Abstract
Over 100 chemical types of RNA modifications have been identified in thousands of sites in all three domains of life. Recent data suggest that modifications function synergistically to mediate biological function, and that cells may coordinately modulate modification levels for regulatory purposes. However, this area of RNA biology remains largely unexplored due to the lack of robust, high-throughput methods to quantify the extent of modification at specific sites. Recently, we developed a facile enzymatic ligation-based method for detection and quantitation of methylated 2′-hydroxyl groups within RNA. Here we exploit the principles of molecular recognition and nucleic acid chemistry to establish the experimental parameters for ligation-based detection and quantitation of pseudouridine (Ψ) and N6-methyladenosine (m6A), two abundant modifications in eukaryotic rRNA/tRNA and mRNA, respectively. Detection of pseudouridylation at several sites in the large subunit rRNA derived from yeast demonstrates the feasibility of the approach for analysis of pseudouridylation in biological RNA samples.
Since the discovery of the first non-canonical nucleosides in RNA in the 1950s, more than 100 different post-transcriptional modifications have been identified at thousands of sites in biological RNAs from all three domains of life (1,2). These modifications span a large range of chemical diversity from a single methylation to elaborate ring addition. A substantial array of enzymatic machinery mediates their installation, representing 1–2% of all genes in bacterial genomes (3). In eukaryotes, hundreds of guide RNAs and numerous proteins direct modifications in ribosomal RNA (rRNA) alone (4,5). Accumulated data from decades of study have suggested important roles for some of these modifications in fine-tuning RNA structure and stability and in promoting the accuracy and efficiency of gene expression (3). While these investigations have largely focused on modification at single sites, it has become increasingly apparent that modifications can act collectively to modulate RNA function, and may serve regulatory roles by changing their levels in response to physiological conditions (6,7). Defining such collective and regulatory action requires knowledge of the global pattern of modifications within an organism and their variations on a genome-wide scale. These aspects of RNA biology remain largely unexplored, however, due to lack of robust methods to evaluate quantitatively the degree of modification of a given nucleotide within an RNA. Recently, we developed a ligation-based approach for detection and quantitation of methylated 2′-hydroxyl groups within RNA. Here we establish the experimental parameters for application of this method to two additional abundant modifications within biological RNA, pseudouridine and N6-methyladenosine.
Pseudouridine (Ψ) ranks among the most prevalent post-transcriptional modifications in RNA. Ψ occurs at many sites in rRNA (e.g. 46 sites in yeast), tRNA (e.g. 72 sites in Escherichia coli) and spliceosomal snRNA (e.g. 24 sites in human), tuning the activity and stability of these RNAs (8). For example, gene deletion studies that block pseudouridylation at specific sites in functionally important domains of rRNA indicate that Ψ in the translating ribosome directly and synergistically affects the subunit interactions and the peptidyl transferase activity (9–11).
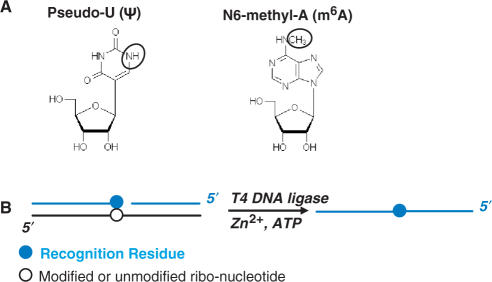
Pseudouridine arises from the isomerization reaction catalyzed by Ψ-synthases, in which a uracil base is detached from its N1 ribose linkage, rotated 120° and reattached to the ribose via its C5 position (Figure 1A). Ψ and U present the same acceptor–donor–acceptor pattern of hydrogen bonding groups on the Watson-Crick face of the nucleobase, but on the opposite face Ψ has an N1H imino group whereas U has a C5H group. Pseudouridylation therefore engenders a very modest chemical change, and as a uridine isomer, Ψ has the same mass as uridine, rendering its detection and the quantitation of the extent of pseudouridylation at a given site within a cellular RNA particularly challenging. Existing methods for Ψ detection involve either chemical modification of the N1H group followed by primer extension (12), or nuclease cleavage of purified cellular RNAs followed by thin-layer chromatography (13). While the latter method has been used on occasion to quantify the extent of Ψ modification at particular sites, the extensive processing required and numerous steps have limited its wider use.
Figure 1.
(A) Chemical structures of Ψ and m6A. (B) Scheme for T4 DNA ligase-catalyzed joining of two DNA substrates. In the ternary RNA/DNA complex, the black line corresponds to the 30-mer RNA template with the modified nucleotide (open circle) located at the 15th position. Blue lines correspond to the ligation substrates with the recognition residue shown as a filled blue circle.
