Abstract
Bacterial DNA gyrase and topoisomerase IV are selective targets of fluoroquinolones. Topoisomerase IV versus gyrase and Gram-positive versus Gram-negative behavior was studied based on the different recognition of DNA sequences by topoisomerase–quinolone complexes. A careful statistical analysis of preferred bases was performed on a large number (>400) of cleavage sites. We found discrete preferred sequences that were similar when using different enzymes (i.e. gyrase and topoisomerase IV) from the same bacterial source, but in part diverse when employing enzymes from different origins (i.e. Escherichia coli and Streptococcus pneumoniae). Subsequent analysis on the wild-type and mutated consensus sequences showed that: (i) Gn/Cn-rich sequences at and around the cleavage site are hot spots for quinolone-mediated strand breaks, especially for E. coli topoisomerases: we elucidated positions required for quinolone and enzyme recognition; (ii) for S. pneumoniae enzymes only, A and T at positions −2 and +6 are discriminating cleavage determinants; (iii) symmetry of the target sequence is a key trait to promote cleavage and (iv) the consensus sequence adopts a heteronomous A/B conformation, which may trigger DNA processing by the enzyme–drug complex.
INTRODUCTION
Type II topoisomerases are ubiquitous ATP-dependent enzymes that catalyze the transport of one DNA segment through a transient double-stranded break in a second segment (1–3). Their role in the cell is to control the supercoiling of DNA and to untangle the catenanes that arise during replication or recombination (4–7). Bacteria express two type IIA topoisomerases, DNA gyrase (Gyr) and DNA topoisomerase IV (Topo IV) with distinct and unique functions (1,8). Gyr introduces negative supercoils into DNA, maintaining bacterial chromosomes in an underwound state to promote compaction and unwinding (9,10). In contrast, Topo IV is an efficient decatenase, separating tangled daughter chromosomes following replication (11–13). Both Gyr and Topo IV are heterotetramers, comprising two subunits each of GyrA and GyrB, and ParC and ParE, respectively (14). The GyrA and ParC subunits are homologous in their N-terminal domain that carries the DNA breakage-reunion function, but diverge in their C-terminal regions that also contribute to DNA binding (1). The GyrB and ParE proteins contain the ATPase site involved in energy transduction and are closely homologous, in both primary sequence and structure.
Transient double-stranded DNA breakage by Gyr and Topo IV proceeds via an enzyme–DNA intermediate termed the ‘cleavage complex’ involving a 4-bp staggered break and covalent attachment of GyrA or ParC subunits to each 5-prime (5′) DNA-end through a phosphotyrosine linkage (3,14). Protein–DNA linkage preserves the energy of the DNA phosphodiester bond and allows resealing of the DNA backbone by attack of the 3-prime (3′) OH-ends of the broken DNA. The breakage-reunion equilibrium is overwhelmingly in favor of the intact nucleic acid chain to avoid fortuitous lethal events in vivo. However, the cleavage complex can be stabilized by drugs such as quinolones producing DNA breaks, which can be detected after enzyme denaturation with sodium dodecyl sulfate. Generally, DNA cleavage is sequence-selective and is characterized by covalent bonding of the nucleic acid to GyrA or ParC (14–18). Identification of DNA breakage site specificity is important for the physiological functions of bacterial topoisomerases and for their roles as quinolone targets.
Cleavage determinants have been recently determined for Gyr and Topo IV from a Gram-positive bacterial species, Streptococcus pneumoniae, in the presence of various clinically relevant quinolones (19). The cleavage consensus sequences were GN4G(G/c)(A/c)G*GNNCtTN (C/a) for Gyr and G(G/c)(A/t)a*GNNCt(T/a)N(C/a) for Topo IV (where the asterisk denotes the cleavage site, capital letter indicates preferred base, lower case letter denotes unfavored base and N indicates no base preference). Similar cleavage preferences were observed among the quinolones tested. Even though the analysis was extremely comprehensive, some aspects remain to be answered (19): (i) The consensus sequence obtained with both enzymes is strikingly symmetric: does this property derive from actual enzyme requirements or does it arise from asymmetric sites as a consequence of analyzing just one strand of the double helix? (ii) Base prevalence was analyzed independently at each position: we do not know yet if the full consensus sequence is required for efficient DNA cleavage or if only part of it is necessary. (iii) If the entire consensus sequence, which spans a 12-base-long DNA tract, is needed for efficient DNA processing, this relatively long DNA segment should be statistically represented. Statistical analysis would hence benefit from using a more assorted DNA template repertoire.
For Gram-negative bacteria, site-specific DNA breakage details are much less exhaustive. Even for Escherichia coli topoisomerases, the data are scattered and discordant; for Gyr the most extensive analysis to date involved only 19 sites induced in E. coli on plasmid pBR322 and generated a 20-bp consensus (20–22); for Topo IV just one study has been performed, which mapped a weak 2-bp consensus (15).
This study aimed to address the following points: (i) investigation and comparison of the cleavage consensus sequence exhibited by topoisomerases IIA from Gram-positive (S. pneumoniae) and -negative species (E. coli) on a large selection of DNA templates; (ii) comparison of cleavage sites generated by Gyr and Topo IV enzymes; (iii) assessment of the minimal sequence requirements and (iv) symmetric/asymmetric properties for effective DNA recognition and scission by bacterial topoisomerases in the presence of quinolones.
We found discrete preferred sequences for each enzyme analyzed. In particular, consensus sequences were very similar when comparing different enzymes (i.e. Gyr and Topo IV) from the same bacterial source, but partly different using enzymes from diverse origins (i.e. E. coli and S. pneumoniae). The chemical nature of the fluoroquinolones used to stabilize the DNA/enzyme complex in each case did not play a significant role. We proved that symmetry was a crucial property of the recognized DNA sequence and that an extended Gn/Cn-rich sequence improved efficient DNA processing. Specific bases inside the 4-bp staggered break were required, possibly because of their interaction with quinolones, however, an important role in enzyme recognition was also played by bases outside the enzymatic cleavage site, Finally, we provided evidence that the peculiar consensus sequence suggested by our study adopts an unusual double-stranded DNA conformation which could trigger enzyme-DNA recognition.
MATERIALS AND METHODS
Chemicals and reagents
Ciprofloxacin and Gemifloxacin were provided by GlaxoSmithKline (Verona, Italy). Stock solutions were made in mQ-grade water and diluted to the working concentration in the desired buffer. Buffer components were purchased from Sigma-Aldrich (St. Louis, MO, USA). Electrophoretic reagents, dNTPs and Taq polymerase were from Amersham Biosciences Europe (Freiburg, Germany). [γ-32P]ATP was from Perkin Elmer (MA, USA); T4 polynucleotide kinase and DNA ligase were purchased from Invitrogen (Paisley, UK). Restriction enzymes were purchased from New England Biolabs (MA, USA).
Topoisomerase enzymes
Topo IV and Gyr from S. pneumoniae were purified as previously described (19,23,24). Topo IV and gyrase from E. coli were purified as described in (25,26).
