000) and on a population of box turtles (Terrapene ornata; Kuo & Janzen 2004), while the cause of the low genetic diversity in several greater Carnivores remain controversial (Merola 1994; Hedrick 1996; Amos & Balmford 2001). Impoverished genetic diversity in North Atlantic right whales (Eubalaena glacialis) was initially attributed to mass killing during extensive whaling that took place from the seventeenth to mid-twentieth centuries. But this interpretation has been challenged by historical mtDNA samples and right whales were possibly genetically depleted well before the seventeenth century (Rastogi et al. 2004). However, microsatellite variation in the more numerous South Atlantic right whales (E. australis) was about twice as high as in the E. glacialis (Waldick et al. 2002). Likewise, four microsatellite markers that were used in the wandering and in at least one other albatross species showed that heterozygosity is clearly lower in the former species for some of these markers (see electronic supplementary material). These observations indicate that comparable life histories do not necessarily lead to similar levels of genetic diversity. Given current knowledge, it is thus hard to tell whether the wandering albatross represents a unique genetic case due to a conjunction of particular life-history traits and population history, or if such a scenario has arisen in other species having similar traits.
(c) Surviving with a low genetic diversity
If species like albatrosses have thrived for nearly 1 Myr despite an extremely poor genetic diversity, they must have been able to avoid excessive inbreeding depression and erosion of evolutionary potential. To date, we have no clear indication that inbreeding depression may be occurring in albatrosses. Average reproductive (fledging) success of wandering albatrosses is generally high, even higher than that of many other Procellariiformes (Weimerskirch & Jouventin 1998). The quick recovery of the Amsterdam albatross colony despite minute genetic variation is also remarkable.
At least two mechanisms may contribute to the reduction of inbreeding depression. First, a species may develop inbreeding avoidance behaviours. Selection favouring mating between partners with dissimilar genotypes might be in action in wandering albatrosses (Amos et al. 2001). Whether these birds can recognize and avoid their kin is unknown, but odour-based individual recognition exists in other Procellariiformes (Bonadonna & Nevitt 2004). Sex-biased dispersal is generally the rule in birds (Greenwood 1980), and may also limit the occurrence of inbred mating. However, if the whole albatross population is highly inbred owing to long-term small Ne, inbreeding avoidance mechanisms might not be sufficient to explain their reproductive success. Second, inbreeding depression should be more severe in species having high genetic variation, that suddenly decrease dramatically in population size or become inbred for other reasons, as mutation load is expected to be higher for populations of large effective size. In small historically inbred populations, deleterious alleles may be purged faster after their first appearance, thus limiting the burden of mutational meltdown (Crnokrak & Barrett 2002; but see Ballou 1997). For example, in experimentally purged populations of Drosophila, inbreeding depression was only one-third of that observed in the original source population, after one generation of full-sib mating (Swindell & Bouzat 2006). A striking finding possibly attributable to purging was that genetic uniformity in a small herd of cattle does not impair on animal fertility and viability (Visscher et al. 2001).
Albatrosses appear to challenge the classical view about the negative impacts of genetic depletion on populations. Still, our findings do not dispute the importance of maintaining genetic diversity in natural populations. Several studies, especially those regarding inbreeding depression, have provided convincing evidence that impoverished diversity may cause serious reduction in fitness traits and increase extinction risk. Nonetheless, in the context of interpreting the consequences of genetic diversity with regards to conservation issues, our results show that some species may behave differently that previously appreciated.
Acknowledgments
We thank Richard Philips and Peter Ryan for samples from Bird Island and Marion Island, Cédric Marteau, Marie-Hélène Burle and Jean-Marie Vuanat for field assistance. Stéphanie Dano extracted DNA from feathers. This research was supported by Institut Polaire Paul-Émile-Victor program v. 109 (H.W.), the Canadian Research Chair in Genomics and Conservation of Aquatic Resources (L.B.), as well as a NSERC scholarship to E.M.