N6-methyladenosine (m6A) represents the major modification present in mammalian mRNA. The functions of m6A remain largely unknown, though it may influence mRNA processing (14–17). The modification arises from an S-adenosylmethionine derived, single methylation of adenosine at N6, which leaves the Watson–Crick face unaltered (Figure 1A). Primer extension reactions therefore are unable to detect its presence (18). The only established method for detecting m6A at specific sites involves nuclease cleavage of purified cellular RNA followed by thin-layer chromatography or mass spectrometry (19,20). Thus, the existing methods for detecting N6-adenosine methylations and pseudouridylations are laborious and low throughput, precluding analysis on a genome-wide scale that will ultimately be necessary to reveal the biological purposes of the modifications.
MATERIALS AND METHODS
Syntheses of the phosphoramidites of N6-aryl-2′-deoxyadenosine analogs and their incorporation into oligonucleotides
The installation of various aryl groups at the N6-position of 2′-deoxyadenosine was accomplished by the direct copper catalyzed arylation of 2′-deoxyadenosine (21) with the corresponding aryl iodide (or bromide) in dimethylsulfoxide in the presence of copper (I) iodide (catalyst), ethylenediamine (ligand) and potassium phosphate (base) and sodium iodide (in the case of arylbromide) to give 1a-f (Figure 2B). Dimethoxytritylation of 1a-f in the presence of 4,4′-dimethoxytrityl chloride in pyridine afforded intermediates 2a-f in 80–92% yields. Phosphitylation of the 3′-OH of 2a-f in dichloromethane in the presence of diisopropylethylamine generated the phosphoramidites 3a-f in 85–93% yield. The coupling of 3a-f into DNA oligonucleotides was quantitative. Detailed characterization of the synthesis products and oligonucleotides are provided in the Supplementary Data section.
Figure 2.
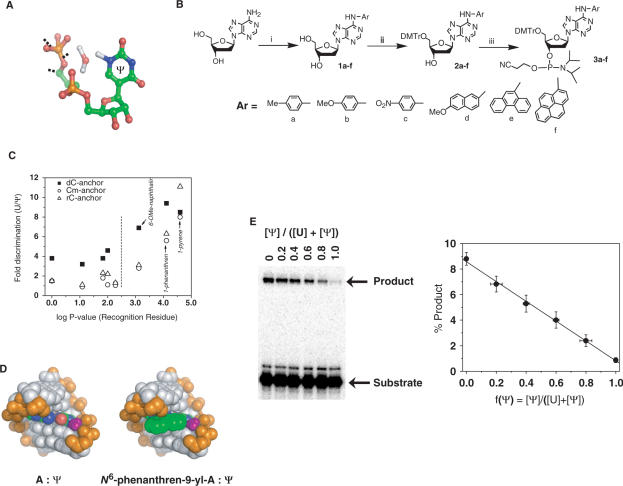
Identifying a recognition residue for Ψ. (A) Within pseudouridine-containing RNA duplexes, a water molecule localizes in the major groove, forming hydrogen bonds to the phosphate backbone and to Ψ via N1H (25–27). (B) Synthetic scheme for N6-aryldeoxyadenosine analogs. (C) Relative efficiency for U versus Ψ-directed ligation correlates with the hydrophobicity of the recognition residue. The yield of ligation product using the unmodified 30-mer RNA template (U15) relative to the yield of ligation product using the Ψ 15 template gives the ‘fold discrimination’ plotted on the y-axis. Log P values (x-axis) for the deoxyadenosine substituent at the recognition position were calculated using CS Chem 3D (version 5.0). The 3′ nucleotide at the ligation junction bears at the 2′-position either a hydrogen atom (dC-anchor), a methoxyl group (Cm-anchor) or a hydroxyl group (rC-anchor). (D) Molecular model of the base pair N6-phenanthren-9-yl-A: Ψ in an A-form helix. The phenanthrene ring is in green and the N1H-coordinated water is in purple. (E) Ligation yield correlates linearly with the fraction of pseudouridylation f(Ψ). Ligation reactions contained U and Ψ 30-mer RNA templates mixed together in defined ratios as indicated. Linear fit has a r-value of 0.998 and P-value of <0.0001.
2a-f
To N6-aryl-2′-deoxyadenosine (1a-f) (0.3 mmol) in pyridine (5 ml) was added 4,4′-dimethoxytrityl chloride (1.2 eq.). After being stirred overnight at room temperature, the reaction was quenched with methanol (1 ml) and stirred for an additional 5 min. The reaction mixture was then concentrated to dryness under vacuum. The resulting residue was dissolved in dichloromethane (50 ml) and washed consecutively with 5% sodium bicarbonate, water and brine, and then dried over sodium sulfate. After the organic phase was concentrated to dryness, the residue was purified by silica gel chromatography, eluting with 3% methanol in dichloromethane containing 0.2% triethylamine to give 2a-f as foam.