DNAs
Plasmid pBluescript II KS(+) and SV40 DNA were purchased from MBI Fermentas (MD, USA) and Invitrogen (Paisley, UK), respectively. Primers were purchased from Eurogentec (Liege, Belgium). Primers for sequencing analysis used to amplify 239-bp PCR fragments on SV40 DNA template are described in (23). The amplified fragments encompass ∼27% of the complete SV40 sequence. Consensus oligos cloned in pBluescript II KS(+) are: AGGG SYM F: 5′-GCC GGG ATC CTT ACT TCA TGA ATT ATA GGG CCC TGA TAA TAC CTT AAG AAT TCG CGC-3′ and its complementary (AGGG SYM R); AGGG N-SYM F: 5′-GCC GGG ATC CTT ACT TCA TGA ATT ATA GGG TGT AGA TAA TAC CTT AAG AAT TCG CGC-3′ and its complementary (AGGG N-SYM R); GGAGGG F: 5′-GCC GGG ATC CTT ACT TCA TGA ATT GGA GGG CCC TCC TAA TAC CTT AAG AAT TCG CGC-3′ and its complementary (GGAGGG R); GGATGG F: 5′-GCC GGG ATC CTT ACT TCA TGA ATT GGA TGG CCA TCC TAA TAC CTT AAG AAT TCG CGC-3′ and its complementary (GGATGG R); GGAGTG F: 5′-GCC GGG ATC CTT ACT TCA TGA ATT GGA GTG CAC TCC TAA TAC CTT AAG AAT TCG CGC-3′ and its complementary (GGAGTG R); GGAGGT F: 5′-GCC GGG ATC CTT ACT TCA TGA ATT GGA GGT ACC TCC TAA TAC CTT AAG AAT TCG CGC-3′ and its complementary (GGAGGT R). Consensus sequences are underlined.
Primer sequences used to amplify a 215-bp PCR fragment on each p(consensus oligo) Bluescript II KS(+) DNA template are as follows: prBSF 5′-GTAA TACGA CTCACTATAGGGC-3′ and prBSR 5′-GACCA TGA TTACGCCAAGCG-3′. Consensus sequence oligos used for CD experiments are: reference: 5′-GGGGCCCC-3′; sample1: 5′-GGAGGGCCCTCC-3′; sample2: 5′-ATA GGGCCCTAT-3′; control: 5′-GCGACGCGTCGC-3′.
Cloning
All consensus sequence oligos (57 bp) contained BamHI and EcoRI restriction sites at their 5′- and 3′-end, respectively, for insertion into EcoRI- and BamHI-cut pBluescript II KS(+). Consensus oligos contained an additional AflII site flanking the 3′-end of BamHI site to facilitate screening of mutant colonies. Restricted oligos and plasmid were purified with Nucleotide Removal kit and Gel Extraction kit (Qiagen, CA, USA), respectively, and ligated at 16°C overnight. Ligation mixtures were transformed into competent E. coli DH5α cells. Bacteria were grown in the presence of IPTG and X-Gal, and white colonies were selected to confirm the presence of plasmid with the inserted oligo of interest [p(consensus oligo)Bluescript II KS(+)].
5′-end labeling of primers and PCR reaction
For primer labeling, 100 pmol of primer were incubated with 2 μl (10 μCi/μl) of [γ-32P]ATP and 10 units of T4 polynucleotide kinase in 50 mM Tris–HCl (pH 7.5), 7 mM MgCl2 and 10 mM DTT, at 37°C for 30 min. After incubation, DNA was purified through MicroSpin G-25 columns (Amersham Biosciences Europe). The labeled primers were used in PCR to amplify 239- or 215-bp fragments of SV40 or each p(consensus oligo)Bluescript II KS(+), respectively. Each PCR reaction was prepared by mixing 200 μM dNTPs, the labeled primer, 100 pmol of the cold primer, 50 ng of template DNA and 5 units of Taq polymerase in PCR reaction buffer [10 mM Tris–HCl (pH 9.0), 50 mM KCl and 1.5 mM MgCl2] to a final volume of 100 μl. PCR cycles for both SV40 and p(consensus oligo)Bluescript II KS(+) templates were 94°C for 30 s, 55°C for 30 s and 72°C for 30 s (30 cycles). DNA fragments were then purified with a QIAquick PCR purification kit (Qiagen, CA, USA).
Topoisomerase cleavage assays
Topo IV from S. pneumoniae was reconstituted with 0.45 μg ParC and 1.7 μg ParE in reaction buffer [40 mM Tris–HCl (pH 7.5), 6 mM MgCl2, 10 mM DTT, 200 mM potassium glutamate, 50 µg/ml bovine serum albumin (BSA)]. Reaction buffer for Gyr from S. pneumoniae (1U/μl) was 35 mM Tris–HCl (pH 7.5), 24 mM KCl, 4 mM MgCl2, 2 mM DTT, 6.5% glycerol, 0.1 mg/ml BSA. Topo IV from E. coli was diluted to a final concentration of 0.22 mg/ml ParC and 0.44 mg/ml ParE in dilution buffer [50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM DTT, 40% (w/v) glycerol, 1 mM EDTA]. Reaction buffer consisted of 40 mM Tris–HCl, pH 7.5, 20 mM KCl, 6 mM MgCl2, 1 mM DTT, 10 mM spermidine, 0.5 mg/ml BSA. Gyr from E. coli was diluted to 0.35 μM (0.1 U/μl) in dilution buffer [50 mM Tris–HCl (pH 7.5), 100 mM KCl, 2 mM DTT, 50% glycerol, 1 mM EDTA]. Reaction buffer consisted of 7 mM Tris–HCl (pH 7.5), 5 mM KCl, 0.8 mM MgCl2, 0.4 mM DTT, 1.3% (w/v) glycerol, 0.1 mg/ml BSA. One unit of Gyr is defined as the amount of enzyme that supercoils 0.5 μg of relaxed pBR322 DNA in 30 min at 37°C; one unit of Topo IV is the amount of enzyme that relaxes 0.4 μg of supercoiled pBR322 DNA in 30 min at 37°C.
For DNA cleavage assays, reconstituted topoisomerase enzymes were incubated with labeled PCR product DNA in 25 µl reaction mixtures in the presence of different drug concentrations. After a 30-min incubation at 37°C, 1 µl of 10% SDS and 2 µl of a 20 mg/ml stock of proteinase K were added, and incubation continued for 30 min at 45°C. Samples were precipitated with ethanol, resuspended in formamide gel loading buffer [95% formamide, 200 mM EDTA pH 8.0, 0.1% (w/v) xylene cyanol and 0.1% (w/v) bromophenol blue] and heated at 90°C for 2 min. Cleavage products were resolved in 8% polyacrylamide, 7 M urea gels, alongside DNA markers obtained with the dideoxynucleotide chain termination protocol. Briefly, G, C, A and T dideoxy-nucleotide ladders were obtained by incubating 1 μl of labeled primer, 4 μl of ddNTP/dNTPs extension/termination mixture (27), 50 ng template DNA, 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris–HCl (pH 7.5), 5 U Taq DNA polymerase in a volume of 10 μl at 95°C for 30 s, 54°C for 30 s, 72°C for 1 min for 25 cycles and at 95°C for 30 s, 72°C for 2 min for 10 cycles. Reactions were purified by ethanol precipitation. Gels were then transferred to Whatman 3MM filter paper, dried under vacuum at 80°C, and bands were visualized by phosphorimaging analysis (Molecular Dynamics, Amersham Biosciences Europe). Quantification was performed by ImageQuant software (Molecular Dynamics). Each cleavage site was quantified as the ratio of the intensity of the cleavage band over the intensity of the corresponding band/position in the control lane (enzyme without drug) (intensity ratio = IR). Only sites with IR > 40 were considered in the statistical analysis.
