Abstract
The gene encoding dARC1, one of three Drosophila homologs of mammalian activity-regulated cytoskeleton-associated protein (ARC), is upregulated in both seizure and muscular hypercontraction mutants. In this study we generate a null mutant for dArc1 and show that this gene is not involved in synaptic plasticity at the larval neuromuscular junction or in formation or decay of short-term memory of courtship conditioning, but rather is a modifier of stress-induced behavior. dARC1 is expressed in a number of neurosecretory cells and mutants are starvation resistant, exhibiting an increased time of survival in the absence of food. Starvation resistance is likely due to the fact that dArc1 mutants lack the normal hyperlocomotor response to starvation, which is almost universal in the animal kingdom. dARC1 acts in insulin-producing neurons of the pars intercerebralis to control this behavior, but does not appear to be a general regulator of insulin signaling. This suggests that there are multiple modes of communication between the pars and the ring gland that control starvation-induced behavioral responses.
Keywords: Synaptic plasticity, learning, insulin, adipokinetic hormone, locomotion
INTRODUCTION
Gene expression changes in response to acute alterations in a cell's environment or metabolic state is a common feature of all organisms. The rapid, and usually transient, activation of immediate early genes is the first response to many cellular stimuli. The expression of immediate early genes does not require new protein synthesis and is believed to initiate secondary processes which mediate longer-lasting changes that allow cells to adapt to new conditions. In the mammalian brain, immediate early genes are induced in response to high levels of synaptic activity. In Drosophila, seizure activity upregulates a diverse set of genes that function in adhesion, signaling and cell excitability (Guan et al., 2005). While many of these gene are involved in synaptic plasticity and activity-dependent modification of circuits, it is also important to note that high levels or activity are metabolically costly, decreasing neuronal ATP/ADP ratio (Crotty et al., 2006). The brain, like the rest of the body, has to deal with the energy costs of high levels of activity and a number of immediate early genes have been shown to be involved in metabolic regulation (Koo et al., 2005; Suo et al., 2006; Yamagata et al., 1994).
Activity-regulated cytoskeleton associated protein, (ARC), is a mammalian neuronal immediate early gene that shows robust and cell-specific upregulation with activity (Lyford et al., 1995). ARC has roles in several types of plasticity and appears to regulate vesicle trafficking and receptor insertion in mice (Tzingounis and Nicoll, 2006). ARC has also been shown to be induced by insulin signaling (Kremerskothen et al., 2002). In Drosophila, the dArc1 gene (CG12505), the closest homolog of mammalian Arc, is upregulated by seizure (Guan et al., 2005). To investigate the function of dARC1 in Drosophila we have generated mutants and characterized them electrophysiologically and behaviorally. We find that the dipteran homolog of ARC is also an activity-inducible gene. Mutational analysis of dArc1 indicates its main role appears to be in the behavioral responses to metabolic stress, and that it does not function in synaptic plasticity in Drosophila.
RESULTS
Drosophila Arc homolog genes are clustered and encode proteins homologous to the C-terminal of mammalian ARC
Homology searching against the Drosophila genome using the mammalian Arc sequence identifies three predicted genes located on chromosome 2: CG12505, CG13941 and CG10102. CG12505 (dArc1) and CG13941 (dArc2) were both identified on microarrays of seizure (Guan et al., 2005) and muscle hypercontraction mutants (Montana and Littleton, 2006), but only dArc1 was upregulated by seizure activity. CG10102 (dArc3) message has not been detected in any of our microarray screens (J.T. Littleton et al., unpublished data) and is not represented in any of the cDNA libraries that are publicly available for screening in silico, indicating that it may be a pseudogene. Alignment of the protein products of dArc1, dArc2 and mammalian Arc (Figure 1A) reveals that the Drosophila genes have homology only to the C-terminal half of mammalian Arc, missing the entire N-terminus (218 amino acids) and encoding proteins approximately half the size of mammalian ARC. Within the C-terminal, the homology of dARC1 to ARC consists of 23% identity and 40% similarity.
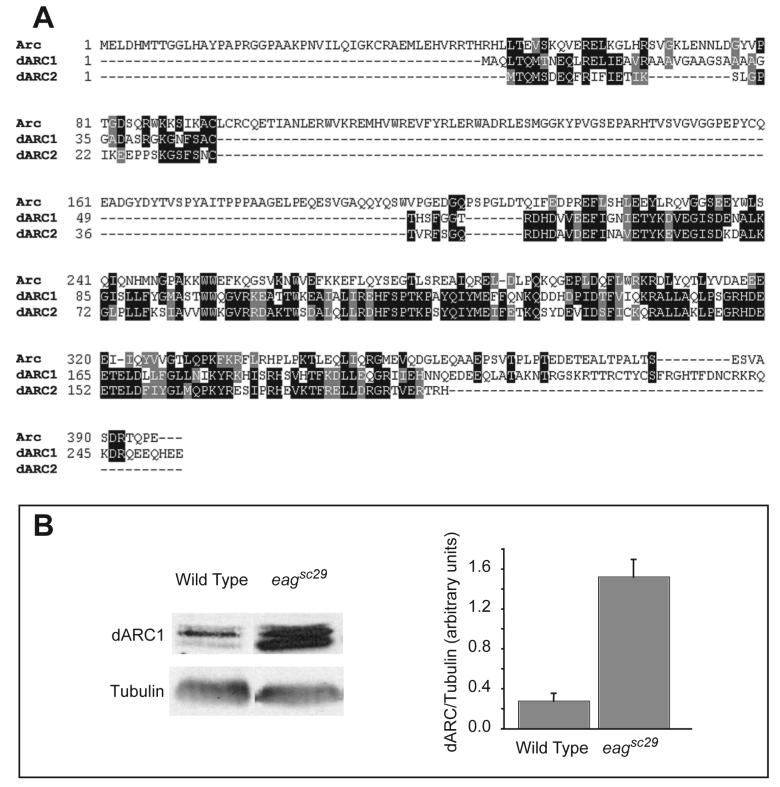
Figure 1. Drosophila has Arc homologs.
A) Alignment of rat ARC with the Drosophila dARC1 and dARC2 proteins. The fly proteins are smaller and homologous primarily to the C-terminus of the mammalian protein. B) Drosophila dARC1 is induced by neuronal hyperexcitability. Immunoblots of dARC1 and tubulin (as a loading control) from wild type Canton S and eagsc29 heads are shown at left. Quantification of four experiments shows that dARC1 is significantly (P < 0.001, Student's T-test) induced by the seizure activity of the eag potassium channel mutant.
We generated antibodies against the N-terminus of either dARC1, a region which should not cross-react with dARC2 or dARC3. Western analysis of fly head extracts demonstrated immunoreactive proteins of the predicted MW for dARC1 (Figure 1B). The induction by activity seen by microarray analysis also holds true for the protein product of this gene. dARC1 levels are significantly increased in head extracts from the hyperexcitable mutant eagsc29 (Wu et al., 1983) compared to Canton S wild type. These results indicate that the upregulation of the ARC family of gene products in response to enhanced neuronal activity is conserved from invertebrates to mammals.
dARC1 is expressed in the nervous system and neurosecretory tissue
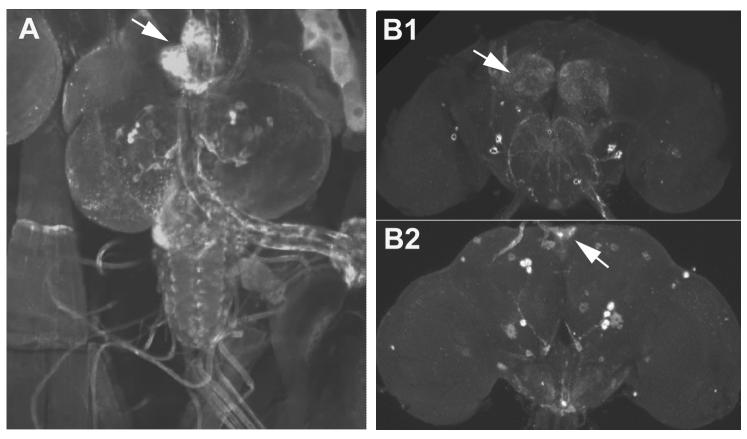
To determine the in vivo expression pattern of dARC1, we stained third instar larval brains using anti-dARC1 antibody. Examination of whole mount brains with anti-dARC1 antisera revealed immunoreactivity in a number of large neurons that innervate the ring gland (Figure 2A), a neurosecretory organ responsible for regulation of glucose metabolism (Wu and Brown, 2006) and ecdysteroid and juvenile hormone synthesis (Flatt et al., 2005). The ring gland itself also appears to express high levels of dARC1, and there is also expression in the ventral ganglion. In the adult brain, dARC1 is expressed in large neurons of the central brain that are located in the pars intercerebralis and in the lateral brain (Figure 2B). dARC1 can also be seen in the antennal lobes and in the subesophageal ganglion, areas associated with olfactory and taste processing respectively.