3a-f
To N6-Aryl-5′-O-(4,4′-Dimethoxytrityl)-2′-deoxyguanosine (2a-f) (0.2 mmol) in dry dichloromethane (10 ml) and N,N-diisopropylethylamine (0.4 ml) was added 2-cyanoethyl N,N-diisopropylchlorophosphoramidite (3.0 eq.). After being stirred at room temperature for 0.5 h, the reaction mixture was diluted with dichloromethane (40 ml) and washed with 5% aqueous sodium carbonate and brine, dried over sodium sulfate and concentrated. The residue was purified by silica gel chromatography, eluting with 10–12% acetone in dichloromethane containing 0.2% triethylamine to give 3a-f as foam.
Incorporating phosphoramidites 3a-f into oligonucleotides
Upon a 10-min reaction, all phosphoramidites were coupled into the 5′ position of a 15-mer (sequence 5′-ZGC GTT ACA GCG GAT) as efficiently as wild-type DNA phosphoramidite based on quantification of DMTr cation release. After deprotection with ethanolic ammonia and gel purification, all oligonucleotides were 5′-phosphorylated using T4 polynucleotide kinase.
Synthesis of phosphoramidite of N6-methyladenosine 7 and its incorporation into RNA oligonucleotides
N6 methylation of adenosine was accomplished by following Hobartner's procedure to yield intermediate 4 (22). Selective protection of the 2′-OH of 4 with t-butyldimethylsilylchloride was accomplished by adopting Beigelman's strategy to give intermediate 5 (23). Dimethoxytritylation of 5 in the presence of 4,4′-dimethoxytrityl chloride in pyridine afforded intermediate 6. Phosphitylation of the 3′-OH of 6 in dichloromethane in the presence of diisopropylethylamine generated phosphoramidite 7 (Figure 3B) in 82% yield. Detailed characterization of the synthetic products and oligonucleotides are provided in the Supplementary Data section.
Figure 3.
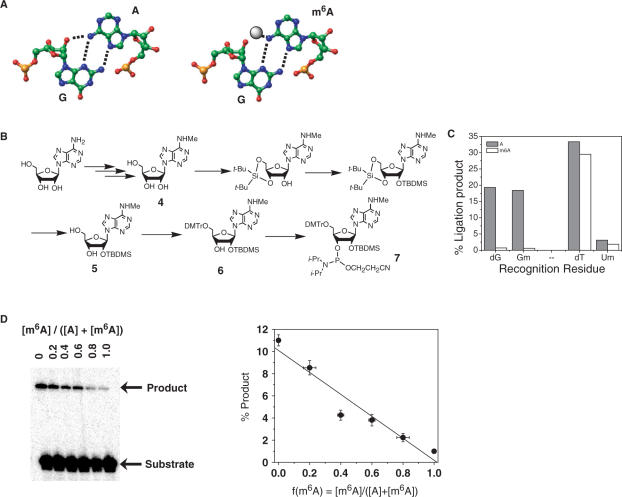
Identifying a recognition residue for m6A. (A) The sheared G–A base pair found in many RNA structures (left). Within this purine–purine base pair N6H(A) and 2′OH(G) reside in close proximity to each other. The methyl group of m6A (right) might sterically clash with the phosphate backbone of G. (B) Synthetic scheme for N6-methyl-rA phosphoramidite. (C) Efficiency of A and m6A-directed ligation using different recognition residues. The 30-mer RNA templates contained at position 15 either unmodified adenosine (A) or N6-methyladenosine (m6A). Substrates contained one of the following recognition residues: dG (2′-deoxyguanosine), Gm (2′-methoxyguanosine), dT (thymidine) or Um (2′-methoxyuridine). (D) Ligation yield correlates linearly with the fraction of adenosine methylation f(m6A). Ligation reactions contained adenosine and N6-methyladenosine 30-mer RNA templates mixed together in defined ratios as indicated. Linear fit has a r-value of 0.964 and P-value of 0.002.
Compound 5
To a solution of N6-methyladenosine 4 (550 mg, 1.96 mmol) in dimethylformamide (20 ml) was added di-tert-butylsilyl bis(trifluoromethanesulfonate) (810 μl, 1.1 eq.) at 0°C under argon. After 15 min, imidazole (0.700 g) and t-butyldimethylsilyl chloride (0.700 g) were added, and the mixture was heated to 60°C for 2 h. After removing the solvent under reduced pressure, the residue was dissolved in tetrahydrofuran (15 ml). Diluted hydrofluoric acid in pyridine was added, and the mixture was stirred at room temperature for 2 h. After removing the solvent under reduced pressure, the residue was dissolved in dichloromethane (100 ml) and washed with 5% sodium bicarbonate solution followed by brine and dried over magnesium sulfate. After evaporation of the solvent under reduced pressure, the residue was purified by silica gel chromatography, eluting with 3–5% methanol in dichloromethane to give 5 (555 mg, 75%) as a white foam.