Appendix A. Estimating ancestral polymorphism
Suppose we are provided with a sample of genotypes for species A and B over some common set of AFLP loci. Fixed absence or fixed presence at a given locus are represented by scores 0 and 1, respectively. We consider only those loci for which there is fixation within each of species A and B, possibly to two different scores. We retain the largest subset for which a single species shows fixation for presence (1s) at all loci. For instance, from the following arrays of frequencies:
| species A: | 0.25 | 1 | 0 | 1 | 1 | 0.47 | 1 | 0.12 | 1 | 0.73 | 0.17 | 1 | 1 |
| species B: | 0.25 | 1 | 1 | 0 | 1 | 0.47 | 0 | 0 | 1 | 1 | 0.19 | 1 | 1 |
we would keep the subarrays:
| species A: | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| species B: | 1 | 0 | 1 | 0 | 1 | 1 | 1 |
Assuming that these fixed loci in A and B are derived from a set of ancestral loci, the goal is to estimate the total number of ancestral loci (ALtotal) and, among the latter, the number of loci which were already fixed (as 1s) at time of speciation (ALfixed). In the above two subarrays, species A contains seven 1s while species B has five. Let us refer to seven and five as the N1 and n1 fixation statistics, respectively. In other words, N1 and n1 represent the largest and the smallest number of loci fixed for presence within the retained subarrays, respectively.
A Monte Carlo simulator was built to estimate the probability that, given a pair of parameter values (ALtotal, ALfixed), the output random variables N1 and n1 would take on the observed values, i.e. 185 and 184, respectively in the wandering and Amsterdam albatrosses.
The simulator produces fixation statistics for a range of starting values of ALtotal and ALfixed. The originally fixed loci are scored as 1 in each species. The distribution of the dominant allele frequency p among originally polymorphic loci is assumed to be U [0,1], i.e. uniform over the (0,1) range. For each of the two species, each polymorphic locus is turned into a fixed locus, with probability of fixation for presence (1) proportional to p and probability of fixation for absence (0) proportional to 1−p. Note that even though p is the same for both species, X may be fixed for distinct values in the two species. More precisely, the simulator performs the following algorithm:
For each species and each polymorphic locus X:
a frequency p is selected at random based on U [0,1]
based on U [0,1], a number x is selected at random
if x<p then X is allocated score 1
else if x>p then X is allocated score 0
Then N1 and n1 statistics are computed as above. This completes a single iteration. The number of iterations for which N1 and n1 match the observed values (185, 184) is the likelihood of the pair of the parameter starting values (ALtotal, ALfixed). Standardized likelihoods are obtained by dividing each likelihood by the sum of likelihoods. The number of originally polymorphic loci for the set of currently fixed loci is thus ALtotal−ALfixed. Polymorphism in the ancestral species is estimated as described in §2. Confidence intervals can be computed by considering most likely continuous sets of (ALtotal, ALfixed) values.
A.1 Worked out example of a single simulation iteration
Suppose ALtotal=10, ALfixed=5 and observed N1, n1 are 8, 5, respectively,
- Since the number of originally polymorphic loci is assumed to be ALtotal−ALfixed=5, five polymorphic loci are made to drift until fixation:
- U [0,1] randomly allocates ancestral p frequencies for each locus
- 0.43 0.72 0.21 0.85 0.67
- a fixation score is randomly allocated to each species/locus following p
- species A: 1, 1, 0, 0, 0
- species B: 0, 1, 0, 1, 1
- scores of originally fixed loci:
- species A: 1, 1, 1, 1, 1
- species B: 1, 1, 1, 1, 1
- complete scores for contemporary species from (1) and (2):
- species A: 1, 1, 0, 0, 0, 1, 1, 1, 1, 1
- species B: 0, 1, 0, 1, 1, 1, 1, 1, 1, 1
- subarrays:
- 1, 0, 0, 1, 1, 1, 1, 1
- 1, 1, 1, 1, 1, 1, 1, 1
- N1=8, n1=6
Match the observed N1, n1? No, therefore the iteration is scored a 0.
