Abstract
H/ACA RNP complexes change uridines to pseudouridines in target non-coding RNAs in eukaryotes and archaea. H/ACA RNPs are comprised of a guide RNA and four essential proteins: Cbf5 (pseudouridine synthase), L7Ae, Gar1 and Nop10 in archaea. The guide RNA captures the target RNA via two antisense elements brought together to form a contiguous binding site within the pseudouridylation pocket (internal loop) of the guide RNA. Cbf5 and L7Ae interact independently with the guide RNA, and here we have examined the impacts of these proteins on the RNA in nucleotide protection assays. The results indicate that the interactions observed in a fully assembled H/ACA RNP are established in the sub-complexes, but also reveal a unique Cbf5–guide RNA interaction that is displaced by L7Ae. In addition, the results indicate that L7Ae binding at the kink (k)-turn of the guide RNA induces the formation of the upper stem, and thus also the pseudouridylation pocket. Our findings indicate that L7Ae is essential for formation of the substrate RNA binding site in the archaeal H/ACA RNP, and suggest that k-turn-binding proteins may remodel partner RNAs with important effects distant from the protein-binding site.
INTRODUCTION
In all organisms, post-transcriptional modifications play an important role in the maturation and function of cellular RNAs, especially stable non-coding RNAs (1–4). The human ribosome is estimated to contain over 200 modified nucleotides and these fall primarily in functionally important regions of the rRNAs (1,5,6). In eukaryotes and archaea, rRNAs and other non-coding RNAs are modified by two classes of RNA-guided modification enzymes: C/D and H/ACA RNPs (7–9). C/D RNPs methylate the 2′-O-hydroxyl group of ribose rings in target nucleotides (10,11). H/ACA RNPs isomerize target uridine residues to pseudouridines by base rotation (12,13). These modification enzymes are comprised of a set of three or four core proteins and a cognate guide RNA that determines the target nucleotide by base pairing with the substrate RNA (7–9).
Some key aspects of the mechanism of H/ACA RNP function have been well defined (3,14). Seminal studies revealed that the substrate recognition site is formed by juxtaposing two antisense sequences within an internal loop of the conserved hairpin structure of the guide RNA (Figure 1B) (13,15). This loop that comprises the substrate recognition site is termed the pseudouridylation pocket. The antisense elements recognize substrate sequences flanking the target uridine, resulting in placement of the uridine to be modified at the apex of the pseudouridylation pocket. It is quite clear based on sequence and structure homology, and mutational analysis that Cbf5 is the pseudouridine synthase (16,17). The functions of the other three proteins, Gar1, Nop10 and L7Ae (or Nh2p in eukaryotes) are not established, but are known to be essential for the function of the complex (18,19).
Figure 1.
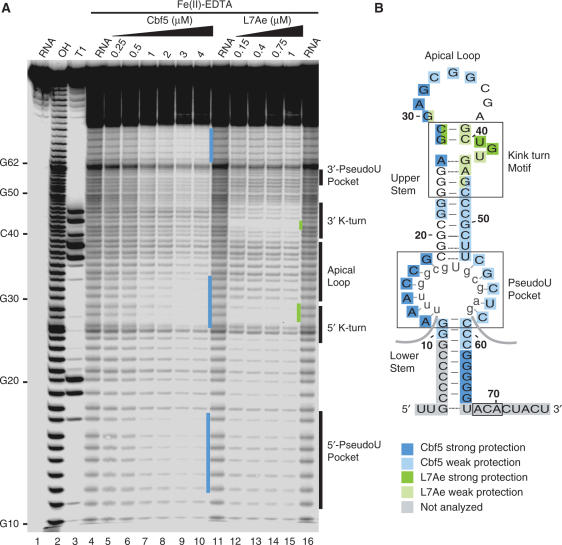
Hydroxyl radical footprinting of Cbf5–Pf9 and L7Ae–Pf9 complexes. (A) 5′-end labelled Pf9 was incubated in the absence (lanes 4, 11, 16) or presence of increasing concentrations of Cbf5 (lanes 5–10) or L7Ae (lanes 12–15) and subjected to hydroxyl radical cleavage. Lane 1 is undigested RNA and lanes 2 and 3 are size markers generated by alkaline hydrolysis (OH) and RNase T1 digestion (T1) of the free RNA, respectively. Nucleotides corresponding to secondary structure landmarks are indicated to the right. Blue and green bars indicate regions of strong Cbf5 and L7Ae protection, respectively. (B) Summary of protections in the context of a functional secondary structure model of Pf9 RNA. Box ACA, the pseudouridylation pocket and k-turn are boxed. Apical loop, upper and lower stems are labelled. The rRNA target of Pf9 is shown in grey lowercase letters. Cbf5 and L7Ae protections observed in A are shown as indicated in the legend. The regions shaded grey were not assessed due to the resolution limits of the gel.
Our laboratory and the Branlant laboratory successfully reconstituted and characterized functional H/ACA RNPs using components from Pyrococcus furiosus and Pyrococcus abyssi, respectively (18,19). These studies established that the four core proteins and a guide RNA are necessary and sufficient for full activity in vitro. We found that both Cbf5 and L7Ae interact directly with the guide RNA in the absence of other proteins. The remaining proteins, Gar1 and Nop10, bind to independent sites on Cbf5.
L7Ae belongs to a family of proteins that interact with RNA kink (k)-turns (20). The k-turn binding proteins also include components of the ribosome, proteins involved in the assembly of spliceosomes, mRNA binding proteins and components of the RNase P and MRP complexes that function in tRNA and rRNA processing (21–24). L7Ae appears to be important for the kinetics of pseudouridylation by the H/ACA RNP (19). The primary interaction of L7Ae within the H/ACA RNP is with the k-turn of the guide RNA; no substantial interaction with the other proteins is observed in the absence of the RNA (18). Moreover, L7Ae binding is not required for association of the other three proteins with the guide RNA, though it may enhance their binding (18,19). L7Ae binding sites are located either near (canonical k-turn) or overlapping (non-canonical k-turn) the apical loop of archaeal H/ACA RNAs (16,25). The essential role of L7Ae in H/ACA RNP function was not apparent, but seemed likely to be accomplished through its interaction with the RNA component.