Compound 6
To 5 (500 mg, 1.26 mmol) in pyridine (10 ml) was added 4,4′-dimethoxytrityl chloride (1.2 eq.) under argon. After stirred overnight at room temperature, the reaction was quenched with methanol (1 ml) and stirred for an additional 5 min. The reaction mixture was then concentrated to dryness under vacuum. The resulting residue was dissolved in dichloromethane (100 ml) and the solution was washed consecutively with 5% sodium bicarbonate, water and brine, and then dried over sodium sulfate. After the organic phase was concentrated to dryness, the residue was purified by silica gel chromatography, eluting with 1–2% methanol in dichloromethane containing 0.2% triethylamine, to give 6 as a white foam.
Compound 7
To 6 (175 mg 0.251 mmol) in dry dichloromethane (10 ml) and N,N-diisopropylethylamine (0.4 ml) was added 2-cyanoethyl N,N-(diisopropylamino)-chlorophosphoramidite (3 eq.). After being stirred at room temperature for 2 h, the reaction mixture was diluted with dichloromethane (40 ml) and washed with 5% aqueous sodium carbonate and brine, dried over sodium sulfate and concentrated. The residue was purified by silica gel chromatography, eluting with 8–10% acetone in dichloromethane containing 0.2% triethylamine to give 7 as a white foam.
Incorporating phosphoramidite 7 into oligonucleotide
Phosphoramidite 7 was coupled into the 15th position of a 30-mer (sequence 5′-AUC CGC UGU AAC GC7 GAG CAA UGC CUG GUA) as efficiently as wild-type RNA phosphoramidite based on quantification of DMTr cation release. After standard deprotection and gel purification, Maldi-TOF Ms: 9598 (found), 9606 (calcd.)
Ligation reactions to study RNA modifications
The ligation substrates consist of two oligonucleotides. The oligo containing the recognition residue at its 5′ end residue (e.g. N6-phenanthren-dA for Ψ or dG for m6A) is referred to as the ‘floater’, the other oligo substrate is referred to as the ‘anchor’.
A 30-mer model RNA (5′-AUCCGCUGUAACGCXGAGCAAUGCCUGGUA, X = U, Ψ, A from Dharmacon Research, Inc., X = m6A was synthesized as described above) was used to identify the recognition residue for RNA modifications (24). The optimized ligation reactions were carried out with 0.15 µM 30-mer RNA with or without Ψ or m6A modifications, 0.5 µM floater and 0.38 µM of 5′-32P-labeled anchor in 66 mM Tris–HCl, pH 7.6, 0.5 mM ZnCl2, 10 mM DTT, 66 µM ATP, 15% DMSO and 0.25 U/µl T4 DNA ligase (USB Inc.). All components were mixed and incubated at 16°C for 16 h, and the ligation products separated on denaturing polyacrylamide gels containing 7 M urea.
The analysis of Ψ in the rRNA present in yeast total RNA was performed as follows. A total of 0.4 µM anchor and 0.5 µM 5′-32P-labeled floater first hybridized with yeast total RNA in 20 mM Tris–HCl, pH 7.5, 50 mM NaCl, 0.4 µg/µl total yeast RNA, plus 10 nM model 30-mer RNA, its 30 nM 32P-labeled floater, and 60 nM anchor as the control for ligation efficiency and loading. Hybridization was performed by placing the tubes in a 95°C heat block for 1 min, followed by immediately placing the heat block at 4°C for 45 min before placing on ice for 5 min. Following hybridization, the ligation reaction was initiated by the addition of a 2× ligation mixture containing 132 mM Tris–HCl, pH 7.5, 1 mM ZnCl2, 20 mM DTT, 132 µM ATP, 30% DMSO and 1 U/µl T4 DNA ligase. The ligation proceeded at 27°C for up to 120 min. In order to remove excessive background of the unreacted, 5′-32P-labeled oligos (the 32P-label is always in the middle of the ligation product), 5 µl aliquots of the ligation mixture were treated with 0.1 U/µl calf-intestine alkaline phosphatase (Boehringer-Mannheim) and 0.1 U/µl RNase H (Epicenter Technologies) at 37°C for 10 min. Alkaline phosphatase was used to reduce the background derived from the unligated 32P-oligo substrates, and RNase H was used to reduce the interference of rRNA in the subsequent gel analysis. The reaction was quenched with the addition of an equal volume of 9 M urea/50 mM EDTA. The mixture was boiled for 2 min and rapidly cooled on ice prior to its loading on denaturing polyacrylamide gels containing 7 M urea.