Statistical tests
The probability (P) of deviation from expectation of each nucleotide at each position around the cleavage site (positions −20/+20) was calculated as previously described in (23,28).
Circular dichroism
Reference, sample1, sample2 and control oligos were dissolved in 10 mM Na.Cacodylate (29) to a final concentration of 90 μM per residue. Circular dichroism spectra from 220 to 320 nm were recorded at 4°C using 10 mm path length cells on a Jasco J810 spectropolarimeter equipped with a NESLAB temperature controller and interfaced to a PC100. Observed ellipticities were converted to mean residue ellipticity [θ] = deg cm2dmol−1 (Molar Ellipticity).
RESULTS
Statistical analysis of DNA cleavage sites induced by quinolone–prokaryotic topoisomerase complexes reveals discrete consensus sequences
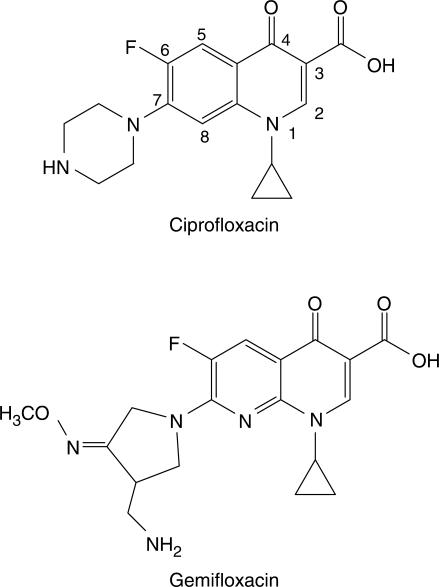
To analyze and compare DNA sequence determinants of fluoroquinolones and prokaryotic topoisomerase complexes, we used (i) two clinically approved fluoroquinolones, i.e. ciprofloxacin and gemifloxacin (Figure 1). Ciprofloxacin, a second-generation fluoroquinolone, was used as a reference compound with assessed activity against Gram-negative and -positive bacteria; gemifloxacin was included as a new IIIb generation fluoroquinolone with increased activity against Gram-positive species. (ii) Both Gyr and Topo IV from E. coli and S. pneumoniae were employed as representative of Gram-negative and -positive species, respectively. (iii) SV40 DNA was used as a generic nucleic acid template to avoid bias in species-specific enzyme/DNA recognition.
Figure 1.
Chemical structures of ciprofloxacin and gemifloxacin. Numbering of the fluoroquinolone ring system is shown for ciprofloxacin.
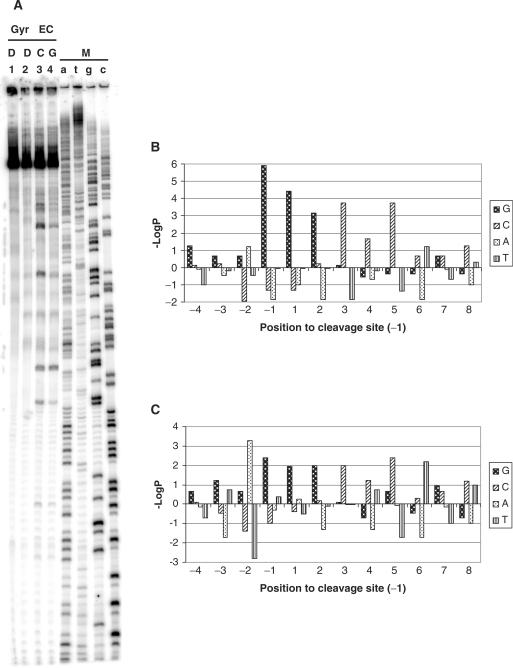
Six 239-bp long SV40 segments were 5′-end 32P-labeled and amplified by PCR reaction to obtain significant coverage of the whole SV40 sequence and processing of a total of 1434 bp. The forward or reverse strand were labeled and analyzed independently to gain information on the symmetry of enzyme-mediated DNA cleavage. All DNAs were incubated with E. coli- or S. pneumoniae-derived Gyr or Topo IV in the presence of ciprofloxacin or gemifloxacin, and the cleavage patterns were analyzed by loading reaction mixtures on denaturing urea-polyacrylamide gels to separate DNA fragments (Figure 2A). For each enzyme/drug combination, we were able to collect an adequate amount of cleavage sites (i.e. around 30–50 per each strand). Site number was similar between ciprofloxacin and gemifloxacin with E. coli enzymes, while it was in favor of gemifloxacin with S. pneumoniae proteins. Sequences of cleavage sites were aligned at the point of the phosphodiester bond break in the 5′ → 3′ orientation, and bases immediately 5′ and 3′ to the cut were numbered −1 and +1, respectively.
Figure 2.
Sequence analysis of fluoroquinolone-stimulated DNA cleavage by Topo IV and Gyr from E. coli and S. pneumoniae. (A) Example of sequencing gel. A 239-bp SV40 DNA fragment, labeled with 32P at one 5′ end, was incubated with E. coli Gyr (GyrEC) and fluoroquinolones. reaction mixtures were loaded and run on a denaturing urea–8% polyacrylamide gel. DNA fragments were visualized by phosphorimaging. Lanes 3 and 4 are treated with ciprofloxacin (C) (25 μM) and gemifloxacin (G) (25 μM), respectively. Lanes 1 and 2 are controls for non-treated DNA and DNA incubated with the enzyme without drugs. Lanes a, c, g, t are marker lanes (M) obtained by the dideoxynucleotide chain termination method. (B and C) Probabilities of the observed base frequency deviations from expectation at positions −4/+8 observed (B) for Topo IV from E. coli and ciprofloxacin-mediated DNA cleavage or (C) for Gyr from S. pneumoniae and gemifloxacin-mediated DNA cleavage. Position –1 indicates the base 5′ of the cleaved phosphodiester bond identified in sequencing gels. In the ordinate, P is the probability of observing that deviation or more, as either an excess (above base line) or deficit (below base line) relative to the expected frequency of each base. The expected frequencies were calculated on 416 sites using the overall base frequency of SV40 DNA (G = C = 20.4%; A = T = 29.6%).
The deviation of base distribution from the expected SV40 DNA base frequencies was evaluated at each position (±20 from the cleavage site) by χ2 analysis: a core region of non-random base composition was found between positions −4 and +8. Probabilities of the observed base frequency deviations from expectation at each position were assessed for all cleavage sites on the forward strand, and non-complementary sites on the reverse strand, and vice versa, for each enzyme/drug combination (statistical analysis for Topo IV/ciprofloxacin from E. coli and Gyr/gemifloxacin from S. pneumoniae, positions −4/+8, are reported in Figure 2B and C, as examples). The complete list of favored and disfavored bases (−LogP > 2 and −LogP < −2, respectively) is reported in Table 1.