Mutational analysis in combination with RNA–protein-binding assays indicated that Cbf5 requires several important elements of the guide RNA for its interaction, including sequences in the apical loop, pseudouridylation pocket and box ACA (Figure 1B), suggesting that Cbf5 may interact with these regions of the RNA (18). A subsequent crystal structure of the P. furiosus H/ACA RNP (including the four proteins and a guide RNA) indicates that in the context of the complete complex, Cbf5 interacts with box ACA and nucleotides in the lower stem, and to a lesser extent with the apex of the pseudouridylation pocket (26). Similar interactions were mapped in RNA footprinting studies with yeast Cbf5 (27). No interaction of Cbf5 with the apical loop was observed in the holoenzyme (26).
In this work, we have examined the impacts of Cbf5 and L7Ae, both individually and in combination, on a guide RNA by enzymatic and chemical footprinting. The influences of the proteins on the RNA footprinting patterns substantiate and clarify the RNA–protein interactions predicted by the previous mutational analysis and observed in the crystal structure of the full complex (18,26). In addition, the results indicate that L7Ae plays an important role in formation of the pseudouridylation pocket (i.e. substrate recognition site). Finally, we observed an interaction of Cbf5 with the apical loop of the RNA that is disrupted by the binding of L7Ae. Our results indicate that RNA remodelling events triggered by binding of specific components of the H/ACA RNP govern the ability of the RNP to function in target RNA recognition and nucleotide modification.
MATERIALS AND METHODS
Protein expression and purification
Cbf5 and L7Ae genes were amplified by PCR from P. furiosus genomic DNA and sub-cloned into a modified version of pET21D expression vector as previously described (18). The resultant recombinant proteins containing N-terminal 6 × histidine tags were purified by affinity chromatography on Ni-NTA agarose (Qiagen), eluted with buffer A (20 mM sodium phosphate, pH 7.0, 1 M NaCl, 350 mM imidazole) and quantified using BCA protein assay (Pierce). Prior to use in RNA-binding assays, the proteins were dialysed against 40 mM HEPES–KOH, pH 7.0, 1 M KCl (or K-acetate).
End labelling of H/ACA RNA
The single hairpin, P. furiosus H/ACA RNA Pf9 was transcribed in vitro from PCR-amplified DNA product containing a SP6 promoter using SP6 RNA polymerase (Epicentre Biotechnologies) as previously described (18). RNA was gel purified by electrophoresis through a 15% polyacrylamide/7 M urea gel. Purified RNA was ethanol precipitated and washed with 70% ethanol. Purified RNA was dephosphorylated with calf intestinal alkaline phosphatase according to the manufacturer's protocol (Ambion). The dephosphorylated RNA was 32P labelled with T4 polynucleotide kinase (Ambion) and [γ-32P]ATP (7000 Ci/mmol, MP Biomedicals). 5′-end labelled RNA was then gel purified as described above.
Gel mobility shift assay
Reconstitution of RNP complexes was performed as described previously (18). Briefly, 5′-end radiolabelled RNA (0.05 pmol) was incubated in buffer B (20 mM HEPES–KOH, pH 7.0, 500 mM KCl, 1.5 mM MgCl2, 5 μg Escherichia coli tRNA) alone or with various concentrations of protein in a final volume of 20 μl for 1 h at 65°C. RNP complexes were analysed on an 8% non-denaturing polyacrylamide gel and visualized by autoradiography.
Enzymatic and chemical probing
32P-end labelled Pf9 RNA (0.05–0.1 pmol) was incubated in the absence (free RNA) or presence (RNPs) of increasing concentrations of purified Cbf5 or L7Ae proteins for 1 h at 65°C in buffer B (described above) in a final volume of either 20 or 50 μl. For ribonuclease cleavage, the reactions were initiated by addition of 0.1 or 0.2 U RNase T1 (Sigma), or 1 or 2 ng RNase A (Sigma) and incubated for 15 min at 37°C. The enzymatic reactions were stopped by extraction with phenol/chloroform/isoamyl alcohol. Hydroxyl radical footprinting experiments were performed essentially as described (28). Briefly, the cleavage reactions were initiated by adding freshly prepared 18 μM ethylenediaminetetraacetic acid (EDTA) iron (III) sodium salt dihydrate (Aldrich), 2 mM sodium ascorbate (Sigma) and 0.14% (v/v) H2O2 (Sigma). The reactions were carried out at 65°C for 30 s and stopped by addition of 1 mM thiourea (Aldrich) followed by phenol/chloroform/isoamyl alcohol extraction. For lead (II) footprinting, the reactions were carried out in a modified buffer B where the KCl was substituted with 200 mM K acetate. Lead cleavage was performed essentially as previously described (29) with 15 mM Pb(II) acetate (Merck) freshly prepared in sterile water. The reactions were performed at room temperature for 10 min and were stopped by adding EDTA to final concentration of 20 mM before ethanol precipitation. As sequence markers, RNA alkaline hydrolysis ladders (cleavage after each nucleotide) were generated by incubating RNA with 5 μg E. coli tRNA in 50 mM sodium carbonate at pH 9.5, 1 mM EDTA for 5 min at 90°C. RNase T1 ladders (ΔT1) (cleavage after each guanosine) were generated by incubating the RNA in 20 mM sodium citrate at pH 4.5, 1 mM EDTA, 7 M urea for 10 min at 50°C. For both enzymatic and chemical probing reactions, the treated RNA samples were then ethanol precipitated in the presence of 0.3 M sodium acetate at pH 5.2 followed by washing with 70% ethanol. The dried RNA pellets were resuspended in RNA loading dye [10 M urea, 2 mM EDTA, 0.5% (w/v) SDS, 0.02% (w/v) each bromophenol blue and xylene cyanol]. The cleavage products were separated on 15 or 20% polyacrylamide (acrylamide:bis ratio 19:1) 7 M urea-containing gel and visualized by autoradiography.