Supplementary Material
Map of the breeding distribution of albatrosses and sampling
Genotyping errors and homoplasy
Allocations tests, single-locus simulations, microsatellite diversity in albatrosses
References
- Alderman R, Double M.C, Valencia J, Gales R.P. Genetic affinities of newly sampled populations of wandering and black-browed albatross. Emu. 2005;105:169–179. doi:10.1071/MU04034 [Google Scholar]
- Amos W, Balmford A. When does conservation genetics matter? Heredity. 2001;87:257–265. doi: 10.1046/j.1365-2540.2001.00940.x. doi:10.1046/j.1365-2540.2001.00940.x [DOI] [PubMed] [Google Scholar]
- Amos W, Harwood J. Factors affecting levels of genetic diversity in natural populations. Phil. Trans. R. Soc. B. 1998;353:177–186. doi: 10.1098/rstb.1998.0200. doi:10.1098/rstb.1998.0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos W, Worthington Wilmer J, Fullard K, Burg T.M, Croxall J.P, Bloch D, Coulson T. The influence of parental relatedness on reproductive success. Proc. R. Soc. B. 2001;268:2021–2027. doi: 10.1098/rspb.2001.1751. doi:10.1098/rspb.2001.1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou J.D. Ancestral inbreeding only minimally affects inbreeding depression in mammalian populations. J. Hered. 1997;88:169–178. doi: 10.1093/oxfordjournals.jhered.a023085. [DOI] [PubMed] [Google Scholar]
- Bellinger M.R, Johnson J.A, Toepfer J, Dunn P. Loss of genetic variation in greater prairie chickens following a population bottleneck in Wisconsin, USA. Conserv. Biol. 2003;17:717–724. doi:10.1046/j.1523-1739.2003.01581.x [Google Scholar]
- Bello N, Francino O, Sanchez A. Isolation of genomic DNA from feathers. J. Vet. Diag. Invest. 2001;13:162–164. doi: 10.1177/104063870101300212. [DOI] [PubMed] [Google Scholar]
- Bensch S, Akesson M. Ten years of AFLP in ecology and evolution: why so few animals? Mol. Ecol. 2005;14:2899–2914. doi: 10.1111/j.1365-294X.2005.02655.x. doi:10.1111/j.1365-294X.2005.02655.x [DOI] [PubMed] [Google Scholar]
- Bensch S, Helbig A.J, Salomon M, Siebold I. Amplified fragment length polymorphism analysis identifies hybrids between two subspecies of warblers. Mol. Ecol. 2002;11:473–481. doi: 10.1046/j.0962-1083.2001.01455.x. doi:10.1046/j.0962-1083.2001.01455.x [DOI] [PubMed] [Google Scholar]
- Beutler E, Gelbart T, Han J.H, Koziol J.A, Beutler B. Evolution of the genome and the genetic code: selection at the dinucleotide level by methylation and polyribonucleotide cleavage. Proc. Natl Acad. Sci. USA. 1989;86:192–196. doi: 10.1073/pnas.86.1.192. doi:10.1073/pnas.86.1.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonadonna F, Nevitt G.A. Partner-specific odor recognition in an Antarctic seabird. Science. 2004;306:835. doi: 10.1126/science.1103001. doi:10.1126/science.1103001 [DOI] [PubMed] [Google Scholar]
- Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P. How to track and assess genotyping errors in population genetics studies. Mol. Ecol. 2004;13:3261–3273. doi: 10.1111/j.1365-294X.2004.02346.x. doi:10.1111/j.1365-294X.2004.02346.x [DOI] [PubMed] [Google Scholar]
- Burg T.M, Croxall J.P. Global population structure and taxonomy of the wandering albatross species complex. Mol. Ecol. 2004;13:2345–2355. doi: 10.1111/j.1365-294X.2004.02232.x. doi:10.1111/j.1365-294X.2004.02232.x [DOI] [PubMed] [Google Scholar]
- Busch J.D, Miller M.P, Paxton E.H, Sogge M.K, Keim P. Genetic variation in the endangered southwestern willow flycatcher. Auk. 2000;117:586–595. doi:10.1642/0004-8038(2000)117[0586:GVITES]2.0.CO;2 [Google Scholar]
- Campbell D, Bernatchez L. Generic scan using AFLP markers as a means to assess the role of directional selection in the divergence of sympatric whitefish ecotypes. Mol. Biol. Evol. 2004;21:945–956. doi: 10.1093/molbev/msh101. doi:10.1093/molbev/msh101 [DOI] [PubMed] [Google Scholar]