Table 1.
Summary of favored and non-favored bases obtained by statistical analysis of cleavage sites with Topo IV and Gyr from E. coli (Ec) and S. pneumoniae (Sp) and ciprofloxacin (Cipro) or Gemifloxacin (Gemi) quinolones on DNA alternatively labeled on the forward (F) or reverse (R) strand
| Species | Enzyme | Drug | DNA strand | Position with respect to cleavage site (−1) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −4 | −3 | −2 | −1 | +1 | +2 | +3 | +4 | +5 | +6 | +7 | +8 | ||||
| Ec | Topo IV | Cipro | F | Gc | G | G | C | C | |||||||
| R | G | Ga | C | Cg | Ct | ||||||||||
| Ec | Topo IV | Gemi | F | G | G | Ga | Ga | Ct | Ct | ||||||
| R | G | G | G | G | Ga | C | C | C | g | a | |||||
| Ec | Gyr | Cipro | F | G | G | G | C | ||||||||
| R | G | Gt | C | C | |||||||||||
| Ec | Gyr | Gemi | F | G | G | C | |||||||||
| R | G | C | C | ||||||||||||
| Sp | Topo IV | Cipro | F | A | A | A | G | C | C | C | Ta | C | |||
| R | A | G | C | Ta | |||||||||||
| Sp | Gyr | Gemi | F | At | G | G | G | C | C | T | |||||
| R | At | G | C | C | C | T | |||||||||
Preferred and non-preferred bases were selected based on their –LogP value (>2 and <−2, respectively). Preferred bases are shown in capital letters and non-preferred bases are shown in lowercase letters.
For the E. coli enzymes, a strong and consistent preference was observed for positions −1G, +1G, +2G, +3C, +5C (Table 1). Position +4 also exhibited a consistent preference for C (6 out of 8 enzyme/drug/DNA strand combinations), but with a slightly lower statistical significance (1.5 < −LogP < 2). Positions −2/−4 and +6/+8 indicated a very mild preference for G and C, respectively (0.5 < −LogP < 1.5). No essential cleavage differences were found between ciprofloxacin and gemifloxacin. As for the two enzymes, the requirements for Topo IV appeared to be somewhat more stringent than for Gyr, especially for positions −1 and +5. Positions outside the −4/+8 interval did not show distinct preferences. Disfavored bases were much less regularly repeated: inside the 4-bp staggered break only position +2A showed an indication of non-preference (Table 1).
A thorough statistical analysis for the S. pneumoniae enzymes has already been reported by Leo et al. (19). Nonetheless, we performed cleavage analysis on SV40 DNA substrate to: (i) compare results obtained with S. pneumoniae and E. coli enzymes using the same drugs and DNA template, (ii) compare results with the same enzymes on different DNA templates [i.e. SV40 and S. pneumoniae genome (19)] and (iii) address the issue of consensus sequence symmetry by analyzing both cut DNA strands. Only one drug was tested with each enzyme (ciprofloxacin with Topo IV and gemifloxacin with Gyr) because it had already been shown that fluoroquinolone identity does not play a critical role in DNA recognition (19). In accordance with the previous analysis (which used −LogP > 3 and −LogP < −3 as significant), we found that highly preferred bases for both Topo IV and Gyr were −2A and +6T; +1G and +4C, and −1G (gyrase only) were also significantly represented. In addition, +3C and +5C were favored (at −LogP > 2) (Table 1). No steady preference was observed for other positions, even though positions −4, −3 and +7, +8 showed a mild inclination for G and C, respectively (0.5 < −LogP < 1.5). Non-preference data were again less stringent: consistency and significant –LogP values (−LogP < −2) were found just for –2T and +6A (Table 1).
By collecting data on both forward and reverse DNA strands, we were able to differentiate complementary and non-complementary sites. Analysis of just non-complementary sites should clarify if DNA symmetry is a requirement for enzyme recognition. Indeed, symmetry was maintained for most positions in this set of data (data not shown). In particular, the most symmetrically conserved positions were −1 and +5, and +2 and +3.
Symmetry is required for efficient cleavage at the DNA consensus sequence
Our statistical analysis indicated a clear-cut consensus sequence that was in part different between E. coli- and S. pneumoniae-derived enzymes. In particular, GGGCCC sequence at the −1/+5 interval was obtained with the Gram-negative enzymes, with enhanced preference for positions −1 and +5, +2 and +3. On the opposite, the most preferred bases with S. pneumoniae proteins were −2A and +6T. Positions –1/+5 were GGGCCC as with E. coli topoisomerases, but this preference was less statistically represented. In neither case, a major difference was noticed between Topo IV and Gyr or between ciprofloxacin and gemifloxacin.
Since these data were obtained independently at each position, we did not know if the full consensus sequence or any other base combination was in fact required for efficient topoisomerase/fluoroquinolone DNA processing. Further, based on the indication that symmetry afforded efficient DNA cleavage even at non-complementary sites, we wanted to verify to what extent this condition affected enzyme/drug activity on the nucleic acid.
To answer these questions, we focused on the E. coli enzymes and their preferred sequences: we cloned the −1/+5 GGGCCC consensus sequence into a plasmid vector and subsequently amplified a 215-bp DNA fragment by PCR. Sequences upstream (−8/−2) and downstream (+6/+13) the consensus sequence were arbitrary chosen to be A/T rich (see Materials and Methods section). A 215-base long nucleic acid was employed to achieve optimal topoisomerase processivity: in fact DNA Gyr is reported to protect as much as 150 bp on binding to DNA (16,17).
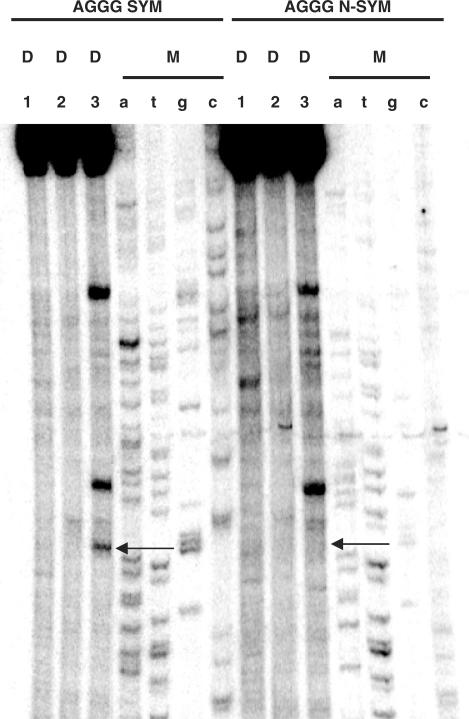
To test for symmetry, a non-symmetric sequence was assayed, which maintained only positions −1, +1 and +2 with reference to the established consensus sequence (i.e. GGGTGT).