RESULTS
RNA–protein interactions in Cbf5–guide RNA and L7Ae–guide RNA complexes
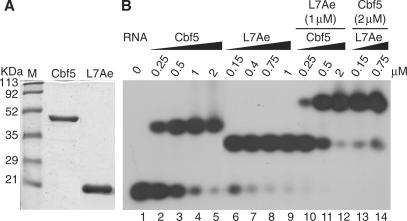
To assess the interactions of Cbf5 and L7Ae with P. furiosus H/ACA guide RNA Pf9, we analysed the two RNA–protein sub-complexes (Cbf5-Pf9 and L7Ae-Pf9) by hydroxyl radical nucleotide protection assays (Figure 1). Hydroxyl radicals cleave the RNA backbone independent of RNA sequence or secondary structure (30,31). Thus, in the absence of the proteins, cleavages were observed at all ribose moieties (Figure 1A, lanes 4, 11, 16). Protection of a ribose from hydroxyl radical cleavage upon addition of a protein generally indicates a direct association with the protein (32). RNA–protein complex formation (with 5′ end-labelled Pf9 RNA and purified recombinant proteins) was verified by gel shift analysis (Figure 2). The majority of the Pf9 is shifted into RNA–protein complexes at 2 μM Cbf5 and 1 μM L7Ae. Both proteins provided some global protection of the RNA, however, as can be seen in Figure 1A, distinct RNA protections were observed with increasing Cbf5 or L7Ae concentrations (see regions indicated with blue and green bars).
Figure 2.
Reconstitution of Cbf5–Pf9, L7Ae–Pf9 and Cbf5–L7Ae–Pf9 sub-complexes. (A) Coomassie blue staining of purified recombinant Cbf5 and L7Ae following PAGE. M lane contains protein standards. (B) Gel mobility shift analysis of 5′-end labelled Pf9 RNA, alone (lane 1) or with increasing amounts of Cbf5 and/or L7Ae as indicated. Complexes were separated on an 8% native gel and visualized by autoradiography.
Figure 1B shows the protection results in the context of a secondary structure model of Pf9 RNA that is based on the well-defined, functional features of the H/ACA RNA family (3). The pseudouridylation pocket of the H/ACA RNA is the bipartite target recognition site, established and bounded by the upper and lower stems. The predicted pseudouridylation pocket of Pf9 is complementary to sequences that flank 16S rRNA U910 (i.e. nts 905–917), and consistent with this model, we have confirmed that U910 is modified in rRNA extracted from P. furiosus (Marshburn,S., Terns,R. and Terns,M., unpublished data). Box ACA, the signature sequence element, is located 3′ of the lower stem. The k-turn of Pf9 is found within the upper stem, adjacent to the apical loop. Canonical k-turns are helix-bulge-helix structures that produce an ∼120° bend between the axes of the two adjacent RNA helices (20). The bulge of a k-turn is bounded by two G–A base pairs that terminate the first helix and a G–C base pair that initiates the second helix. The motif generally includes several flanking base pairs (Figure 1B).
In multiple studies, L7Ae and its close homologues have been found to interact directly with sequences in the k-turn of partner RNAs (18,25,33–35), and as expected, L7Ae provides strong protection on both strands of the k-turn (green shading, Figure 1). In particular, the k-turn-binding proteins are consistently found to contact nucleotides in the bulge of the k-turn (18,25,33–35) and L7Ae's protection of Pf9 includes the bulge (Figure 1).
The interaction of Cbf5 with H/ACA RNAs is less well studied. Previous gel shift analysis suggested that box ACA, the pseudouridylation pocket and sequences in the apical loop of the RNA, may be involved in the interaction; alterations in these elements affect the stability of the Cbf5–guide RNA complex (18). In the crystal structure of a complex that includes Nop10, Gar1 and L7Ae as well as Cbf5 and a guide RNA, contacts were observed between nucleotides in box ACA, the lower stem and the pseudouridylation pocket of the RNA and Cbf5 (26). As can be seen in Figure 1 (blue shading), Cbf5 provides extensive protection of Pf9 from hydroxyl radical cleavage. Cbf5 significantly reduces cleavage of the 3′ strand of the lower stem, the 5′ strand of the pseudouridylation pocket and the 5′ half of the k-turn and the apical loop. Additional weak protection was observed along the 3′ strand of the pseudouridylation pocket and the upper stem. The 5′ strand of the lower stem and the 3′ single-stranded region that contains box ACA were not assessed in this experiment due to resolution limitations. The results suggest that Cbf5 interacts directly with the lower stem, pseudouridylation pocket, apical loop and 5′ strand of the k-turn in the absence of other proteins.
The observed protections were specific to the individual proteins with the notable exception of the 5′ strand of the k-turn (Figure 1). Interestingly, the results reveal strong protection of the 5′ strand of the k-turn by both Cbf5 and L7Ae, suggesting that both proteins interact with this region of the RNA in the sub-complexes. Direct interactions of Cbf5 and L7Ae with the 5′ side of the k-turn would be expected to be mutually exclusive.