- Crnokrak P, Barrett S.C.H. Perspective: purging the genetic load: a review of the experimental evidence. Evolution. 2002;56:2347–2358. doi: 10.1111/j.0014-3820.2002.tb00160.x. doi:10.1554/0014-3820(2002)056[2347:PPTGLA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Curtis J.M.R, Taylor E.B. The genetic structure of coastal giant salamanders (Dicamptodon tenebrosus) in a managed forest. Biol. Conserv. 2004;115:45–54. doi:10.1016/S0006-3207(03)00092-2 [Google Scholar]
- Duchesne P, Bernatchez L. Aflpop: a computer program for simulated and real population allocation, based on AFLP data. Mol. Ecol. Notes. 2002;2:380–383. doi:10.1046/j.1471-8286.2002.00251.x [Google Scholar]
- England P.R, Osler G.H.R, Woodworth L.M, Montgomery M.E, Briscoe D.A, Frankham R. Effects of intense versus diffuse population bottlenecks on microsatellite genetic diversity and evolutionary potential. Conserv. Genet. 2003;4:595–604. doi:10.1023/A:1025639811865 [Google Scholar]
- Frankham R, Ballou J.D, Briscoe D.A. Cambridge University Press; Cambridge, UK: 2002. Introduction to conservation genetics. [Google Scholar]
- Gales R. Albatross populations: status and threats. In: Robertson G, Gales R, editors. Albatross biology and conservation. Surrey Beatty & Sons; Chipping Norton, Australia: 1997. pp. 20–45. [Google Scholar]
- Greenwood P.J. Mating systems, philopatry and dispersal in birds and mammals. Anim. Behav. 1980;28:1140–1162. doi:10.1016/S0003-3472(80)80103-5 [Google Scholar]
- Groombridge J.J, Jones C.G, Bruford M.W, Nichols R.A. ‘Ghost’ alleles of the Mauritius kestrel. Nature. 2000;403:616. doi: 10.1038/35001148. doi:10.1038/35001148 [DOI] [PubMed] [Google Scholar]
- Hedrick P.W. Bottleneck(s) or metapopulation in cheetahs. Conserv. Biol. 1996;10:897–899. doi:10.1046/j.1523-1739.1996.10030897.x [Google Scholar]
- Hoelzel A.R, Fleischer R.C, Campagna C, Le Boeuf B.J, Alvord G. Impact of a population bottleneck on symmetry and genetic diversity in the northern elephant seal. J. Evol. Biol. 2002;15:567–575. doi:10.1046/j.1420-9101.2002.00419.x [Google Scholar]
- Inchausti P, Weimerskirch H. Dispersal and metapopulation dynamics of an oceanic seabird, the wandering albatross, and its consequences for its response to long-line fisheries. J. Anim. Ecol. 2002;71:765–770. doi:10.1046/j.1365-2656.2002.00638.x [Google Scholar]
- Jouventin P, Lequette B, Dobson F.S. Age-related mate choice in the wandering albatross. Anim. Behav. 1999;57:1099–1106. doi: 10.1006/anbe.1999.1083. doi:10.1006/anbe.1999.1083 [DOI] [PubMed] [Google Scholar]
- Kuo C.H, Janzen F.J. Genetic effects of a persistent bottleneck on a natural population of ornate box turtles (Terrapene ornata) Conserv. Genet. 2004;5:425–437. doi:10.1023/B:COGE.0000041020.54140.45 [Google Scholar]
- Lecomte, N., Gauthier, G., Bernatchez, L. & Giroux, J.-F. In preparation. Fine-scale genetic structure inside colonies: balancing sex-biased dispersal and rearing site fidelity of greater snow geese.
- Lynch M, Milligan B.G. Analysis of population genetic structure with rapid markers. Mol. Ecol. 1994;3:91–99. doi: 10.1111/j.1365-294x.1994.tb00109.x. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, O'Brien S.J. Dating the genetic bottleneck of the African cheetah. Proc. Natl Acad. Sci. USA. 1993;90:3172–3176. doi: 10.1073/pnas.90.8.3172. doi:10.1073/pnas.90.8.3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merola M. Reassessment of homozygosity and the case for inbreeding depression in the cheetah, Acinonyx jubatus: implications for conservation. Conserv. Biol. 1994;8:961–971. doi:10.1046/j.1523-1739.1994.08040961.x [Google Scholar]
- Mock K.E, Theimer T.C, Rhodes O.E, Greenberg D.L, Keim P. Genetic variation across the historical range of the wild turkey (Meleagris gallopavo) Mol. Ecol. 2002;11:643–657. doi: 10.1046/j.1365-294x.2002.01467.x. doi:10.1046/j.1365-294X.2002.01467.x [DOI] [PubMed] [Google Scholar]