Cleavage analysis was performed on the symmetric and non-symmetric sequences in the presence of E. coli Topo IV or Gyr and ciprofloxacin or gemifloxacin. Data for the E. coli Topo IV/ciprofloxacin combination are shown in Figure 3. Cleavage was indeed obtained corresponding to −1G of the inserted consensus sequences in the symmetric oligonucleotide (lane 3, AGGG SYM oligo, Figure 3), but this was relatively mild compared to cleavage sites on other portions of the tested sequence. In contrast, no cleavage was observed at −1G in the non-symmetric consensus sequence (lane 3, AGGG N-SYM, Figure 3). It should be noted that all cleavage bands were shared between the symmetric and non-symmetric DNA. Similar results were gained with the remaining enzyme/drug patterns (data not shown).
Figure 3.
Sequence analysis of topoisomerase/fluoroquinolone-mediated DNA cleavage on the symmetric (AGGGCCCT) and non-symmetric (AGGGTGTA) consensus sequences. A 215 bp DNA fragment containing the consensus sequences of interest was labeled with 32P at one 5′ end, and incubated with Topo IV from E. coli in the presence of ciprofloxacin (C). Reaction mixtures were loaded and run on a denaturing urea–8% polyacrylamide gel. Lanes 3 are treated with ciprofloxacin, respectively. Lanes 1 are controls for non-treated DNA; lanes 2 are controls for DNA incubated with the enzyme without drugs. Lanes a, t, g and c are marker lanes obtained by the dideoxynucleotide chain termination method.
Determinants of quinolone-induced E. coli topoisomerase-mediated DNA cleavage
While it is now clear that symmetry is crucial to improve topoisomerase/fluoroquinolone-mediated DNA cleavage, the GGGCCC consensus sequence did not stand out as a hot spot for induced DNA scission. The consensus sequence found in the present study was limited to positions between −1 and +5, and the region just outside this interval (i.e. −8/−2 and +6/+13) was arbitrary chosen to be extremely A/T rich (14 out of 15 bases were A or T, see Materials and Methods section) in the DNA oligonucleotide used for the cleavage assay. We hypothesized that these flanking non-random regions could decrease the cleavage efficiency of the consensus sequence. In fact, our current statistical analysis on both E. coli and S. pneumoniae topoisomerases showed a preference for −4G, −3G, −2A/G, +6T/C, +7C, +8C, which was not considered before because at a low statistical significance (i.e. −LogP < 2) (Figure 2B and C). In addition, it had previously been demonstrated that S. pneumoniae enzymes tested on a different DNA template (19), preferred G/C-rich tracts outside the cleavage site (positions −4, −3 and +8). We hence introduced this additional information in our current consensus sequence, obtaining the GGAGGGCCCTCC DNA stretch (named GGAGGG oligo) spanning positions −4/+8.
Further, to investigate the relative importance of each position from −1 to +2 (and symmetric bases accordingly) we systematically and independently substituted G with T from −1 to +1 (and consequently C with A from +3 to +5), obtaining the following modified consensus sequences: GGATGGCCATCC (named GGATGG), GGAGTGCACTCC (named GGAGTG), GGAGGTA CCTCC (named GGAGGT).
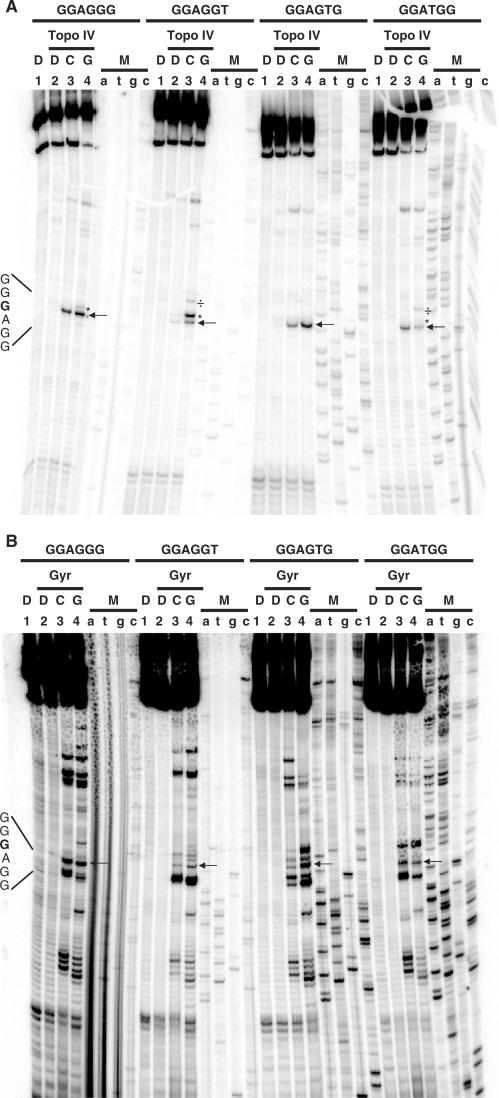
The new four extended and modified consensus oligonucleotides were inserted into plasmid vectors, amplified to obtain 215 bp DNA tracts and processed as described above. It should be noted that DNA tracts outside positions −4 and +8 remained unchanged compared to the previous AGGG SYM oligo. Cleavage experiments with E. coli Topo IV and Gyr are shown in Figure 4A and B, respectively.
Figure 4.
Sequence analysis of topoisomerase/fluoroquinolone-mediated DNA cleavage on the extended and modified symmetric consensus sequences (GGA(GorT)(GorT)(GorT)(CorA)(CorA)(CorA) TCC). A 215-bp DNA fragment containing the consensus sequences of interest was labeled with 32P at one 5′ end, and incubated with Topo IV (A) or Gyr (B) from E. coli in the presence of ciprofloxacin (C) or gemifloxacin (G). Reaction mixtures were loaded and run on a denaturing urea–8% polyacrylamide gel. Lanes 3 and 4 are treated with ciprofloxacin and gemifloxacin, respectively. Lanes 1 are controls for non-treated DNA; lanes 2 and 5 are controls for DNA incubated with the indicated enzyme without drugs. Lanes a, t, g and c are marker lanes obtained by the dideoxynucleotide chain termination method. Asterisk and division symbols indicate additional cleavage sites around position −1.
In the presence of the new oligo substrates, unique and extremely strong cleavage bands corresponding to position −1G of the consensus sequences were produced by Topo IV and fluoroquinolones (lanes 3 and 4, Figure 4A), indicating that A/T at positions −4, −3 and +7, +8 in fact decrease DNA processing. In particular, the −1G cut site was the most intense with both fluoroquinolones in the case of the wild-type consensus-derived oligo. In the mutated sequences, gemifloxacin-induced cleavage was more prominent with GGAGGT and GGAGTG sequences, while ciprofloxacin was more efficient with the GGATGG oligo (compare lanes 3 and 4, all oligo, Figure 4A). Gemifloxacin induced an extra band at position +2 in oligos GGAGGG, GGAGGT and GGATGG, respectively (lanes 4, * symbol, Figure 4A). This cleavage site was particularly intense in the case of oligo GGAGGT, where T, instead of G, is present at position +2. An additional band was produced by gemifloxacin at position +5T in oligos GGAGGT and GGATGG, respectively (lanes 4, ÷ symbol, Figure 4A). A few minor sites were present towards the DNA 3′-end, which were shared by all oligos.