Figure 2. dARC1 expression in larval and adult CNS.
Dissected larvae or adult brains were stained with rabbit anti-dARC1 (1:1000) and imaged using confocal microscopy. A) Third instar larval dissection. Several pairs of large neurons are seen in the central brain as well as in the ventral ganglion. Arrow indicates the larval ring gland. B) Adult brain. Top panel is an anterior confocal stack. Arrow indicates antennal lobes. Bottom is a posterior view showing large paired central brain neurons. Arrow indicates the pars intercerebralis.
Generation of a dArc1 mutant
Mammalian ARC has been implicated in synaptic plasticity in a variety of paradigms, but its biochemical function has been obscure. To analyze dARC1 function, we generated null mutations by imprecise excision of a P-element located between the intronless dArc1 and dArc2 genes (Figure 3A). By PCR, dArc1esm18 and dArc1esm113 deleted the entire coding region of dArc1, but did not affect dArc2. dArc1esm295 and dArc1esm115 were precise excisions from the P-element screen and were used as controls in subsequent experiments. Two lines, dArc1esm18 and dArc1esm115 were chosen for full characterization. Figure 3B shows an immunoblot probed with anti-dARC1, indicating that dArc1esm18 does not express dARC1 protein, but still has some non-specific higher molecular weight bands. Immunohistochemical staining of dArcesm18 third instar larval brains demonstrates loss of staining in the cell bodies of large central brain and pars intercerebralis cells, as well as most cell bodies in the ventral ganglion. Residual staining in the ventral ganglion likely reflects non-specific staining. Using antisera generated against a region of dARC2 that has low homology to dARC1 and dARC3 (see Experimental Procedures), we find that dARC2 is still present in dArc1esm18, and its levels are not changed compared to the revertant control (Figure 3C), indicating that lack of dARC1 does not result in an alteration of dARC2 regulation. It is likely that dARC2 would not be able to compensate for dARC1 function in any case since it's expression pattern does not overlap with that of dARC1 and is predominantly glial (M.D.M., unpublished data).
Figure 3. Generation of a null mutation in dArc1.
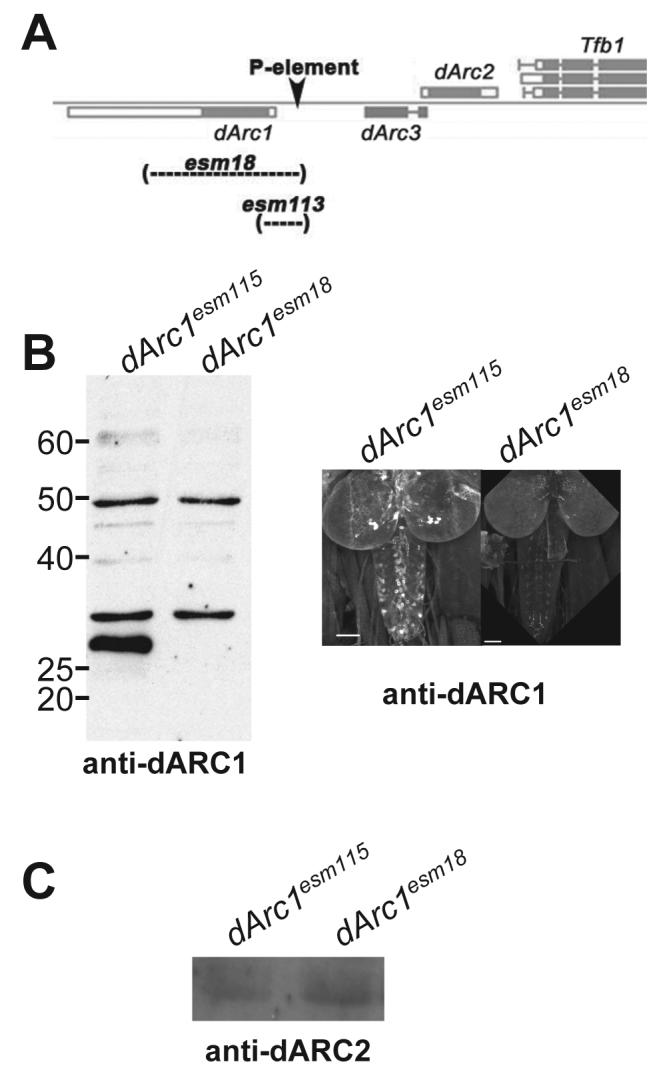
A) Schematic map of the dArc region showing exon structure. Transcription units that go left to right are above the line, while units that go right to left are below. A P-element inserted between dArc1 and dArc3 (position indicated by arrowhead) was mobilized to generation precise excisions (control lines dArc1esm115 and dArc1esm295) and deletions that take out all (dArc1esm18) or part (dArc1esm113) of the dArc1 locus. Deleted regions are indicated by a dotted line. B) dArc1esm18 is a null allele. Extracts of mutant and precise excision flies were immunoblotted with anti-dARC1 (left) and third instar larval brains of the same genotypes were stained (right). A protein of the predicted molecular weight for dARC1 is missing in the mutant flies, while high molecular weight background bands remain. Mutant brains lack staining in central brain and in the region of pars neurosecretory cells, but show some residual background staining in the ventral ganglion. Scale bar = 50 μm. C) Deletion of dArc1 does not significantly affect levels of dARC2. Head extracts from deletion and precise excision flies were immunoblotted with anti-dARC2. Both lines show normal dARC2 expression.
dARC1 does not contribute to basal function or activity-dependent plasticity at the larval neuromuscular junction
Mammalian ARC protein accumulates at recently activated synaptic regions (Moga et al., 2004) and it has been implicated in memory consolidation (Guzowski et al., 2000) and AMPA receptor trafficking in mammals (Chowdhury et al., 2006). The ability of activity to induce expression of dArc1 and its homology to the mammalian Arc gene suggested that it might be involved in synaptic plasticity. Development of the NMJ in Drosophila requires normal levels of synaptic activity (for review see Keshishian et al., 1994). If dARC1 is important for neuronal function, loss of activity-dependent dARC1 accumulation during development might therefore be expected to disrupt both basal and stimulation-dependent functions of the third instar NMJ. To assess this possibility we measured spontaneous and evoked activity at the muscle 6 NMJ in segment A3. No significant differences were found between amplitude and frequency of dArc1esm18 and control dArc1esm115 mEJPs recorded in the presence of 1.5 mM external calcium (Kolmogorov-Smirnov test, P > 0.05, Figure 4A). Evoked activity was also normal in the dArc1esm18 null as measured by EJP amplitude after single pulse stimulation in 0.1, 0.2, 0.4 and 1.5 mM calcium (Figure 4B, Student's t-test, P > 0.05 compared to dArc1esm115). We also examined the effects of overexpression of dARC1 in either the presynaptic motor neurons or the muscle and saw no effects on spontaneous or evoked EJPs (E.S.M. and J.T.L., data not shown).