RESULTS AND DISCUSSION
Recently, we reported a ligation-based method for the detection and quantification of 2′-O-methyl modifications in RNA that can overcome the limitations of the current methods (16). Our approach uses the RNA as a template to direct the enzymatic ligation of two adjunct oligodeoxynucleotides (Figure 1B blue strands) designed to form Watson–Crick base pairs with sequence elements immediately upstream and downstream of the modification site (Figure 1B, open circle). The residue that opposes the modification site in the template/substrate ternary complex (herein referred to as the ‘recognition residue:’ Figure 1B, blue circle) influences the ligation efficiency. An oligonucleotide bearing an appropriate recognition residue for a 2′-O-methylnucleotide (2′-OMe-A, G, C or U) ligates with an efficiency that depends significantly on the presence or absence of the modification. The ligation yield then correlates with the fraction of RNA bearing the modification. Because the ligation reaction for each modification site produces a DNA oligonucleotide of unique sequence, we can adapt this approach to a microarray platform to enable analysis on a genome-wide scale (for example, all known Ψ-modification sites in rRNA). The central challenge in extending this approach to other modification types therefore involves the identification of recognition residues for specific modification types.
In previous work, we screened empirically a collection of 24 oligonucleotide pairs to identify recognition residues for all four 2′-O-methyl nucleotides (2′-OMe-A, C, G, U) within RNA (24). We found that when the 2′-OMe-nucleotide opposes the complementary 2′-deoxynucleotide, T4 DNA ligase works much less efficiently compared to when the corresponding unmodified (2′-OH) nucleotide opposes the recognition residue. In hindsight, we could have rationalized these findings by the distortion in A-form helix backbone geometry caused by the 2′H–2′OMe (modified RNA) base pair as compared to the 2′H–2′OH (unmodified RNA) base pair.
Identification of recognition residues
Pseudouridine (Ψ)
An analogous empirical screen using an expanded set of 83 oligonucleotide pairs failed to identify a ‘recognition residue’ for Ψ (Table S1), perhaps reflecting less distortion of base-pair geometry by Ψ compared to 2′-OMe-U. The N1H imino group that Ψ presents in the major groove of an A-form duplex represents the one obvious feature that distinguishes Ψ from U. Crystallographic and computational studies suggest that an ordered water molecule bridges the N1H and the phosphate backbone of the preceding nucleotide, possibly accounting for the stabilizing effect that Ψ exerts in helical regions of RNA (25–27)(Figure 2A). The polarity of the N1H requires that it remain solvated or hydrogen bonded upon duplex formation (28). Disruption of this water bridge would be expected to destabilize or perturb the duplex structure relative to the corresponding U-containing duplex. Therefore modified nucleotides that disrupt this ‘water bridge’ could enable T4 DNA ligase to distinguish Ψ from U. To this end, we constructed as potential recognition residues a series of 2′-deoxyadenosine derivatives bearing at the N6 position functional groups of varying sizes and electronic properties (Figure 2B). These nucleotide analogs introduce steric and/or electronic perturbations in the deep and narrow major groove of the A-form helix. In particular, large hydrophobic groups positioned in this major groove could disrupt the N1H-water coordination and as a consequence decrease the ligation efficiency of T4 DNA ligase. This rationale suggests that large, hydrophobic groups at the N6-position should favor ligation opposite U over ligation opposite Ψ in RNA.
The series of N6-modified 2′-deoxyadenosine derivatives (Figure 2B) contains groups with varying size (Ar = Me, < p-Tol < 6-MeO-naphthen-2-yl < phenanthren-9-yl < pyren-1-yl), inductive properties (electron donating Ar = p-methoxyphenyl, electron withdrawing Ar = p-nitrophenyl), and hydrophobicity. Installation of these aryl groups at the N6-position of 2′-deoxyadenosine gave 2′-deoxynucleosides 1a-f (21). Dimethoxytritylation and phosphitylation of 1a-f according to standard procedures gave the corresponding phosphoramidites 3a-f in excellent yields (Figure 2B, Supplementary Data). We constructed 15-mer DNA oligonucleotides containing 3a-f at the 5′-end (recognition residue). After deprotection and gel purification, the 5′-phosphorylated oligonucleotides were further characterized by MALDI-TOF mass spectrometry (Table S2).