When employing E. coli Gyr (Figure 4B), the results were quite different. Most strikingly, while cutting at position −1G in the consensus sequence was still present; this was not the most intense band along the DNA sequence. As observed for Topo IV, the relative intensity of strand scission at position −1 was higher in the wild-type oligo (GGAGGG) compared to the mutant sequences (lanes 3 and 4, Figure 4B). Ciprofloxacin-induced cleavage was stronger than gemifloxacin-mediated breakage in the case of oligo GGAGGG and GGATGG, while the reverse was found for oligos GGAGGT and GGAGTG. Additional bands were found both inside (downstream position −1) and outside the consensus sequence (both upstream and downstream). The wild-type oligo showed fewer cleavage bands inside the consensus sequence.
The GGAGGGCCCTCC sequence adopts an A-like guanine–guanine stacking in solution
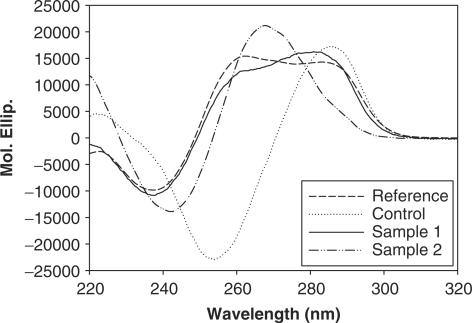
Next, we wondered if the above described peculiar consensus sequence could adopt a definite structure in solution. It is reported that the DNA octamer GGGGCCCC forms in aqueous solution a stable heteronomous DNA duplex where guanine–guanine stacking is A-like, whereas cytosine bases stack in a B-like fashion (30). To examine the actual conformation of the consensus sequence primarily recognized by E. coli Topo IV (Figure 4A), we compared the CD spectrum of our sample1 oligo (GGAGGGCCCTCC), with that of the reference oligo (GGGGCCCC) and of a control DNA (GCGACGCGTCGC), which shares the same base composition of the sample1 oligo, but displays a different sequence. In addition, sample1 oligo was compared to sample2 oligo (ATAGGGCCCTAT) where flanking regions were A/T rich such as those used in the cleavage assay of Figure 3. Under native conditions (T well below Tm, i.e. 4°C), the spectrum of the reference oligo exhibited two strong overlapping positive bands at 285 nm and at 260 nm, and a negative peak at 237 nm (Figure 5). The CD of our sample1 oligo was essentially superimposable to the reference oligo, showing the same bands with very similar intensities. Conversely, the control and sample2 oligos displayed substantially different CD patterns with one positive band at 285 or 265 nm, respectively, and a negative peak at 255 or 240 nm, respectively. We conclude that the native consensus sequence adopts in solution a peculiar conformation, which closely resembles the heteronomous conformation of the reference oligo [i.e. a combination of B-type and A-type base stacking (30)], whereas the control sequence is organized in a more classical B form. Sample2 oligo, on turn, exhibits a spectrum which reflects a lower propensity to adopt the heteronomous conformation thus favoring a B form. This is in agreement with the shift of the CD cross-over towards longer wavelength as an indication of lower A-like character and with the crystal structure of the dodecamer duplex CATGGGCCCATG showing a structural continuum along the transition between A- and B-DNA (31,32).
Figure 5.
CD spectra of sample1 oligo GGAGGGCCCTCC, sample2 oligo ATAGGGCCCTAT, reference oligo GGGGCCCCC and control oligo GCGACGCGTCGC measured at 4°C. Observed ellipticities were converted to mean residue ellipticity [θ] = deg cm2 dmol−1 (Mol. Ellip.).
DISCUSSION
Type IIA DNA topoisomerases have concerted and complementary functions in controlling bacterial chromosome topology: Gyr catalyzes the negative supercoiling of closed-circular DNA, thus, it removes the positive supercoils that arise during DNA unwinding; Topo IV relaxes positive and negative supercoils, and unlinks daughter chromosomes at cell division (33). In addition, topoisomerases are crucial targets for fluoroquinolone-based therapy. In this instance, Gyr or Topo IV can be alternative selective targets depending on the species of origin: Gyr is often the primary target in Gram-negative species, such as E. coli, while Topo IV is principally affected in Gram-positive species, such as S. pneumoniae (14). However, it has also been reported that topo IV and/or gyrase can be alternative intracellular target depending on quinolone structure (34,35).
To investigate whether this double biased behavior (Gyr versus Topo IV and S. pneumoniae versus E. coli) could arise from a distinctively specific recognition of DNA sequences by topoisomerases (± quinolones), we comprehensively studied and compared the DNA sequence specificity of Gyr and Topo IV from E. coli and S. pneumoniae in the presence of clinically used fluoroquinolones. In view of the key role played by these proteins, the resulting information would be important from both physiological and therapeutic standpoints.
A consensus sequence for the two topoisomerases from S. pneumoniae has been recently reported using S. pneumoniae ParE-ParC gene sequences as DNA templates (19). Here, to impartially compare DNA sequence specificity of enzymes from different species, we used non-specific DNA fragments derived from the SV40 genome.
By statistical analysis of preferred bases around the cleavage position (−20/+20) of a large set of sites (over 400), we found that the GGGCCC sequence at positions −1/+5 was favored with both Gyr and Topo IV from the two bacterial species considered. However, this preference was much more prominent for the E. coli enzymes. Conversely, positions −2 and +6 were non-randomly selected only with S. pneumoniae enzymes, which revealed a striking preference for A and T, respectively.
Overall, statistical analysis of consensus sequences reported no major differences between Topo IV and Gyr, although the requirements for the latter appear to be less stringent in the Gram-negative species. By contrast, discrimination of Gram-positive and -negative enzymes apparently resides on different DNA sequence requirements for efficient cleavage, in particular at positions −2 and +6.
The high structural similarity of the N-terminal binding regions of ParC and GyrA from both S. pneumoniae and E. coli species (36,37) may explain the comparable DNA recognition properties shared by the tested topoisomerases in the presence of the two quinolone drugs. As an added proof, the aminoacidic residues reported to be essential in the protein catalytic activity and implicated in quinolone resistance, hence, likely involved in the concurrent binding of topoisomerases to DNA and quinolones in the cleavage complex (i.e. Arg32, Arg47, His78, His80, Ser83 and Asp/Glu87 in E. coli GyrA and corresponding positions in the other topoisomerases) (38–40), are identical in the four tested enzymes. It is intriguing to note that the only consistent difference in the aminoacidic sequence between E. coli and S. pnemoniae N-terminal region of GyrA/ParC subunits is at position 84 of E. coli GyrA (and corresponding positions in the other enzymes), where an Ala is substituted with a Ser in the Gram-positive proteins. Since position 84 is just adjacent to other positions involved in enzyme/quinolone/DNA interactions (see above), it could be speculated that the introduction of an H-bond donor site (i.e. Ser) modulates S. pneumoniae topoisomerases DNA recognition properties, driving sequence specificity towards A/T at positions −2/+6.