- Nunn G.B, Stanley S.E. Body size effects and rates of cytochrome b evolution in tube-nosed seabirds. Mol. Biol. Evol. 1998;15:1360–1371. doi: 10.1093/oxfordjournals.molbev.a025864. [DOI] [PubMed] [Google Scholar]
- O'Brien S.J. Genetic and phylogenetic analyses of endangered species. Annu. Rev. Genet. 1994;28:467–489. doi: 10.1146/annurev.ge.28.120194.002343. doi:10.1146/annurev.ge.28.120194.002343 [DOI] [PubMed] [Google Scholar]
- Ogden R, Thorpe R.S. The usefulness of amplified fragment length polymorphism markers for taxon discrimination across graduated fine evolutionary levels in Caribbean Anolis lizards. Mol. Ecol. 2002;11:437–445. doi: 10.1046/j.0962-1083.2001.01442.x. doi:10.1046/j.0962-1083.2001.01442.x [DOI] [PubMed] [Google Scholar]
- Penhallurick J, Wink M. Analysis of the taxonomy and nomenclature of the Procellariiformes based on complete nucleotide sequences of the mitochondrial cytochrome b gene. Emu. 2004;104:125–147. doi:10.1071/MU01060 [Google Scholar]
- Polyakov A, Beharav A, Avivi A, Nevo E. Mammalian microevolution in action: adaptive edaphic genomic divergence in blind subterranean mole-rats. Proc. R. Soc. B. 2004;271:S156–S159. doi: 10.1098/rsbl.2003.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompanon F, Bonin A, Bellemain E, Taberlet P. Genotyping errors: causes, consequences and solutions. Nat. Rev. Genet. 2005;6:847–859. doi: 10.1038/nrg1707. doi:10.1038/nrg1707 [DOI] [PubMed] [Google Scholar]
- Questiau S, Eybert M.C, Taberlet P. Amplified fragment length polymorphism (AFLP) markers reveal extra-pair parentage in a bird species: the bluethroat (Luscinia svecica) Mol. Ecol. 1999;8:1331–1339. doi: 10.1046/j.1365-294x.1999.00703.x. doi:10.1046/j.1365-294X.1999.00703.x [DOI] [PubMed] [Google Scholar]
- Rastogi T, Brown M.W, McLeod B.A, Frasier T.R, Grenier R, Cumbaa S.L, Nadarajah J, White B.N. Genetic analysis of 16th-century whale bones prompts a revision of the impact of Basque whaling on right and bowhead whales in the western North Atlantic. Can. J. Zool. 2004;82:1647–1654. doi:10.1139/z04-146 [Google Scholar]
- Robertson C.J.R, Nunn G.B. Towards a new taxonomy for albatrosses. In: Robertson G, Gales R, editors. Albatross biology and conservation. Surrey Beatty & Sons; Chipping Norton, Australia: 1998. pp. 13–19. [Google Scholar]
- Rogers A, Prügel-Bennett A. Evolving populations with overlapping generations. Theor. Popul. Biol. 2000;57:121–129. doi: 10.1006/tpbi.1999.1446. doi:10.1006/tpbi.1999.1446 [DOI] [PubMed] [Google Scholar]
- Russello M.A, Gladyshev E, Miquelle D, Caccone A. Potential genetic consequences of a recent bottleneck in the Amur tiger of the Russian far east. Conserv. Genet. 2004;5:707–713. doi:10.1007/s10592-004-1860-2 [Google Scholar]
- Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I. Inbreeding and extinction in a butterfly metapopulation. Nature. 1998;392:491–494. doi:10.1038/33136 [Google Scholar]
- Sæther B.-E, et al. Life-history variation predicts the effects of demographic stochasticity on avian population dynamics. Am. Nat. 2004;164:793–802. doi: 10.1086/425371. doi:10.1086/425371 [DOI] [PubMed] [Google Scholar]
- Sherwin W.B, Moritz C. Managing and monitoring genetic erosion. In: Young A.G, Clarke G.M, editors. Genetics, demography and viability of framented populations. Cambridge University Press; Cambridge, UK: 2000. pp. 9–34. [Google Scholar]
- Spielman D, Brook B.W, Briscoe D.A, Frankham R. Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv. Genet. 2004;5:439–448. doi:10.1023/B:COGE.0000041030.76598.cd [Google Scholar]
- Swindell W.R, Bouzat J.L. Reduced inbreeding depression due to historical inbreeding in Drosophila melanogaster: evidence for purging. J. Evol. Biol. 2006;19:1257–1264. doi: 10.1111/j.1420-9101.2005.01074.x. doi:10.1111/j.1420-9101.2005.01074.x [DOI] [PubMed] [Google Scholar]