RNA–protein interactions in Cbf5-L7Ae–guide RNA complexes
Cbf5 and L7Ae do not interact directly, either independently (18) or in the context of the fully assembled complex (26), and therefore should not directly influence the interaction of the other protein with the guide RNA; however, we were interested in the possibility of effects translated through the RNA (for example, via changes in RNA structure). Moreover, we were interested in examining the footprint in the presence of both proteins on the 5′ side of the k-turn (where both proteins were observed to bind, Figure 1). To test for any impact of one protein on the interaction of the other protein with the RNA, we examined the hydroxyl radical footprints of combinations of Cbf5 and L7Ae on Pf9 (Figure 3). In these experiments, the RNA was mixed with increasing concentrations of one protein in the presence of a constant concentration of the other protein in final amounts that promote nearly complete incorporation of the RNA into complexes (Figure 2, lanes 10–14).
Figure 3.
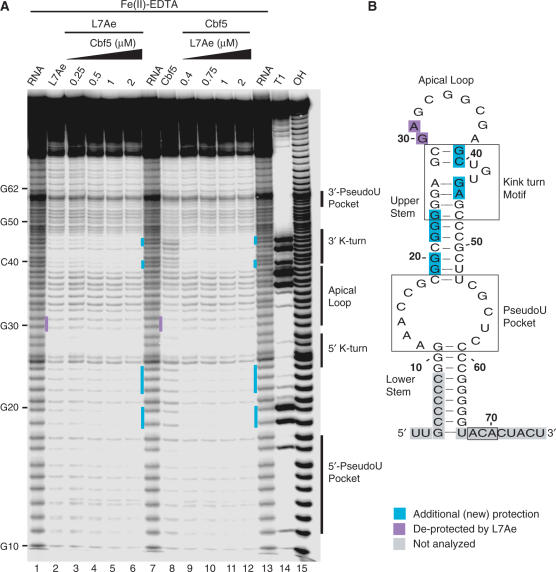
Hydroxyl radical footprinting of Cbf5–L7Ae–Pf9 complexes. (A) 5′-end labelled Pf9 was incubated in the absence (lanes 1, 7, 13) or presence of either 1 μM L7Ae (lane 2) with increasing concentrations of Cbf5 (lanes 3–6), or 2 μM Cbf5 (lane 8) with increasing concentrations of L7Ae (lanes 9–12), and subjected to hydroxyl radical cleavage. Lanes 14 and 15 are size markers generated by alkaline hydrolysis (OH) and RNase T1 digestion (T1). Turquoise and purple bars indicate new sites of protection observed in the presence of both proteins and of Cbf5 protections lost upon addition of L7Ae, respectively. (B) Summary of changes in protection in the context of Pf9 RNA secondary structure model (as in Figure 1).
For the most part, the guide RNA protection pattern in the presence of both proteins (Figure 3, lanes 6 and 12) appears to be the simple sum of the patterns obtained with the individual proteins (Figure 3, lanes 2 and 8), however there are several interesting exceptions. Examination of multiple experiments and exposures revealed two regions where greater protection is observed than would be expected from the individual protections (Figure 3, turquoise shading). In the 3′ strand of the k-turn, several nucleotides (nts 39, 40, 44 and 45) are partially protected by L7Ae, and not significantly protected by Cbf5, but are nearly completely protected in the presence of both proteins (Figure 3A, lanes 6 and 12, and indicated in Figure 3B). Similarly, little protection of sequences in the 5′ strand of the upper stem was observed with either L7Ae or Cbf5 binding, however cleavage of most nucleotides in this region (19,20,22–24) is reduced by the combination of the two proteins (Figure 3). These increased protections likely reflect enhanced interaction of the proteins with these regions when the other protein is bound to the RNA.
In contrast, protection of nucleotides in the apical loop by Cbf5 (nts 30 and 31) is lost upon introduction of L7Ae (Figure 3A, lane 8 versus lane 9, purple shading). The de-protected nucleotides are part of a larger, contiguous Cbf5-binding site that also includes the 5′ strand of the k-turn, the region where both Cbf5 and L7Ae interact with the RNA (Figure 1). The loss of the Cbf5 protection pattern suggests that L7Ae disrupts or prevents the interaction of Cbf5 with the 5′ k-turn/apical loop region. This is consistent with the absence of an interaction between the k-turn/apical loop of the RNA and Cbf5 in the crystal structure of the full complex (26).
Interaction of Cbf5 with the conserved ACA sequence
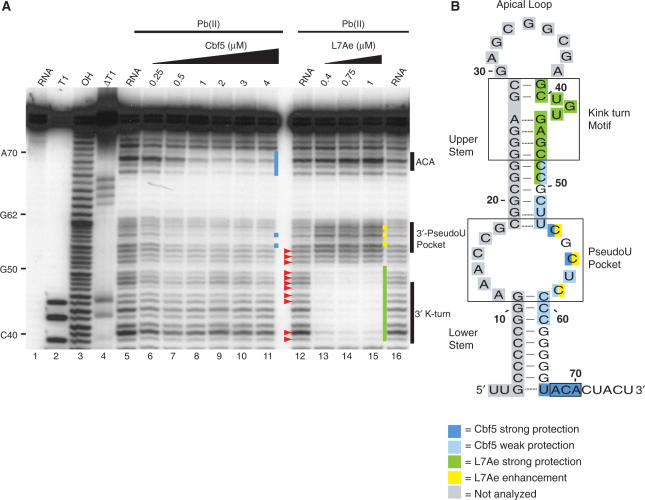
Box ACA is the signature sequence of the H/ACA guide RNAs, and RNA–protein-binding studies and crystal structure data indicate that Cbf5 specifically binds and recognizes this family feature (18,19,26). We examined the individual impacts of Cbf5 and L7Ae on the 3′ half of the RNA including box ACA by lead (II) acetate cleavage. Lead (II) acetate induces cleavage preferentially at single-stranded and dynamic regions of RNAs such as bulges and loops (29,36). As expected from the predicted secondary structure, the 3′ strand of the lower stem of Pf9 is inaccessible to lead-induced cleavage in the absence of proteins (see lack of cleavage between 3′ pseudouridylation pocket and ACA in Figure 4A, lanes 5, 12, 16). However, the 3′ strand of the upper stem was unexpectedly sensitive to cleavage (indicated with red arrowheads in Figure 4A, lane 12).