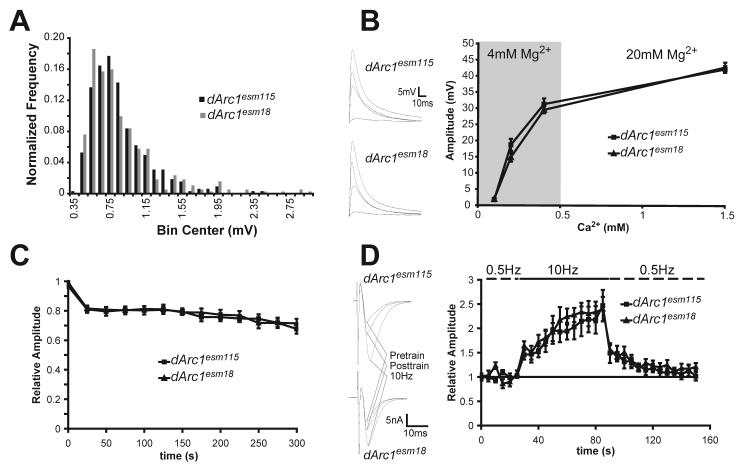
Figure 4. Mutation of dArc1 does not alter basal transmission or short-term synaptic plasticity at the larval neuromuscular junction.
Recordings were made from muscle 6 from abdominal segments A3-5 in HL3 with indicated ion concentrations. A) Miniature EJPs were recorded from mutant and precise excision controls in 1.5 mM calcium with 3 μM TTX. No difference was seen in either frequency or amplitude of mEJPs. B) Evoked EJPs were recorded in mutant and precise excision animals in varying calcium. Higher calcium levels required elevation of magnesium to suppress muscle contraction. Averages of 10 traces from each calcium concentration are shown at left. Amplitude increased as a function of extracellular calcium, but did not differ between genotypes. C) High frequency stimulation does not differentially alter vesicle recycling in mutant and precise excision controls. 10 Hz stimulation in 1.5 mM calcium and 20 mM magnesium reduced release by about 30% in both genotypes measured in current clamp. D) Post-tetanic potentiation is identical in mutant and precise excision control animals as measured in voltage clamp. Averages of 10 traces from the 0.5 Hz pre-tetanus test pulses, the 10 Hz tetanus, and the post-tetanus 0.5 Hz test pulses are shown at left.
To test the requirement for dARC1 in vesicle recycling, we tested the response of mutant and control NMJs to high-frequency (10 Hz) stimulation. Such stimulation alters transmission via a number of mechanisms, including by modulation of actin cytoskeleton-dependent processes (for review see Bader et al., 2004) with changes in vesicle recycling dynamics leading to a reduced EJP amplitude after prolonged stimulation. Both mutant and control NMJs, showed a similar reduction of about 30% from baseline after 5 min of stimulation (Figure 4C). These results suggest that dARC1 is not required for normal basal transmission at the NMJ.
Short-term plasticity at the NMJ was tested by measuring paired-pulse facilitation and post-tetanic potentiation. Paired-pulse facilitation occurs in low calcium on a msec timescale and is characterized by an increased EJC amplitude after the second stimulation in a closely spaced pair. The enhancement of the second EJC is believed to be a result of residual presynaptic calcium (Zucker and Regehr, 2002). No difference in the ratio of the first and second EJCs was seen (A2/A1 for 25 msec interpulse interval: dArc1esm18, 1.30 ± 0.08; dArc1esm115, 1.21 ± 0.05; P > 0.05, Student's t-test). Post-tetanic potentiation occurs on a time scale of minutes and is also believed to be due to residual calcium (Zucker and Regehr, 2002). In this paradigm, the amplitude of EJCs evoked by low-frequency stimulation is increased for several minutes after a high-frequency tetanus. Both mutant and control animals show a greater than 2-fold increase in EJC amplitude which decays over the course of about a minute (Figure 4D). There were no obvious differences in either the development of potentiation or its decay. These data suggest that dARC1 does not have a role in short-term plasticity at the NMJ.
dARC1 does not affect synaptic structure at the neuromuscular junction
Although dARC1 does not appear to regulate synaptic transmission at the third instar NMJ, it may have a role in determining the structure of the synapse. To assess this possibility we quantified the number of boutons at the muscle 6/7 NMJ in segment A3 in third instar larvae. Presynaptic terminals were visualized by staining with anti-synaptotagmin 1 antibody. The number of boutons at the NMJ of the null mutant was not significantly different than a precise excision line (dArc1esm18, 72.7 ± 5.4; dArc1esm295, 71.6 ± 4.0 boutons; P > 0.05, Student's t-test). Presynaptic overexpression similarly had no effect on bouton number or morphology compared to controls (w; UAS-dArc1, 107.4 ± 4.7; C155-GAL4/w, 100.9 ± 3.5; C155-GAL4/w;UASdArc1/+, 107.3 ± 3.5; P > 0.05, ANOVA). These data suggest that dARC1 does not have a role in the structural development of the third instar NMJ.
dARC1 is not required for memory of courtship conditioning
Larval NMJ studies did not detect a role for dARC1 in cellular plasticity, but the expression of dARC1 at the NMJ is very low and quite variable between animals (M.D.M and L.C.G., unpublished results). In contrast, dARC1 is expressed robustly in the pars intercerebralis, an adult brain structure known to be involved in courtship learning (Joiner and Griffith, 1999), as well as in a small number of other cells that appear to be neurosecretory (Figure 2). Exposure of a male to a mated female leads to decreased courtship of a subsequently presented virgin female (Siegel and Hall, 1979). This suppression reflects association of an aversive cue with the female stimulatory pheromone (Tompkins et al., 1983). dArc1esm18 null males and dArc1esm115 precise excision control males were paired with a mated female for 1 h, then tested for their ability to be conditioned by testing with a virgin either 10 min or 2 h after training. No difference between mutant and control males was seen in either initial memory or in its decay (Figure 5, P > 0.05).
Figure 5. Mutation of dArc1 does not disrupt learning or short-term memory of courtship conditioning.
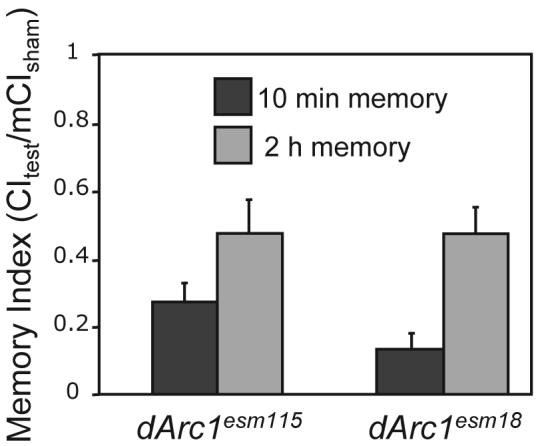
Male flies were exposed to a mated female for one hour. Behavior during the training period was indistinguishable between the null dArc1esm18 and precise excision control dArc1esm115 flies (data not shown). Sham trained (1 h in an empty chamber) controls were done for each genotype. Memory was assessed at 10 min and 2 h after training by measuring courtship of a virgin female. Data are expressed as CItest/meanCIsham where a value of 1 indicates no memory. Both genotypes have normal initial memory and normal decay.
dARC1 is not required for circadian rhythms
The pars intercerebralis has also been implicated in circadian rhythms (Jaramillo et al., 2004). To determine if dARC1 was required for locomotor rhythms we entrained flies to a 12 h:12 h light:dark (LD) cycle for 5 days, then transferred them to the dark (DD) for 13 days. Activity was measured continuously in a trikinetics monitor. Both mutants and controls exhibit normal anticipation of light transitions, a key feature of the clock (Stoleru et al., 2004). Even after 13 days in DD, both mutant and control flies showed normal rhythmicity (data not shown). The period of dArc1 mutant flies in DD was 24.3 h, which was not different than the revertant control flies period of 24.0 h. We conclude that dARC1 is not a critical component of the core circadian clock.
dARC1 is involved in the response to metabolic stress
The pars intercerebralis in Drosophila has also been shown to be involved in the regulation of metabolism. This brain region consists of several groups of neurons, including median cells that express Drosophila insulin-like peptide genes, specifically dilp2, dilp3 and dilp5 (Ikeya et al., 2002; Rulifson et al., 2002). These cells project to the ring gland which releases adipokinetic hormone (AKH), a peptide hormone that plays a role in metabolism analogous to mammalian glucagon, stimulating glucose/trehalose release into the hemolymph (Kim and Rulifson, 2004; Lee and Park, 2004). dARC1 was expressed in all the dilp+ median cells and in several other pars neurosecretory cells (Figure 6A). dARC1 was also expressed in ring gland cells including those that make AKH (Figure 6B). Fusion of 1.5 kb of the dArc1 upstream region to GAL4 produced a dArc1-GAL4 line which expresses in pars and other neurons that innervate the ring gland (Figure 6C), but appears not to express in the glandular cells themselves.