The fluoroquinolone chemical nature did not play a major role in determining the consensus sequence, at least for the tested compounds. However, it influenced the extent of cleavage stimulation. In particular, sites number was similar between ciprofloxacin and gemifloxacin with E. coli enzymes, while it was clearly in favor of gemifloxacin with S. pneumoniae proteins. These results are in line with the reported activity of the two drugs on bacterial species: gemifloxacin is more effective than ciprofloxacin on S. pneumoniae strains, while the activity of the two drugs is similar on E. coli bacteria (41,42). Therefore, our data indicate that, from a pharmacodynamic point of view, the biased activity of the two drugs may rest on their different ability to poison the molecular target, rather than on recognizing a different sequence determinant. This implies that DNA recognition is initially driven by the topoisomerases and then modulated by quinolone stabilization of the cleavage complex.
Our data, performed on SV40 DNA template, differ slightly from those obtained previously with the S. pneumoniae genome DNA template (19). In the first report, the consensus found between positions −4 and +8 were GGAN*GNNCNTNC and GGAG*GNNCNTNC, asterisk at the cleavage site, for Topo IV and Gyr, respectively (19), compared to NNAN*GNCCCTNN and NNAG*GNCCCTNN, respectively, in the present work. Noteworthy, several preferences match exactly, whereas others are not observed in one case or the other. This difference depends in part on the level of statistical stringency considered: on lowering the −LogP threshold, +2G and +3C bases become preferred in (19) and −4G, −3G and +8C register as slightly preferred with both Gram-positive and -negative enzymes in this work (Figure 2B and C). The combination of different DNA template, enzyme activity and drug concentration could be responsible for the diverse statistical relevance found. In fact, DNA traits consisting of two or more bases can be differently represented within nucleic acids from various origins. For instance, even though SV40 and ParC/ParE DNAs share similar base composition (G/C percentage is 40% in SV40 and 42% in S. pneumoniae), GGG and CCC sequences are 27% more frequent in the SV40 genome than in the ParC/ParE genes.
Since each position is independently taken into account during statistical analysis, we next analyzed E. coli topoisomerase–quinolone cleavage efficiency on the full DNA consensus sequences found in this work or obtained by combining data from the present and previous research (19). Further, mutations at selected preferred positions were also introduced to determine the contribution of each site on the cleavage complex DNA processivity.
We found that the −2/+6 consensus sequence AGGGCCCT environed in G/C-, but not A/T-, rich flanking regions, is able to induce highly efficient DNA cleavage. In fact, the 8-nt long tract AGGGCCCT at positions −2/+6, environed in an A/T-rich region, was able to promote only weak DNA damage (Figure 3). In this instance, cleavage intensity was similar among the two enzymes and the two quinolones employed. In contrast, addition of G and C bases at positions −4, −3 and +7, +8, respectively (19), significantly augmented cleavage, especially with Topo IV (see Figure 4A and B). The presence of the full consensus sequence was still required: in fact, modification of a single G nucleotide in the −1/+5 region generally decreased cleavage, in the same manner for Topo IV and Gyr. In addition, whereas the intensity of cleavage with the wild-type sequence was similar between ciprofloxacin and gemifloxacin, differences in cutting efficiency were observed at the mutated sequences with the two fluoroquinolones. In particular, positions inside the 4-base staggered break seem primarily implicated in quinolone binding. In fact, changes at positions +2, +3 and +1, +4 more dramatically impaired ciprofloxacin activity, whereas gemifloxacin was more severely affected by nucleotide changes at +1 and +5. Therefore, gemifloxacin requirements seem to be somewhat less stringent than those of ciprofloxacin when modifying the quinolone-interacting region of the consensus sequence. These results are also supported by reports showing an increased binding ability of quinolones to Gn/Cn-rich sequences (43) and site-specific analysis of E. coli Gyr with oxolinic acid (21).
Our data further suggest that positions −4, −3 and +7, +8, which must be G/C rich, are likely required for efficient DNA recognition and binding by the topoisomerases, since these positions significantly affect cutting intensity depending upon the enzyme, but not upon the drug. The consensus found is particularly effective for Topo IV, less so in the case of gyrase, for which additional features outside positions −4/+8 might play a key role in specificity. Positions −2 and +6 are also probably involved in enzyme recognition. In particular, they help discriminate between proteins from different bacterial sources (e.g. S. pneumoniae versus E. coli), but not between the two topoisomerases (see above).
Symmetry is an additional feature required for efficient DNA processing, as proven by analyzing cleavage sites on the two complementary DNA strands and by testing symmetric and non-symmetric consensus sequences. This property reasonably arises from the dyadic symmetry of topoisomerase complexes.
The consensus primary sequence found is very peculiar. It is known that selected nucleic acid sequences can confer unusual 3D characteristics to the DNA (29,44). Indeed, it is reported that Gn/Cn DNA sequences can be crystallized in A/B intermediate conformations (32). More interestingly, the GGGGCCCC sequence adopts a partial A-like structure in aqueous solution. In particular, G bases are stacked in a DNA-A form, whereas the Cs maintain a canonical B-form (30). Surprisingly, in the reverse sequence CCCCGGGG, both C and G bases fold in a different A-like and less stable fashion (45). By circular dichroism analysis, we proved that our consensus sequence GGAGGGCCCTCC folds as the reference GGGGCCCC DNA. A partial A-like conformation is likely to protect G/C-rich regions, which are involved in regulating transcription of many human genes (46), against mutations: in fact these are mutated much less than A/T-rich regions (47). According to our findings, this particular A/B DNA conformation could also prompt DNA recognition by topoisomerases. It should be noted that the reverse sequence CCCCGGGG, which folds up in a full-A-like structure and hence presents a different environment for enzyme recognition, was not selected by statistical analysis of cleavage sites. Topoisomerases would likely recognize −4, −3, −2 and +6, +7, +8 nt, permitting cleavage at −1 and +5 positions. Quinolones would then be able to bind between the two loose ends with subsequent inhibition of the enzymatic DNA resealing.
Finally, our DNA specificity results using the test quinolones suggest that the therapeutic efficacy against a bacterial species should be related to the frequency of occurrence of Gn/Cn-rich sequences in the bacterial genome and to the efficiency of cleavage complex stabilization by the drug. Clearly, the observed clinical outcome is also expected to depend upon the ADME properties of each individual quinolone.
In summary, this work provided evidence that (i) a symmetric Gn/Cn consensus sequence at positions −1/+5 allows efficient bacterial topoisomerase-mediated DNA processing; (ii) G/C over A/T-rich flanking sequences (positions −4, −3 and +7, +8) are highly preferred in protein DNA processing, which is particularly efficient for Topo IV; (iii) positions −2 and +6 interact with the enzyme and distinguish S. pneumoniae from E. coli topoisomerases; (iv) positions −1 to +5 participate to drug activity and may modulate cleavage intensity based on the quinolone chemical nature; (v) symmetry of the target sequence is a key trait to promote effective cleavage and (vi) the consensus sequence adopts a heteronomous A/B conformation, which may trigger efficient DNA processing by enzyme/drug complex.