Figure 4.
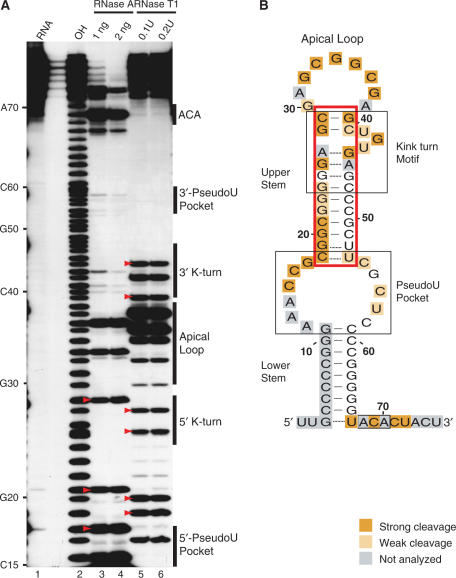
Lead-induced cleavage footprinting of Pf9 RNA, and Cbf5–Pf9 and L7Ae–Pf9 sub-complexes. (A) 5′-end labelled Pf9 was incubated in the absence (lanes 5, 12, 16) or presence of increasing concentrations of Cbf5 (lanes 6–11) or L7Ae (lanes 13–15) and subjected to lead (II)-induced cleavage. Lane 1 is undigested RNA and lanes 3, 2 and 4 are size markers generated by alkaline hydrolysis (OH), and RNase T1 digestion under non-denaturing (T1) and denaturing conditions (ΔT1), respectively. Blue and green bars indicate regions of strong Cbf5 and L7Ae protection. Yellow bars indicate cleavage enhancements observed with L7Ae. Red arrowheads indicate unexpected cleavages in the upper stem of the guide RNA in the absence of protein. (B) Summary of cleavage protections and enhancements in the context of Pf9 RNA secondary structure model (as in Figure 1).
Binding of Cbf5 to Pf9 RNA results in substantial protection of the conserved ACA sequence as well as the nucleotide immediately upstream (Figure 4, blue shading). Protection of most of the lower stem could not be assessed in this experiment (because this region is already insensitive to lead-induced cleavage); however, we observed that Cbf5 also provides some protection to the 3′ strand of the pseudouridylation pocket. In contrast, L7Ae does not protect these regions (Figure 4, lanes 13–15). The addition of L7Ae to the RNA results in reduced cleavage of the 3′ strand of the k-turn and also of the upper stem outside of the k-turn motif (Figure 4, green shading). L7Ae also produced increased sensitivity to lead-induced cleavage in nucleotides in the 3′ half of the pseudouridylation pocket (Figure 4, yellow shading).
Guide RNA secondary structure
The unexpected sensitivity of the 3′ side of the upper stem of Pf9 to lead-induced cleavage (Figure 4) led us to further probe the secondary structure of the RNA in the absence of proteins. We performed partial enzymatic digestions of 5′ end labelled Pf9 RNA using RNase T1 and RNase A (Figure 5). RNases T1 and A cleave accessible phosphodiester backbones following un-base-paired guanines (Gs) and pyrimidines (Cs and Us), respectively. Adenines (As) are not subject to analysis. Base-paired regions are resistant to both enzymes. The results are summarized schematically in the context of the predicted secondary structure in Figure 5B.
Figure 5.
Single-stranded nuclease footprinting of Pf9. (A) 5′-end labelled Pf9 was digested with indicated concentrations of RNase A (lanes 3, 4) or RNase T1 (lanes 5, 6). The cleavage products were separated on a denaturing 20% acrylamide gel. Lane 1 is undigested RNA and lane 2 is a size marker generated by alkaline hydrolysis (OH). Strong cleavages at nucleotides in the upper stem region are indicated with red arrowheads. (B) Summary of Pf9 RNA cleavages by single-stranded nucleases in the context of the predicted secondary structure of Pf9 RNA (as in Figure 1). Upper stem region is boxed in red.
Under the experimental conditions analysed, in the absence of proteins, accessible regions of the RNA include the 3′ tail, the pseudouridylation pocket, the apical loop and the bulge of the k-turn motif as expected (Figure 5B, orange shading). Consistent with the predicted secondary structure, nucleotides in the lower stem are inaccessible to the single-stranded nucleases. In addition, nucleotides in this region are susceptible to RNase V1, a nuclease specific for double-stranded regions (data not shown). However, we found that the upper stem is sensitive to single-stranded nuclease digestion [see strong cleavages in red boxed region (Figure 5B) and indicated by red arrowheads (Figure 5A)]. The results suggest that the upper stem of Pf9 RNA is not stably structured under these experimental conditions.
Effects of L7Ae on the guide RNA outside the k-turn motif
Although the upper stem of the RNA does not appear to be firmly established in the absence of proteins (Figures 4 and 5), the interaction of L7Ae with sequences on both sides of the k-turn motif (Figure 1) strongly implies the existence of a helix within the upper stem of Pf9 RNA in the presence of the protein. Moreover, the L7Ae-induced resistance of the upper stem beyond the k-turn motif (i.e. outside the region where L7Ae has been shown to directly contact RNA) to lead-induced cleavage (Figure 4) suggests the formation of the stem in the presence of the protein. To further investigate the impact of L7Ae on the secondary structure of the RNA, we also analysed partial enzymatic digestions in the presence of the protein (Figure 6).
Figure 6.