Figure 6. dARC1 expression in neuroendocrine cells.
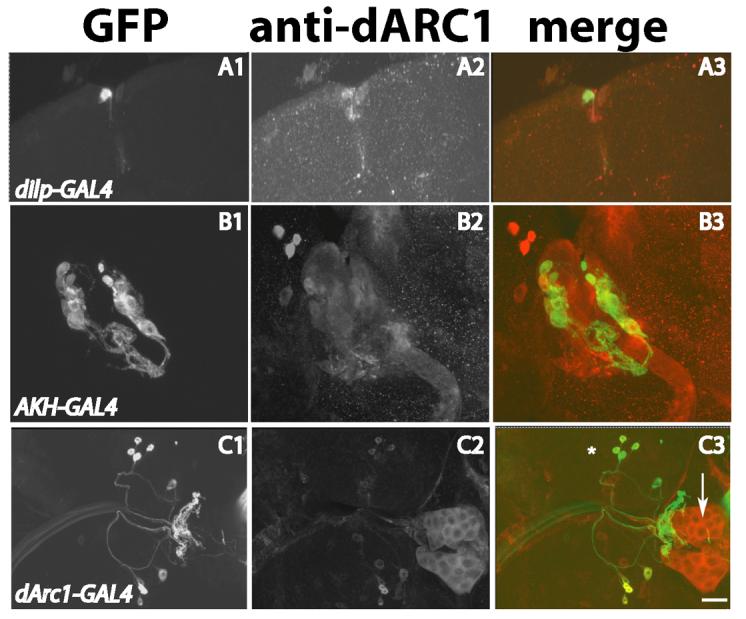
Dissected larvae or adult brains were stained with rabbit anti-dARC1 (1:1000) and imaged using confocal microscopy. Scale bar = 40 μ. A) Adult brain from a dilp-GAL4/+; UAS-mCD8GFP/+ animal. Panel A1 shows GFP expression in a subset of dilp+ pars neurons. Panel A2 shows anti-dARC1 staining. Panel A3 is an overlay showing a subset of the dARC1+ cells are also dilp+. B) Larval brain from an AKHGAL4/+; UAS-mCD8GFP/+ animal. Panel B1 shows GFP expression in AKH cells in the ring gland. Panel B2 shows anti-dARC1 staining. Panel B3 is an overlay showing the AKH-GAL4 cells are dARC1+. C) Larval brain from a dArc1-GAL4/+; UAS-mCD8GFP/+ animal. Panel C1 shows GFP expression in neurons innervating the ring gland. Panel C2 shows anti-dARC1 staining of both neurons and ring gland. Panel C3 is an overlay showing the neuronal subset of dARC1+ cells (indicated by *) which have dArc1-GAL4 expression. Arrow indicates the larval ring gland.
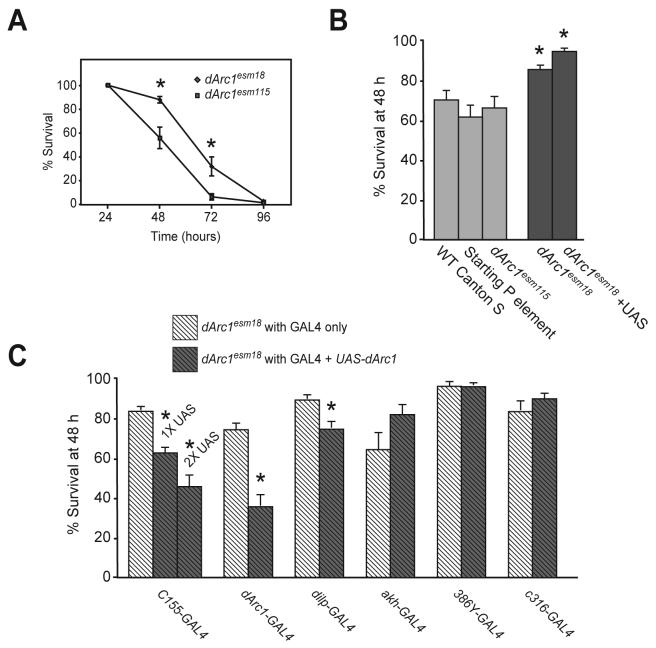
Ablation of the dilp neurons by expression of a cell death gene under control of dilp2-GAL4 and loss of downstream elements of the insulin signaling pathway have been shown to reduce the susceptibility of flies to starvation (Broughton et al., 2005; Clancy et al., 2001). Reduction of AKH has also been shown to enhance starvation resistance (Isabel et al., 2005; Lee and Park, 2004). To determine if dARC1 might be part of the pathway involved in these metabolic effects, we looked at the kinetics of starvation-induced death in dArc1esm18 and dArc1esm115 (Figure 7A). Flies that are null for dARC1 show significantly higher survival rates at both 48 and 72 h after food withdrawal. Survival at 48 h can therefore be used as a measure of starvation resistance. dArc1 precise excision animals show the same sensitivity to starvation as Canton S wild type flies or flies carrying the P-element used for mutagenesis (Figure 7B). All of these genotypes are significantly more sensitive than dArc1esm18. To determine where dARC1 expression was required for a normal starvation response, we used the GAL4/UAS system to express dARC1 in subsets of neurons in dArc1esm18 mutants and measured survival at 48 h (Figure 7C). Expression of dARC1 under control of dilp-GAL4, the panneural driver C155-GAL4 or the dArc1 promoter fused to GAL4 rescued the phenotype. Expression of dARC1 under control of AKH-GAL4, 386Y-GAL4 (a line that expresses in many peptidergic neurons) or c316-GAL4 (a line that expresses in cell that make the amnesiac PACAP-like peptide), did not suppress the resistance phenotype of the dArc1esm18 mutant. Rescue was dependent on the level of dARC1 protein since panneural expression of two copies of the UAS-dArc1 transgene suppresses the resistance phenotype more effectively than one copy. The neuronal overlap in expression pattern in the three lines that rescue defines the anatomical site of action of dARC1 and suggests that only the insulin-producing subset of dARC1-positive pars intercerebralis cells are required for normal regulation of starvation resistance by dARC1. dARC1 does not appear to be required in the ring gland itself or other neurosecretory cells for regulation of this response.
Figure 7. dArc1 mutants are resistant to starvation.
1-3 day old animals were placed in vials containing 1% agar in water in a 25°C, 70% humidity environment room. A) Time course of death from starvation. dArc1esm18 animals show significantly greater survival than WT precise excision dArc1esm115 flies at 48 and 72 h after withdrawal from food. B) Null mutants, with or without the UAS-dArc1 transgene (dark bars) are significantly more resistant to starvation at 48 h compared with Canton S wildtype, the starting P element line or a precise excision control. C) The starvation resistance of dArc1esm18 can be reversed by expression of dARC1 in selected neuronal populations. Panneural expression with C155-GAL4 gives dose-dependent effects, comparing 0, 1 or 2 copies of the UAS-dArc1 transgene. Driving UAS-dArc1 with the dArc1 promoter or dilp-GAL4 significantly decreases survival. Expression in AKH- or Amn-expressing cells does not significantly change survival. In all panels, * indicates P < 0.05, ANOVA with Fisher's PLSD post-hoc analysis.