In a two-step procedure, we tested these dA-analog containing oligonucleotides as ligation substrates along with the corresponding substrate containing dA. First, a wide range of ligation conditions were tested using both unmodified and Ψ-modified 30-mer RNA templates, including ligase concentration (0.125–1 U/µl), temperature (4– 42°C) and divalent metal ions (0.5–10 mM Mg2+, Mn2+, Ca2+, Ba2+, Zn2+, Pb2+ or Cd2+). This initial screen revealed that under conditions optimized for discrimination and overall ligation yield, the N6-phenanthren-9-yl and pyren-1-yl containing oligonucleotides provided the greatest level of discrimination, ligating when directed by the unmodified RNA template with 10-fold greater efficiency than when directed by the Ψ-modified RNA template (Figure 2C). In general, the groups with larger molecular volume tended to provide better discrimination, but the correlation was incomplete (data not shown). However, the degree of discrimination by each substituent did correlate well with log P values, a measure of hydrophobicity related to the partition coefficient of the compound between octanol and water (Figure 2C). It is possible that the N6-phenanthren-9-yl and pyren-1-yl groups alter the stability or geometry of the hybrid Ψ helix by disrupting N1H hydration, thereby rendering Ψ-containing RNA a less efficient template for ligation by T4 DNA ligase. Supporting this hypothesis, molecular modeling of the phenanthren-9-yl group into the N6-position of an A–Ψ base pair within in an A-form helix shows that the group has sufficient size to clash with the N1H-coordinated water molecule (Figure 2D).
In the second step, we examined the ability of the N6-phenanthren-9-yl oligonucleotide to reveal the relative amounts of U and Ψ in the total RNA using mixtures containing defined ratios of unmodified and Ψ-containing 30-mer RNAs (Figure 2E). The ligation yield varied linearly over a 10-fold range with the fraction of Ψ-modified RNA. As the ligation yield shows a statistical variation of less than 1.5-fold, the 10-fold variation provides sufficient discrimination to detect and quantify Ψ modifications in biological RNAs, The N6-pyrene-dA analog provided a similar range of discrimination but gave lower overall ligation yields (data not shown).
N6-methyladenosine (m6A)
In contrast to methylation of the 2′-hydroxyl group, methylation of the adenosine nucleobase engenders minimal backbone conformational distortion that would perturb helix geometry. In addition, the N6-position retains its ability to donate a hydrogen bond, so that m6A can still form Watson–Crick base pairs. Consistent with these expectations, the A and m6A RNA templates directed ligation of substrates bearing T or U as recognition residues with similar efficiency (Figure 3C).
To identify a recognition residue for the detection of m6A, we considered non-Watson–Crick base-pairing arrangements. Many RNA structures contain sheared G–A base pairs, in which guanosine forms hydrogen bonds to the Hoogsteen face of adenosine using N3 and N2H (Figure 3A). In this purine–purine base pair, the adenosine N6H (A) resides within 4 Å of the 2′-OH(G) (29,30). In this crowded environment, a bulky N6-methyl group would sterically clash with the phosphate backbone of G. Consequently, we tested whether guanosine as the ‘recognition residue’ could favor ligation reactions directed by the unmodified RNA template over those directed by the m6A RNA template (Figure 3A).
We synthesized the phosphoramidite for N6-methyladenosine (Figure 3B) and incorporated it into position 15 of the 30-mer RNA ligation template (Supplementary Data). Ligation reactions were performed using oligonucleotide substrates containing dG, 2′-ribo-G, or 2′-OMe-G as the recognition residue (Figure 3C, 2′-ribo-G results not shown due to low overall ligation yield). Consistent with our hypothesis, the unmodified RNA template directed ligation of the G-containing substrates with at least 10-fold greater efficiency than did the m6A template. The discrimination provided by 2′-deoxy-G correlated linearly with the fraction of m6A containing RNA (Figure 3D), suggesting that its application as the recognition residue allows quantitative determination of m6A modification fractions.
Detection of pseudouridine modification in biological RNA samples
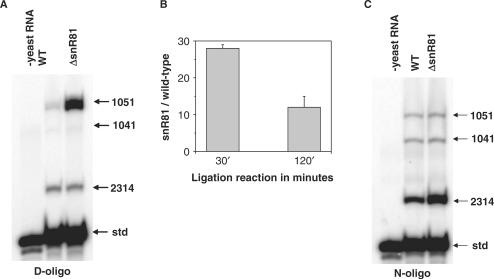
The specific sites of m6A modification within biological mRNAs are largely unknown, but all the positions of pseudouridylation in yeast ribosomal RNA have been mapped, and many of their corresponding pseudouridylation guide snoRNAs have been identified by genetic deletion. Taking advantage of the availability of such deletion strains, we sought to determine whether N6-phenanthren-9-yl-adenosine identified in our experiments could serve as a recognition residue for detecting the presence or absence of Ψ at specific sites in rRNA from samples of total yeast RNA (Figure 4A). Two total RNA samples were used, one isolated from a wild-type yeast strain and the other isolated from an isogenic strain in which the gene encoding the snoRNA, snR81 had been deleted (31). SnR81 directs the pseudouridylation of U1051 in the 3393 residue large subunit rRNA. The rRNA from the snR81 deletion strain contains no Ψ at position 1051, whereas rRNA from the wild-type strain contains only Ψ at position 1051.