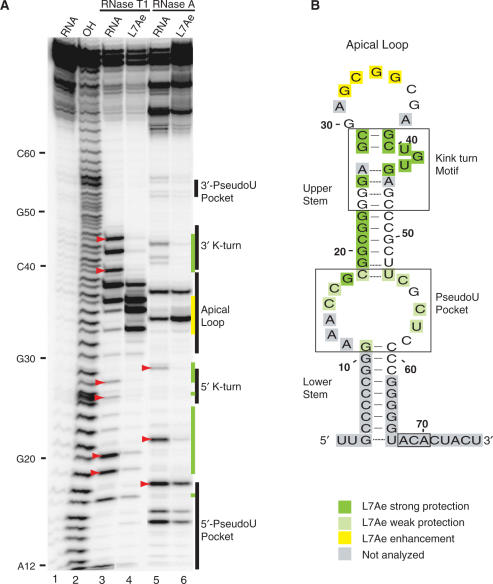
Single-stranded nuclease footprinting of L7Ae–Pf9. (A) 5′-end labelled Pf9 was incubated alone (lanes 3, 5) or with 1 μM L7Ae (lanes 4, 6) and digested with RNase T1 (lanes 3, 4) or RNase A (lanes 5, 6). The cleavage products were separated on a denaturing 15% acrylamide gel. Lane 1 is undigested RNA and lane 2 is a size marker generated by alkaline hydrolysis (OH). Red arrowheads indicate cleavages in the upper stem of the guide RNA in the absence of protein. Green and yellow bars indicate strong L7Ae protections and cleavage enhancements, respectively. (B) Summary of L7Ae cleavage protections and enhancements in the context of the predicted secondary structure of Pf9 (as in Figure 1).
The strong ribonuclease T1 and A cleavages observed in the ‘upper stem’ both within and outside the k-turn motif in the absence of the protein (red arrowheads, Figure 6A, lanes 3 and 5) are significantly reduced upon L7Ae binding (Figure 6A, lanes 4 and 6). At the same time, L7Ae increases cleavage of the apical loop (Figure 6B, yellow shading). The interaction of L7Ae with the k-turn is well documented (18,25,33–35). No extensive interactions outside the k-turn have been described. The strong protection that we observe in the upper stem, as well as the increased sensitivity in the apical loop, is consistent with formation of the upper stem upon L7Ae binding.
DISCUSSION
In this work, we examined the arrangements of the RNAs and proteins in a series of sub-complexes of the H/ACA RNP by chemical and enzymatic footprinting. The combined approach has the potential to detect both physical protein interaction sites and effects on RNA configuration, and we found evidence for both types of impacts in this study. The results provide detailed insight on steps in the assembly of the complex and essential roles of the proteins.
A significant amount is known about the sites of RNA–protein interaction within the fully assembled H/ACA guide RNP from the crystal structure of the P. furiosus holoenzyme (using a modified Afu 46 guide RNA) (26). In the Cbf5–guide RNA and L7Ae–guide RNA sub-complexes that we examined here, we observed footprints consistent with the well-established interaction of L7Ae with the k-turn (18,25,33–35) and with the contacts observed between Cbf5 and the guide RNA in the holoenzyme crystal structure (26). Our results support the extensive interaction of Cbf5 with the guide RNA from box ACA through the lower stem to the pseudouridylation pocket [observed in RNA-protein-binding assays (18,19), holoenzyme crystal structure (26) and RNA footprinting of the eukaryotic complex (27)]. The results indicate that the interactions of both Cbf5 and L7Ae with the guide RNA in the fully assembled enzyme are established in the Cbf5–guide RNA and L7Ae–guide RNA sub-complexes.
In addition, however, we found evidence of an interaction between Cbf5 and the guide RNA that is unique to the sub-complex. In the absence of other proteins, Cbf5 protects the 5′ strand of the k-turn and apical loop from hydroxyl radical cleavage, an effect that generally reflects a physical interaction (Figure 1). Moreover, previous studies showed that mutation of this region of the RNA weakens the binding of Cbf5 (18), supporting the existence of the interaction and suggesting that the interaction is important in formation and stability of the sub-complex. In the crystal structure of the holoenzyme, Cbf5 is not found in proximity with the apical loop and the 5′ strand of the k-turn (26). Our results indicate that L7Ae successfully competes for the site and displaces Cbf5 (Figures 3 and 7). Accordingly, L7Ae is found in close proximity with this region of the RNA in the crystal structure of the holoenzyme (26). [The specific equivalent L7Ae–RNA interactions could not be compared as the guide RNA used in the crystal structure differs significantly from Pf9 in this region (non-canonical k-turn) and its structure is also incomplete in this region (26).]
Figure 7.
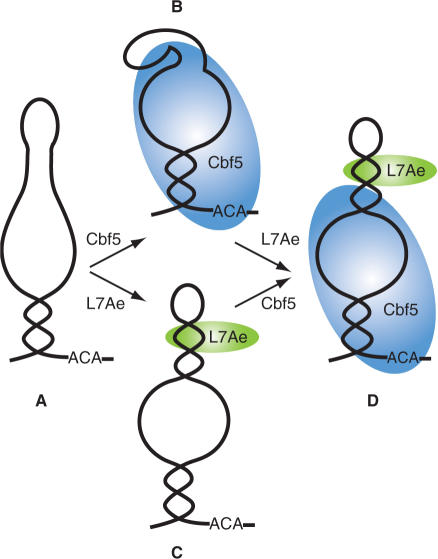
Simplified model depicting the observed conformational changes in the guide RNA and RNA–protein interactions in sub-complexes of the H/ACA RNP. Our footprinting data suggest that the upper stem of the guide RNA is not stably formed in the absence of L7Ae (A and B). Binding of L7Ae at the k-turn induces formation of the upper stem and establishes the pseudouridylation pocket (C). Cbf5 interacts with box ACA, the lower stem, the pseudouridylation pocket, and a unique site that includes the apical loop and the 5′ strand of the k-turn in the absence of L7Ae (B). L7Ae successfully competes with Cbf5 for binding to the 5′ strand of the k-turn and disrupts Cbf5s interaction with this region of the guide RNA (D).