One concern about these rescue experiments is that we are looking for a decrease in survival to demonstrate rescue, and non-specific toxicity might also reduce survival. Four lines of evidence argue that overexpression of dARC1 is not non-specifically toxic. First, we never saw lethality using a wide variety of GAL4 drivers with UAS-dARC1. Second, overexpression of dARC1 at the NMJ did not cause structural defects. Third, using sensitive electrophysiological assays we saw no indications of effects of overexpression in muscle or in neurons on either basal synaptic transmission or synaptic plasticity. Fourth, and most importantly, we did not see rescue with all the GAL4 drivers we used- rescue was specific to a very small number of dilp+ cells and could not be generated by expression in broader patterns that did not include these cells.
The starvation resistance of dArc1esm18 suggests that dARC1 acts at some level in insulin signaling. If it was critical to insulin release or its general actions, it would be expected that dArc1 mutants would show other insulin-related phenotypes. In addition to starvation resistance, reduction in insulin signaling by mutation of the insulin receptor or its downstream effectors is associated with reduced size and weight in adult flies (Bohni et al., 1999; Chen et al., 1996; Tatar et al., 2001) and with increased lifespan (Broughton et al., 2005; Clancy et al., 2001; Tatar et al., 2001). Decreased AKH release has also been associated with increased lifespan (Isabel et al., 2005). We see no changes in longevity of dArc1esm18 flies compared to revertant controls (data not shown) or in the average weight for either males (dArc1esm115 = 6.7 ± 1.0 mg, dArc1esm18 = 6.3 ± 0.5 mg; P > 0.1, Student's t-test) or females (dArc1esm115 = 9.2 ± 0.2 mg, dArc1esm18 = 9.2 ± 0.3 mg; P > 0.1, Student's t-test). We also see no evidence of alterations in trehalose or glucose levels in mutants (dArc1esm115 = 1984 ± 62 mg/dL, dArc1esm18 = 1961 ± 64 mg/dL; P > 0.4, Student's t-test).
These data suggest that dARC1 is not a regulator of general insulin signaling, but rather is involved in a specific output pathway that is critical for survival of flies under starvation conditions. Starvation has been associated with changes in locomotion in Drosophila (Isabel et al., 2005; Lee and Park, 2004) and other organisms (Chen et al., 2005; Nagata and Nagasawa, 2006; Weed et al., 1997; Wicher et al., 2006). AKH is responsible for enhancement of locomotor activity during starvation (Isabel et al., 2005) and for the paroxysmal increase in locomotor activity seen immediately before death from starvation (Lee and Park, 2004). Both of these locomotor responses have been hypothesized to be food-seeking behaviors.
dARC1 has complex effects on basal activity. While total locomotor activity of the mutant is indistinguishable from the revertant (dArc1esm115 = 31.7 ± 0.3 line crossings/30 min, dArc1esm18 = 32.2 ± 0.3 line crossings/30 min averaged over 3 days in L:D; P > 0.1, Student's t-test), the light/dark distribution of this activity was different. dArc1esm18 animals were slightly, but significantly, more active in the night and less active during the day (data not shown). This difference in activity was totally accounted for by differences in the distribution of sleep (Figure 8A). dArc1esm18 flies sleep more during the day, but are awake a greater part of the night. Total sleep is therefore not different between the mutant and revertant, suggesting that the mechanisms of sleep homeostasis are not affected.
Figure 8. dArc1 mutants have alterations in basal and starvation-modulated locomotion.
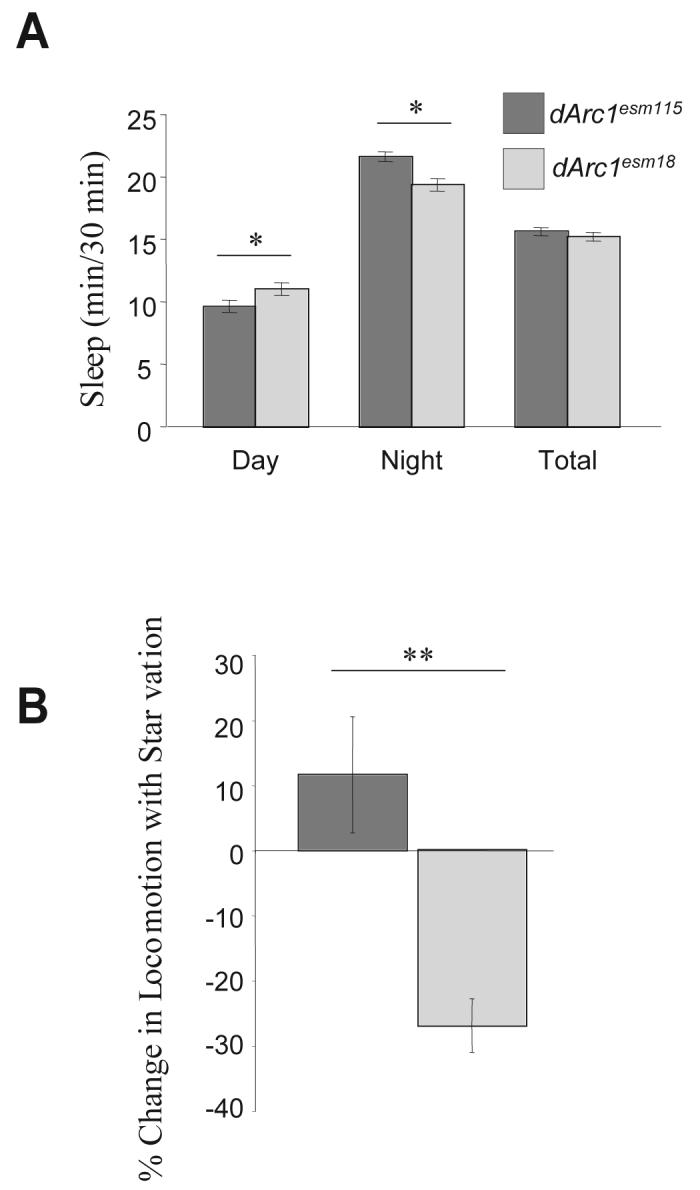
Data from dArc1esm18 are represented by dark gray bars; data from dArc1esm115 are indicated by light gray bars. A) dArc1esm18 animals have altered sleep patterns. Locomotor activity of females of the indicated genotype were collected in a Trikinetics monitor (Waltham, MA) in a 12 h light:dark cycle. Sleep was measured as bouts of 5 min of uninterrupted inactivity, as previously described (Shaw et al., 2000). Both day and night sleep differed between mutant and revertant (* indicates P < 0.05, ANOVA with Tukey-Kramer post hoc test; n > 45). Total sleep was not significantly different between genotypes. B) dArc1esm18 animals have an abnormal locomotor response to starvation. Locomotor activity was averaged for the time window 12-15 h after the initiation of starvation and compared to the average activity of fed flies. The % change from fed flies is shown. dArc1esm18 animals show a significant decrease in locomotor activity with starvation compared to revertant controls (** indicates P < 0.001, Student's t-test, n ≥ 115 for each genotype).
The effect of starvation on locomotion in dArc1 mutants was assessed by monitoring locomotion during starvation. Revertant flies maintained their activity levels until death when many of them showed paroxysmal increases in locomotion. dArc1esm18 animals showed a significant decrease in activity after 12 h of starvation compared to the average activity of fed flies (Figure 8B). Mutants also exhibited episodes of hyperlocomotion immediately before death (data not shown), indicating that their basic AKH pathways are intact. These data suggest that the starvation resistance of dArc1 mutants may be due to an impaired ability to maintain high activity levels during starvation and a consequent reduction of metabolic load.