- Terauds A, Gales R, Baker G.B, Alderman R. Population and survival trends of wandering Albatrosses (Diomedea exulans) breeding on Macquarie Island. Emu. 2006;106:211–218. doi:10.1071/MU06007 [Google Scholar]
- Tickell W.L.N. Yale University Press; New Haven, CT: 2000. Albatrosses. [Google Scholar]
- Visscher P.M, Smith D, Hall S.J.G, Williams J.L. A viable herd of genetically uniform cattle. Nature. 2001;409:303. doi: 10.1038/35053160. doi:10.1038/35053160 [DOI] [PubMed] [Google Scholar]
- Vos P, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldick R.C, Kraus S.S, Brown M.W, White B.N. Evaluating the effects of historic bottleneck events: an assessment of microsatellite variability in the endangered North Atlantic right whale. Mol. Ecol. 2002;11:2241–2250. doi: 10.1046/j.1365-294x.2002.01605.x. doi:10.1046/j.1365-294X.2002.01605.x [DOI] [PubMed] [Google Scholar]
- Wang Z, Baker A, Hill G, Edwards S. Reconciling actual and inferred population histories in the house finch (Carpodacus mexicanus) by AFLP analysis. Evolution. 2003;57:2852–2864. doi: 10.1111/j.0014-3820.2003.tb01526.x. doi:10.1554/03-159 [DOI] [PubMed] [Google Scholar]
- Weimerskirch H, Jouventin P. Population dynamics of the wandering albatross, Diomedea exulans, of the Crozet Islands: causes and consequences of the population decline. Oikos. 1987;49:315–322. doi:10.2307/3565767 [Google Scholar]
- Weimerskirch H, Jouventin P. Changes in population sizes and demographic parameters of six albatross species breeding on the French sub-Antarctic islands. In: Robertson G, Gales R, editors. Albatross biology and conservation. Surrey Beatty & Sons; Chipping Norton, Australia: 1998. pp. 84–91. [Google Scholar]
- Weimerskirch H, Brothers N, Jouventin P. Population dynamics of wandering albatross Diomedea exulans and Amsterdam albatross D. amsterdamensis in the Indian Ocean and their relationships with long-line fisheries: conservation implications. Biol. Conserv. 1997;79:257–270. doi:10.1016/S0006-3207(96)00084-5 [Google Scholar]
- Weimerskirch H, Lallemand J, Martin J. Population sex ratio variation in a monogamous long-lived bird, the wandering albatross. J. Anim. Ecol. 2005;74:285–291. doi:10.1111/j.1365-2656.2005.00922.x [Google Scholar]
- Whitlock M.C. Fixation probability and time in subdivided populations. Genetics. 2003;164:767–779. doi: 10.1093/genetics/164.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisely S.M, Buskirk S.W, Fleming M.A, McDonald D.B, Ostrander E.A. Genetic diversity and fitness in black-footed ferrets before and during a bottleneck. J. Hered. 2002;93:231–237. doi: 10.1093/jhered/93.4.231. doi:10.1093/jhered/93.4.231 [DOI] [PubMed] [Google Scholar]
- Wynen L.P, Goldsworthy S.D, Guinet C, Bester M.N, Boyd I.L, Gjertz J.G, Hofmeyr G.J.G, White R.W.G, Slade R. Postsealing genetic variation and population structure of two species of fur seal (Arctocephalus gazella and A. tropicalis) Mol. Ecol. 2000;9:299–314. doi: 10.1046/j.1365-294x.2000.00856.x. doi:10.1046/j.1365-294x.2000.00856.x [DOI] [PubMed] [Google Scholar]
- Zhang B, Fang S.G, Xi Y.M. Low genetic diversity in the Endangered crested ibis Nipponia nippon and implications for conservation. Bird Conserv. Int. 2004;14:183–190. doi:10.1017/S0959270904000231 [Google Scholar]
- Zhivotovsky L.A. Estimating population structure in diploids with multilocus dominant DNA markers. Mol. Ecol. 1999;8:907–913. doi: 10.1046/j.1365-294x.1999.00620.x. doi:10.1046/j.1365-294x.1999.00620.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Map of the breeding distribution of albatrosses and sampling
Genotyping errors and homoplasy
Allocations tests, single-locus simulations, microsatellite diversity in albatrosses