Because the intermediates in the assembly and function of the H/ACA RNP have not been precisely defined, it is not yet clear what role the newly identified Cbf5–guide RNA interaction may play in H/ACA RNP assembly or function. In eukaryotes, evidence indicates that three of the four core proteins, including Cbf5, assemble on the H/ACA RNA at the site of transcription (and that association of Gar1 occurs at a later point in the temporal and spatial assembly pathway) (3,7,8). Among the core proteins, Cbf5 shows the strongest association with the H/ACA RNA genes, suggesting that Cbf5 could be the first of the H/ACA RNP proteins to associate with the newly made guide RNA in yeast (37). Thus, the Cbf5 interactions defined here may provide for the initial recognition of the guide RNA and subsequent complex assembly. It is also possible that a sub-complex lacking L7Ae is involved in the function of the H/ACA RNP (for example, as a step in substrate release).
Our studies also revealed a substantial effect of L7Ae on the guide RNA configuration beyond the k-turn with significant implications for proper establishment of the target recognition site. Previous studies had shown that the structure of the k-turn motif itself is dynamic in the absence of protein and that formation of the kink (i.e. 120° bend from linear) is induced by the binding of L7Ae and related proteins (38–41). Our results indicate previously undescribed effects on the RNA beyond this region (Figure 7). Our data from both partial enzymatic hydrolysis (Figure 5) and lead-induced cleavage (Figure 4) indicate that the upper stem of the guide RNA is not stably formed in the absence of proteins under the solution conditions used in our work. However, upon addition of L7Ae the upper stem nucleotides become resistant to single-stranded nucleases (Figure 6) and lead-induced cleavage (Figure 4), strongly suggesting L7Ae-induced formation of the upper stem. The observed increase in the sensitivity of nucleotides in the apical loop and pseudouridylation pocket to single-stranded nucleases (Figure 6) and lead-induced cleavage (Figure 4) upon L7Ae binding are also consistent with formation of the upper stem, which defines these loops. While L7Ae provided strong protection of the upper stem against enzymatic cleavage (Figure 6), this region was not significantly protected from hydroxyl radical cleavage outside of the k-turn motif (Figure 1), providing further evidence that the observed protection of this region from enzymatic cleavage reflects induction of base pairing (rather than steric interference). Importantly, the upper stem establishes the pseudouridylation pocket—the H/ACA RNP target RNA-binding site.
Our results reveal that L7Ae plays a significant role in substrate binding and placement in the archaeal H/ACA RNP via formation of the pseudouridylation pocket. These findings may explain the positive impact that L7Ae appears to have on formation of substrate-containing H/ACA RNP complexes and on activity of the complex (18,19). In addition, analysis of a crystal structure of a sub-complex of the H/ACA RNP with a substrate RNA recently obtained by Hong Li's laboratory suggests that both pseudouridylation pocket formation and positioning of the substrate uridine in the Cbf5 active site are defective in the absence of L7Ae (Liang,B., Xue,S., Terns,R., Terns, M. and Li, H., submitted for publication). At the same time, this and several other recent studies describe guide–substrate RNA interactions in the absence of L7Ae (42,43). It is most likely that the difference reflects the high concentrations of molecules used in these structural studies (42,43). In the cell, substrate capture likely depends on a well-formed pseudouridylation pocket established by L7Ae.
Given that the pseudouridine synthase Cbf5 can interact directly with the guide RNA, which has the capacity to capture and present the substrate, it was previously not clear why L7Ae should be needed. Our findings indicate that the importance of L7Ae in the function of the H/ACA RNP is in remodelling the guide RNA to form the substrate-binding site. In eukaryotes, the H/ACA RNP protein homologous to L7Ae is Nhp2, a protein with less well-defined RNA binding properties (44), and it remains to be determined whether Nhp2 will also play a role in definition of the substrate-binding site in the eukaryotic H/ACA RNPs. However, L7Ae is also a component of C/D RNPs and the ribosome in archaea (45), and our findings suggest that L7Ae and other k-turn-binding proteins could play a similar role in important alterations of RNA structure beyond the k-turn in other complexes as well.
ACKNOWLEDGEMENTS
We thank Hong Li for sharing results prior to publication and for stimulating collaborations. We thank Alex Huttenhofer and Pascale Romby for sharing detailed RNA footprinting protocols. This work was supported by NIH grant RO1 GM54682 to M.T. and R.T. and by a fellowship from the Egyptian government to O.Y. Funding to pay the Open Access publication charges for this article was provided by NIH grant RO1 GM54682 to M.T. and R.T.
Conflict of interest statement. None declared.