DISCUSSION
Basic plasticity mechanisms are used by the nervous system to respond to and learn from both positive and negative changes in the environment. Mammalian Arc was first identified as a neuronal immediate-early gene that was upregulated at both the mRNA and protein levels by neuronal activity (Lyford et al., 1995). Since that time, a role for ARC in synaptic plasticity has been established (Tzingounis and Nicoll, 2006), but the molecular basis of its actions has been difficult to dissect. In Drosophila, the dArc1 mRNA is also upregulated by activity (Montana and Littleton, 2006), as is its protein level (Figure 1). These findings indicate an evolutionary conservation not only of dARC1 protein sequence, but also of dARC1 transcriptional regulation by neuronal activity. The genetic tools and wide variety of behavioral paradigms available in the fly allowed us to examine the role of dArc1 in behavior and synaptic structure and function.
We first examined classical learning and synaptic plasticity, since ARC and a number of other activity-regulated genes have important roles in these processes. Surprisingly, we found that dARC1 does not appear to be required for plasticity in these standard learning and electrophysiological assays. dArc1 mutant animals behave normally in the courtship conditioning associative learning paradigm (Figure 5) and have intact synaptic plasticity and morphology at the third instar larval NMJ (Figure 4), indicating that short-term memory and synaptic plasticity, and long-term morphological plasticity are intact. It remains possible that dARC1 is involved in formation of long-term memory, but this has not been tested. We also detect no functional role for dARC1 in vesicle recycling at the third instar NMJ, which might be expected if dARC1 interacted with endophilin and the endocytic machinery in a manner analogous to ARC (Chowdhury et al., 2006). Mutation of dArc1 also did not appear to enhance the lethality of endo1 or endoΔ4 heterozygotes (L.C.G., unpublished results).
While much has been made of their role in learning, activity-regulated genes have also been shown to be involved in a number of stress responses. In the wild, one of the most important tasks an animal has is to find food. CREB, a transcription factor with an important role in long-term memory is induced by starvation in C. elegans (Suo et al., 2006) and also has a central role in regulating liver glucose metabolism (Koo et al., 2005). Rheb, whose gene was originally isolated by the same criteria as Arc, induction in the brain following high levels of activity (Yamagata et al., 1994), is important in signaling changes in nutrient status in mammals (Tee et al., 2003). Indeed, even ARC itself has been shown to be responsive to metabolic state since it can be induced by insulin (Kremerskothen et al., 2002) and is regulated by egr1/zif268 (Li et al., 2005), a transcription factor that is involved in hepatic gluconeogenesis (Berasi et al., 2006). It is not unreasonable to believe that the role of activity-regulated genes in stress predates their role in the nervous system, and the differences between mammalian and fly Arc genes could reflect this evolutionary relationship.
The role of activity-regulated genes in the starvation response clearly must include regulation of nutrient processing, but they may also participate in regulating the behavioral response to starvation. An almost universal response to starvation is to increase locomotor activity. Primates (Weed et al., 1997), rodents (Chen et al., 2005), and a variety of insects (Nagata and Nagasawa, 2006; Wicher et al., 2006), including Drosophila (Isabel et al., 2005; Lee and Park, 2004), all show enhanced locomotor activity during starvation. Humans with eating disorders that produce a starvation-like state also show hyperlocomotion (Beumont et al., 1994). Maintaining high locomotor activity in the face of dwindling energy reserves is believed to be an adaptive response that enhances the probability of the animal finding food, but it is also a costly response, since increasing activity increases energy expenditure.
The loss of this locomotor response in dArc1 null animals is likely to be the reason for their ability to survive longer under laboratory conditions. In long-term breeding experiments where populations of flies were selected based on starvation resistance, resistance correlated with reduced locomotion (Williams et al., 2004). In the laboratory setting, this makes sense since enhanced locomotion will not increase the odds of finding food if the investigator has eliminated it from the flies' environment. In the wild, however, this might well be a maladaptive response, since there is some probability that if the fly moves to a new location, it will be able to find food. One might guess that a similar screen done ‘in the wild’ (if such a thing could be accomplished) might in fact select for flies with higher levels of starvation-induced locomotion. It is clearly important to consider the effect of the laboratory environment on genetic screens that involve behavior.
What is the nature of dARC1's role in regulation of starvation-induced locomotion? In Drosophila (Isabel et al., 2005; Lee and Park, 2004) and in cockroach (Wicher et al., 2006) the locomotor response to starvation requires AKH signaling. Our results would suggest that this role of AKH is controlled by the insulin-producing neurons of the pars intercerebralis. The fact that dArc1 animals have normal lifespan and show no defects in other insulin-related processes such as development, growth and glucose metabolism, suggests that dARC1 is not a regulator of insulin signaling per se. Whether it is a regulator of a particular, locomotion-specific, aspect of insulin release, or whether it is controlling some other signaling pathway that mediates communication between the pars and ring gland is unknown. Our results raise the possibility that there are signals other than the canonical insulin cascade acting in this neuroendocrine axis.
EXPERIMENTAL METHODS
Alignment
Sequence alignments were done using ClustalW alignment with BOXSHADE visualization (http://www.ch.embnet.org/software/BOX_form.html).
Fly stocks
akh-GAL4 (Lee and Park, 2004), dilp2-GAL4 (Rulifson et al., 2002), 386Y-GAL4 (Taghert et al., 2001), c316-GAL4 (Waddell et al., 2000) are all previously described in published literature. Endophilin mutants were obtained from Hugo Bellen (Baylor College of Medicine, Houston, TX). The P-element line P{GT1}BG01371 was obtained from the pEGTB collection and used to generate dArc1 alleles using a standard F3 P-element excision screen. For the generation of dArc1-GAL4 a 1.5 kb fragment of the five prime region of dArc1 was amplified from genomic DNA and cloned into pPGAL. The overexpression lines were made by cloning the dARC1 open reading frame into the pUAST vector. The resulting construct was then used to generate transformant lines by standard procedures (Robertson et al., 1988).
Antibodies, western blotting and immunocytochemistry
Anti-dARC1 and anti-dARC2 are both rabbit polyclonal antibodies. A region corresponding to amino acids 1-42 of dARC1 with low similarity to both dARC2 and dARC3, was cloned into pGEX1zt to create a GST fusion, which was then bacterially expressed, purified and used to inject rabbits. A region corresponding to amino acids 1-27 of dARC2, which has low similarity to dARC1 or dARC3 was also used to generate antiserum in the same way. dARC1 and dARC2 antibodies were used at 1:1000 for Westerns and immunocytochemistry in both the crude serum and nitrocellulose purified forms. Specificity of antisera was confirmed using bacterially expressed proteins and showed that neither the anti-dARC1 or the anti-dARC2 cross-reacted with the other protein (data not shown).
For immunoblots, adult heads were collected and protein concentration was determined using Bio-Rad Protein Assay Reagent (Bio Rad). Equal amounts of protein were boiled in SDS sample buffer for 5 minutes and loaded for SDS-PAGE. Protein gels were transferred to Hybond-P membrane (Amersham Biosciences). Proteins were visualized with chemiluminescence using ECL substrate (Amersham Biosciences). Blots were probed for dARC1 first then stripped and probed with an anti-tubulin antibody used at 1:200,000 (Sigma). Densitometric data were obtained by processing exposed film with Bio-Rad Chemi-Doc using Quantity One software. dARC1 values (arbitrary optical density units) are normalized to tubulin for loading control. Secondary antibodies used include: goat anti-rabbit Cy5 or goat rabbit-FITC (Jackson Laboratories), Donkey anti-rabbit-HRP or Donkey anti-mouse-HRP (Amersham).