Figure 4.
Detecting changes in pseudouridylation levels within ribosomal RNA from yeast. (A) Ligation reactions detect the absence of 1051 pseudouridylation in an snR81 deletion strain. The small nucleolar RNA snR81 specifically guides the pseudouridylation at U1051 in the 3393 nt 25S rRNA from yeast. Ligation reactions contained 30-mer RNA template (std), RNA derived either from a wild-type yeast strain or from the corresponding ▵snR81 deletion strain, and substrates containing N6-phenanthren-9-yl-adenosine in the recognition position designed for ligations at residues 1041, 1051 and 2314 of rRNA and residue 15 of the 30-mer RNA template. (B) Quantitative differences in the amount of ligation products for U/Ψ1051 in rRNA. The two-sided t-test P-value for these data is 0.0885. (C) Ligations as described in (A) but using oligonucleotides substrates containing adenosine at the recognition position.
We constructed three oligonucleotides containing N6-phenanthren-9-yl-adenosine and the corresponding ligation partner directed at U1051 and two other positions (U1041 and U2314) expected to undergo pseudouridylation to similar extents in both strains. The ligation reactions contained in the same vessel the oligonucleotide substrates targeted to all three sites. As an additional control for overall ligation efficiency and loading, we included in each ligation reaction the 30-mer RNA used to establish the recognition residue and the corresponding ligation partner. RNA samples from both strains gave similar yields of ligation products corresponding to positions U1041, U2314 and the control 30-mer RNA (Figure 4A). Strikingly, ligation reactions containing RNA derived from the deletion strain gave >10-fold more U1051 ligation product than did reactions containing RNA derived from the wild-type strain (Figure 4B). In contrast, we observed little difference between the RNA samples for the analogous reactions using substrates containing recognition residues that lack the ability to discriminate between Ψ from U (Figure 4C).
Limitations of the method
For the wild-type RNA sample, we observed different ligation yields for the three sites in the large subunit rRNA (Figure 4A and C), even though these sites supposedly contain the same extent of modification. These variations in ligation yield may reflect the influence of rRNA structure or sequence context of the individual modification sites. Moreover, the relative ratio of discrimination can change as a function of ligation time, as shown for the Ψ1051 site when the modification is completely absent (Figure 4B). These observations suggest that to determine the absolute modification fraction, ligations will have to be calibrated for every individual site of modification. This arduous task precludes high-throughput determination of absolute fractions of modification. Currently, the ligation method is best used to determine the relative differences in the modification fraction between two samples.
SUMMARY AND SIGNIFICANCE
Previously, we established that the reaction catalyzed by T4-DNA ligase can detect and quantitate methylation of the 2′-hydroxyl group at specific sites in RNA if the substrates contained the appropriate recognition residue. In that work, we identified the recognition residue by screening a collection of oligonucleotide ligation substrates bearing commercially available nucleoside analogs at the recognition position. An analogous screen for pseudouridine using a larger collection of oligonucleotide substrates failed to identify a suitable recognition residue. In the current work, we used available structural information pertaining to duplexes containing the modified or unmodified nucleosides together with the principles of molecular recognition to identify recognition residues that enable ligation-based detection and quantitation of Ψ and m6A modifications in RNA. As pseudouridylation and base and hydroxyl group methylation represent the most subtle post-transcriptional RNA modifications, our ligation-strategy appears robust in its ability to discriminate rather subtle chemical changes in RNA. We expect that our approach will be generally applicable for many other, if not all, RNA modifications.