REFERENCES
- 1.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 2.Lane BG, Ofengand J, Gray MW. Pseudouridine and O2′-methylated nucleosides. Significance of their selective occurrence in rRNA domains that function in ribosome-catalyzed synthesis of the peptide bonds in proteins. Biochimie. 1995;77:7–15. doi: 10.1016/0300-9084(96)88098-9. [DOI] [PubMed] [Google Scholar]
- 3.Terns M, Terns R. Noncoding RNAs of the H/ACA family. Cold Spring Harb. Symp. Quant. Biol. 2006;71:395–405. doi: 10.1101/sqb.2006.71.034. [DOI] [PubMed] [Google Scholar]
- 4.Yu YT, Terns RM, Terns MP. Mechanisms and functions of RNA-guided RNA modification. In: Grosjean H, editor. Fine-Tuning of RNA Functions by Modification and Editing. Vol. 12. New York: Topics in Current Genetics; 2005. pp. 223–262. [Google Scholar]
- 5.Maden BE. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 6.Ofengand J. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 2002;514:17–25. doi: 10.1016/s0014-5793(02)02305-0. [DOI] [PubMed] [Google Scholar]
- 7.Kiss T, Fayet E, Jady BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- 8.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 9.Reichow SL, Hamma T, Ferre-D’Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiss-Laszlo Z, Henry Y, Bachellerie J-P, Caizergues- Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 11.Omer AD, Lowe TM, Russell AG, Ebhardt H, Eddy SR, Dennis PP. Homologs of small nucleolar RNAs in Archaea. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 12.Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 13.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 14.Meier UT. How a single protein complex accommodates many different H/ACA RNAs. Trends Biochem. Sci. 2006;31:311–315. doi: 10.1016/j.tibs.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 16.Hamma T, Ferre-D’Amare AR. Pseudouridine synthases. Chem. Biol. 2006;13:1125–1135. doi: 10.1016/j.chembiol.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell Biol. 1999;19:7461–7472. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker DL, Youssef OA, Chastkofsky MI, Dy DA, Terns RM, Terns MP. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev. 2005;19:1238–1248. doi: 10.1101/gad.1309605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charpentier B, Muller S, Branlant C. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res. 2005;33:3133–3144. doi: 10.1093/nar/gki630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allmang C, Carbon P, Krol A. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA. 2002;8:1308–1318. doi: 10.1017/s1355838202020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muslimov IA, Iacoangeli A, Brosius J, Tiedge H. Spatial codes in dendritic BC1 RNA. J. Cell. Biol. 2006;175:427–439. doi: 10.1083/jcb.200607008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenblad MA, Lopez MD, Piccinelli P, Samuelsson T. Inventory and analysis of the protein subunits of the ribonucleases P and MRP provides further evidence of homology between the yeast and human enzymes. Nucleic Acids Res. 2006;34:5145–5156. doi: 10.1093/nar/gkl626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R. Crystal structure of the spliceosomal 15.5 kD protein bound to a U4 snRNA fragment. Mol. Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 25.Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003;31:869–877. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443:302–307. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- 27.Normand C, Capeyrou R, Quevillon-Cheruel S, Mougin A, Henry Y, Caizergues-Ferrer M. Analysis of the binding of the N-terminal conserved domain of yeast Cbf5p to a box H/ACA snoRNA. RNA. 2006;12:1868–1882. doi: 10.1261/rna.141206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tullius TD, Dombroski BA, Churchill ME, Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- 29.Brunel C, Romby P. Probing RNA structure and RNA-ligand complexes with chemical probes. Methods Enzymol. 2000;318:3–21. doi: 10.1016/s0076-6879(00)18040-1. [DOI] [PubMed] [Google Scholar]
- 30.Celander DW, Cech TR. Iron(II)-ethylenediaminetetraacetic acid catalyzed cleavage of RNA and DNA oligonucleotides: similar reactivity toward single- and double-stranded forms. Biochemistry. 1990;29:1355–1361. doi: 10.1021/bi00458a001. [DOI] [PubMed] [Google Scholar]
- 31.Huttenhofer A, Noller HF. Hydroxyl radical cleavage of tRNA in the ribosomal P site. Proc. Natl Acad. Sci. USA. 1992;89:7851–7855. doi: 10.1073/pnas.89.17.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Latham JA, Cech TR. Defining the inside and outside of a catalytic RNA molecule. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 33.Hamma T, Ferre-D’Amare AR. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 A resolution. Structure. 2004;12:893–903. doi: 10.1016/j.str.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 34.Moore T, Zhang Y, Fenley MO, Li H. Molecular basis of box C/D RNA-protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 35.Nolivos S, Carpousis AJ, Clouet-d’Orval B. The K-loop, a general feature of the Pyrococcus C/D guide RNAs, is an RNA structural motif related to the K-turn. Nucleic Acids Res. 2005;33:6507–6514. doi: 10.1093/nar/gki962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindell M, Romby P, Wagner EG. Lead(II) as a probe for investigating RNA structure in vivo. RNA. 2002;8:534–541. doi: 10.1017/s1355838201020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang PK, Hoareau C, Froment C, Monsarrat B, Henry Y, Chanfreau G. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol. Cell Biol. 2005;25:3295–3304. doi: 10.1128/MCB.25.8.3295-3304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goody TA, Melcher SE, Norman DG, Lilley DM. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA. 2004;10:254–264. doi: 10.1261/rna.5176604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marmier-Gourrier N, Clery A, Senty-Segault V, Charpentier B, Schlotter F, Leclerc F, Fournier R, Branlant C. A structural, phylogenetic, and functional study of 15.5-kD/Snu13 protein binding on U3 small nucleolar RNA. RNA. 2003;9:821–838. doi: 10.1261/rna.2130503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mougin A, Gottschalk A, Fabrizio P, Luhrmann R, Branlant C. Direct probing of RNA structure and RNA-protein interactions in purified HeLa cell's and yeast spliceosomal U4/U6.U5 tri-snRNP particles. J. Mol. Biol. 2002;317:631–649. doi: 10.1006/jmbi.2002.5451. [DOI] [PubMed] [Google Scholar]
- 41.Turner B, Melcher SE, Wilson TJ, Norman DG, Lilley DM. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA. 2005;11:1192–1200. doi: 10.1261/rna.2680605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin H, Loria JP, Moore PB. Solution structure of an rRNA substrate bound to the pseudouridylation pocket of a box H/ACA snoRNA. Mol. Cell. 2007;26:205–215. doi: 10.1016/j.molcel.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 43.Wu H, Feigon J. H/ACA small nucleolar RNA pseudouridylation pockets bind substrate RNA to form three-way junctions that position the target U for modification. Proc. Natl Acad. Sci. USA. 2007;104:6655–6660. doi: 10.1073/pnas.0701534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henras A, Dez C, Noaillac-Depeyre J, Henry Y, Caizergues-Ferrer M. Accumulation of H/ACA snoRNPs depends on the integrity of the conserved central domain of the RNA-binding protein Nhp2p. Nucleic Acids Res. 2001;29:2733–2746. doi: 10.1093/nar/29.13.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terns MP, Terns RM. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]