Dissection, staining and imaging of tissue
Larval brains were dissected out in PBS and fixed for 30 min with 4% paraformaldehyde, followed by a 30 min wash in PBS + 0.1% TritonX100 + 1% BSA. Samples were blocked for 60 min in PBS + 0.1% TritonX100 + 1% BSA 2.5% Normal Goat Serum. Primary antibody anti-dARC1 (A8218), either crude serum or nitrocellulose purified, was used at 1:500- 1:1000 diluted in fresh blocking solution. Primary antibody was left on overnight to 48 hours while rotating at 4°. Samples were then washed 6 ×15 minutes in block solution. Secondary antibodies were used at 1:100 diluted in blocking buffer and applied to samples for 4 h. After incubation with secondary antibody, samples were washed 4 × 15 minutes in PBS + 0.1% TritonX100 then cleared in glycerol. Samples were mounted with glycerol and vectashield (Vector Laboratories). All images were acquired using a Leica TCS SP2 confocal scanning microscope (Leica Microsystems, Exton, PA).
For Adult CNS a small tear was made in the eye to allow fix to enter, then heads were fixed for 5 min on ice with 4% paraformaldehyde in PBS, followed by 20 min at room temperature. Samples were rinsed 3x in PBS, brains dissected out and from this point samples were processed as described for larvae.
Structural analysis on third instar neuromuscular junctions was done as previously described using anti-synaptotagmin 1 antiserum at 1:1000 and affinity purified anti-dARC1 at 1:500 (Littleton et al., 1993; Montana and Littleton, 2004). Imaging was done on an AxioScope 2 confocal microscope and processed with LSM software (Zeiss).
Courtship assay
Courtship assays were performed as described (Ejima et al., 2005). Briefly, all courtship behavior was done under dim red lights in a Harris environmental room (25°C, 70% humidity). A 4- or 5-day old male was placed with a trainer in a single-pair-mating chamber for 1 h. Wet filter paper was put in each chamber to maintain humidity. The first and last 10 min of the training period were videotaped with a digital camcorder. Pairs that had an initial CI < 0.1 were eliminated from further analysis. Immediately after training, males were transferred into a clean chamber and paired with a decapitated tester female and videotaped for 10 min either 10 min or 2 h later. Sham-trained males are kept alone in the mating chamber for the first hour and then paired with a tester for 10 min. For each of the 10 min periods, a courtship index (CI) was calculated. CI is the fraction of time a male spent in courtship activity in the 10 min observation period (CI = courtship [s]/observation [s]). Memory index is calculated by dividing CI at test (CItest) by the mean of sham CIs (CIsham): CItest/mCIsham. If CItest/mCIsham = 1, it indicates that there has been no learning because the courtship level of trained males is equivalent to that of sham-trained males. Each CI was subjected to arcsine-square-root transformation to effect an approximation of normal distribution. Statistical analysis was performed using JMP software
Starvation experiments
All experiments were carried out in Harris environmental room which has a constant temperature of 25° C and 70% humidity. Females were collected after eclosion and 1-3 day old flies were placed in groups of 20/vial on 1% agar in order to provide moisture but not food. Flies were transferred to fresh 1% agar vials every other day. Survival was scored every 24 hours from the start of the experiment.
Locomotor activity and sleep data
Female flies were placed in 65 mm × 5 mm glass tubes (Trikinetics, Waltham, MA) containing 2% agarose/5% sucrose. Flies were entrained for 3 days at 25°C in 12:12 light:dark cycles prior to collection of 3 days of locomotor baseline data that was averaged for activity or sleep. Locomotor activity was collected with DAM System monitors (Trikinetics) as previously described (Stoleru et al., 2004). Sleep was measured as bouts of uninterrupted 5 minutes of inactivity, as previously described (Hendricks et al., 2000; Shaw et al., 2000). Overall activity was determined by averaging the activity over a 24 hour period. Flies were transferred to tubes containing 2% agarose only in order to monitor locomotor activity under starvation conditions.
Electrophysiological analysis
Electrophysiological analysis was done as previously described (Montana and Littleton, 2004). Electrode resistances of 20-40 MΩ were used. In analyses using two-electrode voltage clamp, the second electrode was normally 10-20 MΩ on a 1X MGU headstage (Axon Instruments). Only cells which had less than 10 nA of leak current were utilized in the analysis, with most having less than 2 nA. All stimulus signals were generated through the pCLAMP v8.0 program (Axon Instruments). mEJP recordings were done in CNS-intact animals in 1.5 mM Ca2+ and 3 μM TTX. All recordings were done in HL3 using 4 mM or 20 mM Mg2+ at the indicated Ca2+ concentrations (Stewart et al., 1994). Traces in the figures were plotted in Microsoft Excel by exporting the trace values into a spreadsheet.
mEJP analysis was done in a semi-automated manner using the event detection analysis in pCLAMP v9.0 using a template obtained from mEJP recordings done in Canton S wild type based on 1000+ individual events. EJP analysis was done by averaging ten events in each muscle and counting the average amplitude as one measurement for the genotype assayed. High-frequency vesicle depletion was quantified by dividing the EJP at the designated time point by the first EJP amplitude. Paired-pulse facilitation was quantified by averaging ten paired-pulse events from a single muscle, then dividing the average second EJC by the average first EJC. Post-tetanic potentiation was quantified by averaging 30 s of EJCs at 0.5 Hz to determine baseline amplitude. All EJC amplitudes graphed are the EJC at the designated time point divided by the baseline EJC amplitude.
Glucose and trehalose assay
Glucose and trehalose assay was performed as described in Lee et al. 2004 and Rulifson et al., 2004. Briefly, cultures were raised as described for weight determination. Hemolymph was obtained from groups of 10 wandering 3rd instar larvae that had been rinsed with water then patted dry. Glucose was measured by adding hemolymph to Infinity Glucose Reagent (Thermo Electron) and trehalose was converted to glucose by adding porcine trehalose (Sigma). Samples were added to a 96 well plate and incubated at 37° C overnight. N = 6 for each genotype.
Weight determination
Flies were grown at a constant temperature of 25° C and 70% humidity. Groups of 10 age-matched adult flies from cultures seeded at equal density were weighed. Weight is expressed for each group of ten. N = 5 groups for each genotype.
Statistical analysis
Statistical analysis of the data was done in several programs. Student's t-test was done using Microsoft Excel in the Microsoft Office XP Professional Software Suite (Microsoft). KS-test analysis on mEJP distributions was done using the Kolmogorov-Smirnov-test applet at http://www.physics.csbsju.edu/stats/KS-test.n.plot_form.html. Post-hoc analysis of the ANOVA in electrophysiology experiments was done using the Tukey-test applet at http://department.obg.cuhk.edu.hk/ResearchSupport/Least_sig_diff_Tukey.asp in order to determine the least significant difference values. All other ANOVA and posthoc analyses were done in JMP.