The major advantages of the molecular recognition/enzymatic ligation approach to study RNA modifications include the ability to quantify the extent of modification at multiple defined positions simultaneously in the same biological sample and its adaptability to high-throughput study of modifications at all sites. Because the ligation reactions generate DNA oligonucleotides of unique sequence in yields that reflect the fraction of biological RNA modification at each site, microarray technologies can provide a well-established platform for high-throughput analysis. With this technology in hand, we can address questions that require information about the global landscape of RNA modifications within cells. For example, we can begin to assess how the 46 Ψ modifications in yeast rRNA work synergistically for ribosome function, and how the extent of modification at these sites changes as a function of physiological state.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr M. Fournier for insightful discussions. National Institute of Health (R21GM73747 to T.P. and J.A.P.); Howard Hughes Medical Institute of J.A.P. Funding to pay the Open Access publication charges for this article was provided by HHMI.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rozenski J, Crain PF, McCloskey JA. The RNA Modification Database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCloskey JA, Rozenski J. The Small Subunit rRNA Modification Database. Nucleic Acids Res. 2005;33:D135–D138. doi: 10.1093/nar/gki015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grosjean H, editor. Fine-tuning of RNA Functions by Modification and Editing. Berlin: Springer-Verlag; 2005. [Google Scholar]
- 4.Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 5.Kiss T. SnoRNP biogenesis meets Pre-mRNA splicing. Mol. Cell. 2006;23:775–776. doi: 10.1016/j.molcel.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 6.Decatur WA, Fournier MJ. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 2003;278:695–698. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- 7.Agris PF, Vendeix FA, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 8.Grosjean H, editor. Fine-tuning of RNA Functions by Modification and Editing. Berlin: Springer-Verlag; 2005. pp. 1–22. [Google Scholar]
- 9.Ganot P, Bortolin ML, Kiss T. Site specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 10.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell. 2003;11:425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 11.Ni JW, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 12.Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- 13.Zhao X, Yu YT. Detection and quantitation of RNA base modifications. RNA. 2004;10:996–1002. doi: 10.1261/rna.7110804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carroll SM, Narayan P, Rottman FM. N6-methyladenosine residues in an intron-specific region of prolactin pre-mRNA. Mol. Cell. Biol. 1990;10:4456–4465. doi: 10.1128/mcb.10.9.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finkel D, Groner Y. Methylations of adenosine residues (m6A) in pre-mRNA are important for formation of late simian virus 40 mRNAs. Virology. 1983;131:409–425. doi: 10.1016/0042-6822(83)90508-1. [DOI] [PubMed] [Google Scholar]
- 16.Kane SE, Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol. Cell Biol. 1985;5:2298–2306. doi: 10.1128/mcb.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoltzfus CM, Dane RW. Accumulation of spliced avian retrovirus mRNA is inhibited in S-adenosylmethionine-depleted chicken embryo fibroblasts. J. Virol. 1982;42:918–931. doi: 10.1128/jvi.42.3.918-931.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bokar JA. In: Fine-tuning of RNA Functions by Modification and Editing. Grosjean H, editor. Berlin: Springer-Verlag; 2005. pp. 141–178. Hiedelberg, New york. [Google Scholar]
- 19.Kawamura Y, Mizuno Y. Studies on transfer RNAs. II. Modification of Escherichia coli formylmethionine transfer RNA. Biochim. Biophys. Acta. 1972;277:323–334. [PubMed] [Google Scholar]
- 20.Limbach PA, Crain PF, McCloskey JA. Characterization of oligonucleotides and nucleic acids by mass spectrometry. Curr. Opin. Biotechnol. 1995;6:96–102. doi: 10.1016/0958-1669(95)80015-8. [DOI] [PubMed] [Google Scholar]
- 21.Ran C, Dai Q, Harvey RG. N6-arylation of 2′-deoxyadenosine via copper-catalyzed direct coupling with aryl halides. J. Org. Chem. 2005;70:3724–3726. doi: 10.1021/jo040284w. [DOI] [PubMed] [Google Scholar]
- 22.Hobartner C, Kreutz C, Flecker E, Ottenschlager E, Pils W, Grubmayr K, Micura R. The synthesis of 2′-O-[(triisopropylsilyl)oxy] methyl (TOM) phosphoramidites of methylated ribonucleosides (m(1)G, m(2)G, m(2)(2)G, m(1)I, m(3)U, m(4)C, m(6)A, m(2)(6)A) for use in automated RNA solid-phase synthesis. Monatsh. Chem. 2003;134:851–873. [Google Scholar]
- 23.Serebryany V, Beigelman L. An efficient preparation of protected ribonucleosides for phosphoramidite RNA synthesis. Tetrahedron Lett. 2002;43:1983–1985. [Google Scholar]
- 24.Saikia M, Dai Q, Decatur WA, Fournier MJ, Piccirilli JA, Pan T. A systematic, ligation-based approach to study RNA modifications. RNA. 2006;12:2025–2033. doi: 10.1261/rna.208906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnez JG, Steitz TA. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry. 1994;33:7560–7567. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- 26.Yarian CS, Basti MM, Cain RJ, Ansari G, Guenther RH, Sochacka E, Czerwinska G, Malkiewicz A, Agris PF. Structural and functional roles of the N1-and N3-protons of Psi at tRNA's position 39. Nucleic Acids Res. 1999;27:3543–3549. doi: 10.1093/nar/27.17.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auffinger P, Westhof E. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washington, DC: ASM Press; 1998. pp. 103–112. [Google Scholar]
- 28.Kool ET. Roles of Watson-Crick and minor groove hydrogen bonds in DNA replication. Cold Spring Harb. Symp. Quant. Biol. 2000;65:93–102. doi: 10.1101/sqb.2000.65.93. [DOI] [PubMed] [Google Scholar]
- 29.Heus HA, Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 30.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma X, Yang C, Alexandrov A, Grayhack EJ, Behm-Ansmant I, Yu YT. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J. 2005;24:2403–2413. doi: 10.1038/sj.emboj.7600718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.