Acknowledgments
This work was supported by NIH grants GM54408 and grant W81XWH-04-1-0158 from the US Army (LCG) and NS043244 (JTL). We would like to thank Paul Worley for reagents and helpful discussions. We also thank Aki Ejima for help with behavioral assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bader MF, Doussau F, Chasserot-Golaz S, Vitale N, Gasman S. Coupling actin and membrane dynamics during calcium-regulated exocytosis: a role for Rho and ARF GTPases. Biochim Biophys Acta. 2004;1742:37–49. doi: 10.1016/j.bbamcr.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Berasi SP, Huard C, Li D, Shih HH, Sun Y, Zhong W, Paulsen JE, Brown EL, Gimeno RE, Martinez RV. Inhibition of gluconeogenesis through transcriptional activation of EGR1 and DUSP4 by AMP-activated kinase. J Biol Chem. 2006;281:27167–27177. doi: 10.1074/jbc.M602416200. [DOI] [PubMed] [Google Scholar]
- Beumont PJ, Arthur B, Russell JD, Touyz SW. Excessive physical activity in dieting disorder patients: proposals for a supervised exercise program. The International journal of eating disorders. 1994;15:21–36. doi: 10.1002/1098-108x(199401)15:1<21::aid-eat2260150104>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, Hafen E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, Partridge L. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Jack J, Garofalo RS. The Drosophila insulin receptor is required for normal growth. Endocrinology. 1996;137:846–856. doi: 10.1210/endo.137.3.8603594. [DOI] [PubMed] [Google Scholar]
- Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Crotty P, Sangrey T, Levy WB. Metabolic energy cost of action potential velocity. J Neurophysiol. 2006;96:1237–1246. doi: 10.1152/jn.01204.2005. [DOI] [PubMed] [Google Scholar]
- Ejima A, Smith BP, Lucas C, Levine JD, Griffith LC. Sequential learning of pheromonal cues modulates memory consolidation in trainer-specific associative courtship conditioning. Curr Biol. 2005;15:194–206. doi: 10.1016/j.cub.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T, Tu MP, Tatar M. Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. Bioessays. 2005;27:999–1010. doi: 10.1002/bies.20290. [DOI] [PubMed] [Google Scholar]
- Guan Z, Saraswati S, Adolfsen B, Littleton JT. Genome-wide transcriptional changes associated with enhanced activity in the Drosophila nervous system. Neuron. 2005;48:91–107. doi: 10.1016/j.neuron.2005.08.036. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- Isabel G, Martin JR, Chidami S, Veenstra JA, Rosay P. AKH-producing neuroendocrine cell ablation decreases trehalose and induces behavioral changes in Drosophila. American journal of physiology. 2005;288:R531–538. doi: 10.1152/ajpregu.00158.2004. [DOI] [PubMed] [Google Scholar]
- Jaramillo AM, Zheng X, Zhou Y, Amado DA, Sheldon A, Sehgal A, Levitan IB. Pattern of distribution and cycling of SLOB, Slowpoke channel binding protein, in Drosophila. BMC Neurosci. 2004;5:3. doi: 10.1186/1471-2202-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner MA, Griffith LC. Mapping of the anatomical circuit of CaM kinase-dependent courtship conditioning in Drosophila. Learn Mem. 1999;6:177–192. [PMC free article] [PubMed] [Google Scholar]
- Keshishian H, Chang TN, Jarecki J. Precision and plasticity during Drosophila neuromuscular development. Faseb J. 1994;8:731–737. doi: 10.1096/fasebj.8.10.8050672. [DOI] [PubMed] [Google Scholar]
- Kim SK, Rulifson EJ. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature. 2004;431:316–320. doi: 10.1038/nature02897. [DOI] [PubMed] [Google Scholar]
- Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, Takemori H, Montminy M. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- Kremerskothen J, Wendholt D, Teber I, Barnekow A. Insulin-induced expression of the activity-regulated cytoskeleton-associated gene (ARC) in human neuroblastoma cells requires p21(ras), mitogen-activated protein kinase/extracellular regulated kinase and src tyrosine kinases but is protein kinase C-independent. Neurosci Lett. 2002;321:153–156. doi: 10.1016/s0304-3940(01)02532-0. [DOI] [PubMed] [Google Scholar]
- Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167:311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Carter J, Gao X, Whitehead J, Tourtellotte WG. The neuroplasticity-associated arc gene is a direct transcriptional target of early growth response (Egr) transcription factors. Mol Cell Biol. 2005;25:10286–10300. doi: 10.1128/MCB.25.23.10286-10300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littleton JT, Bellen HJ, Perin MS. Expression of synaptotagmin in Drosophila reveals transport and localization of synaptic vesicles to the synapse. Development. 1993;118:1077–1088. doi: 10.1242/dev.118.4.1077. [DOI] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Moga DE, Calhoun ME, Chowdhury A, Worley P, Morrison JH, Shapiro ML. Activity-regulated cytoskeletal-associated protein is localized to recently activated excitatory synapses. Neuroscience. 2004;125:7–11. doi: 10.1016/j.neuroscience.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Montana ES, Littleton JT. Characterization of a hypercontraction-induced myopathy in Drosophila caused by mutations in Mhc. J Cell Biol. 2004;164:1045–1054. doi: 10.1083/jcb.200308158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montana ES, Littleton JT. Expression profiling of a hypercontraction-induced myopathy in Drosophila suggests a compensatory cytoskeletal remodeling response. J Biol Chem. 2006 doi: 10.1074/jbc.M512468200. [DOI] [PubMed] [Google Scholar]
- Nagata S, Nagasawa H. Effects of diet-deprivation and physical stimulation on the feeding behaviour of the larvae of the silkworm, Bombyx mori. J Insect Physiol. 2006;52:807–815. doi: 10.1016/j.jinsphys.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillips RW, Johnson-Schlitz D, Benz WK, Engels WR. A stable genomic source of P-element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- Siegel RW, Hall JC. Conditioned responses in courtship behavior of normal and mutant Drosophila. Proc. Natl. Acad. Sci. USA. 1979;76:565–578. doi: 10.1073/pnas.76.7.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Suo S, Kimura Y, Van Tol HH. Starvation induces cAMP response element-binding protein-dependent gene expression through octopamine-Gq signaling in Caenorhabditis elegans. J Neurosci. 2006;26:10082–10090. doi: 10.1523/JNEUROSCI.0819-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert PH, Hewes RS, Park JH, O'Brien MA, Han M, Peck ME. Multiple amidated neuropeptides are required for normal circadian locomotor rhythms in Drosophila. J Neurosci. 2001;21:6673–6686. doi: 10.1523/JNEUROSCI.21-17-06673.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- Tompkins L, Siegel RW, Gailey DA, Hall JC. Conditioned courtship in Drosophila and its mediation by association of chemical cues. Behav Genet. 1983;13:565–578. doi: 10.1007/BF01076402. [DOI] [PubMed] [Google Scholar]
- Tzingounis AV, Nicoll RA. Arc/Arg3.1: linking gene expression to synaptic plasticity and memory. Neuron. 2006;52:403–407. doi: 10.1016/j.neuron.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell. 2000;103:805–813. doi: 10.1016/s0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- Weed JL, Lane MA, Roth GS, Speer DL, Ingram DK. Activity measures in rhesus monkeys on long-term calorie restriction. Physiology & behavior. 1997;62:97–103. doi: 10.1016/s0031-9384(97)00147-9. [DOI] [PubMed] [Google Scholar]
- Wicher D, Agricola HJ, Sohler S, Gundel M, Heinemann SH, Wollweber L, Stengl M, Derst C. Differential receptor activation by cockroach adipokinetic hormones produces differential effects on ion currents, neuronal activity, and locomotion. J Neurophysiol. 2006;95:2314–2325. doi: 10.1152/jn.01007.2005. [DOI] [PubMed] [Google Scholar]
- Williams AE, Rose MR, Bradley TJ. The respiratory pattern in Drosophila melanogaster selected for desiccation resistance is not associated with the observed evolution of decreased locomotory activity. Physiol Biochem Zool. 2004;77:10–17. doi: 10.1086/381467. [DOI] [PubMed] [Google Scholar]
- Wu CF, Ganetzky B, Haugland FN, Liu AX. Potassium currents in Drosophila: different components affected by mutations of two genes. Science. 1983;220:1076–1078. doi: 10.1126/science.6302847. [DOI] [PubMed] [Google Scholar]
- Wu Q, Brown MR. Signaling and function of insulin-like peptides in insects. Annu Rev Entomol. 2006;51:1–24. doi: 10.1146/annurev.ento.51.110104.151011. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Sanders LK, Kaufmann WE, Yee W, Barnes CA, Nathans D, Worley PF. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J Biol Chem. 1994;269:16333–16339. [PubMed] [Google Scholar]
- Zucker RS, Regehr WG. Short-term synaptic plasticity. Annu Rev Physiol. 2002;64:355–405. doi: 10.1146/annurev.physiol.64.092501.114547. [DOI] [PubMed] [Google Scholar]