ACKNOWLEDGEMENTS
This work was carried out with the financial support of the Italian Ministry for University and Research (MIUR), Rome, Italy and EEC grant #503466. Work in the Fisher laboratory was supported by a project grant from the BBSRC (UK). Work in the Maxwell laboratory was funded by BBSRC (UK). We thank Dr Claudia Sissi for helpful discussion. Funding to pay the Open Access publication charges for this article was provided by the above EEC grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 2.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 3.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 4.Liu LF, Liu CC, Alberts BM. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. 1980;19:697–707. doi: 10.1016/s0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- 5.Roca J, Wang JC. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell. 1992;71:833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 6.Roca J, Wang JC. The probabilities of supercoil removal and decatenation by yeast DNA topoisomerase II. Genes Cells. 1996;1:17–27. doi: 10.1046/j.1365-2443.1996.01001.x. [DOI] [PubMed] [Google Scholar]
- 7.Adams DE, Shekhtman EM, Zechiedrich EL, Schmid MB, Cozzarelli NR. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 8.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 9.Holmes VF, Cozzarelli NR. Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc. Natl Acad. Sci. USA. 2000;97:1322–1324. doi: 10.1073/pnas.040576797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vologodskii AV, Cozzarelli NR. Conformational and thermodynamic properties of supercoiled DNA. Annu. Rev. Biophys. Biomol. Struct. 1994;23:609–643. doi: 10.1146/annurev.bb.23.060194.003141. [DOI] [PubMed] [Google Scholar]
- 11.Zechiedrich EL, Khodursky AB, Cozzarelli NR. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev. 1997;11:2580–2592. doi: 10.1101/gad.11.19.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 13.Deibler RW, Rahmati S, Zechiedrich EL. Topoisomerase IV, alone, unknots DNA in E. coli. Genes Dev. 2001;15:748–761. doi: 10.1101/gad.872301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marians KJ, Hiasa H. Mechanism of quinolone action. A drug-induced structural perturbation of the DNA precedes strand cleavage by topoisomerase IV. J. Biol. Chem. 1997;272:9401–9409. doi: 10.1074/jbc.272.14.9401. [DOI] [PubMed] [Google Scholar]
- 16.Fisher LM, Mizuuchi K, O'Dea MH, Ohmori H, Gellert M. Site-specific interaction of DNA gyrase with DNA. Proc. Natl Acad. Sci. USA. 1981;78:4165–4169. doi: 10.1073/pnas.78.7.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirkegaard K, Wang JC. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell. 1981;23:721–729. doi: 10.1016/0092-8674(81)90435-9. [DOI] [PubMed] [Google Scholar]
- 18.Morrison A, Cozzarelli NR. Contacts between DNA gyrase and its binding site on DNA: features of symmetry and asymmetry revealed by protection from nucleases. Proc. Natl Acad. Sci. USA. 1981;78:1416–1420. doi: 10.1073/pnas.78.3.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leo E, Gould KA, Pan XS, Capranico G, Sanderson MR, Palumbo M, Fisher LM. Novel symmetric and asymmetric DNA scission determinants for Streptococcus pneumoniae topoisomerase IV and gyrase are clustered at the DNA breakage site. J. Biol. Chem. 2005;280:14252–14263. doi: 10.1074/jbc.M500156200. [DOI] [PubMed] [Google Scholar]
- 20.Oram M, Travers AA, Howells AJ, Maxwell A, Pato ML. Dissection of the bacteriophage Mu strong gyrase site (SGS): significance of the SGS right arm in Mu biology and DNA gyrase mechanism. J. Bacteriol. 2006;188:619–632. doi: 10.1128/JB.188.2.619-632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher LM, Barot HA, Cullen ME. DNA gyrase complex with DNA: determinants for site-specific DNA breakage. EMBO J. 1986;5:1411–1418. doi: 10.1002/j.1460-2075.1986.tb04375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lockshon D, Morris DR. Sites of reaction of Escherichia coli DNA gyrase on pBR322 in vivo as revealed by oxolinic acid-induced plasmid linearization. J. Mol. Biol. 1985;181:63–74. doi: 10.1016/0022-2836(85)90324-9. [DOI] [PubMed] [Google Scholar]
- 23.Richter SN, Leo E, Giaretta G, Gatto B, Fisher LM, Palumbo M. Clerocidin interacts with the cleavage complex of Streptococcus pneumoniae topoisomerase IV to induce selective irreversible DNA damage. Nucleic Acids Res. 2006;34:1982–1991. doi: 10.1093/nar/gkl127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan XS, Fisher LM. Streptococcus pneumoniae DNA gyrase and topoisomerase IV: overexpression, purification, and differential inhibition by fluoroquinolones. Antimicrob. Agents Chemother. 1999;43:1129–1136. doi: 10.1128/aac.43.5.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng H, Marians KJ. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 26.Maxwell A, Howells AJ. Overexpression and purification of bacterial DNA gyrase. Methods Mol. Biol. 1999;94:135–144. doi: 10.1385/1-59259-259-7:135. [DOI] [PubMed] [Google Scholar]
- 27.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: a Laboratory Manual. New York: Cold Spring Harbor; 1982. [Google Scholar]
- 28.Capranico G, Kohn KW, Pommier Y. Local sequence requirements for DNA cleavage by mammalian topoisomerase II in the presence of doxorubicin. Nucleic Acids Res. 1990;18:6611–6619. doi: 10.1093/nar/18.22.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mergny JL, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stefl R, Trantirek L, Vorlickova M, Koca J, Sklenar V, Kypr J. A-like guanine-guanine stacking in the aqueous DNA duplex of d(GGGGCCCC) J. Mol. Biol. 2001;307:513–524. doi: 10.1006/jmbi.2001.4484. [DOI] [PubMed] [Google Scholar]
- 31.Lindqvist M, Graslund A. An FTIR and CD study of the structural effects of G-tract length and sequence context on DNA conformation in solution. J. Mol. Biol. 2001;314:423–432. doi: 10.1006/jmbi.2001.5164. [DOI] [PubMed] [Google Scholar]
- 32.Ng HL, Dickerson RE. Mediation of the A/B-DNA helix transition by G-tracts in the crystal structure of duplex CATGGGCCCATG. Nucleic Acids Res. 2002;30:4061–4067. doi: 10.1093/nar/gkf515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zechiedrich EL, Cozzarelli NR. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- 34.Pan XS, Fisher LM. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 1998;42:2810–2816. doi: 10.1128/aac.42.11.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan XS, Fisher LM. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob. Agents Chemother. 1997;41:471–474. doi: 10.1128/aac.41.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 37.Laponogov I, Veselkov DA, Sohi MK, Pan XS, Achari A, Yang C, Ferrara JD, Fisher LM, Sanderson MR. Breakage-Reunion Domain of Streptococcus pneumoniae Topoisomerase IV: Crystal Structure of a Gram-Positive Quinolone Target. PLoS ONE. 2007;2:e301. doi: 10.1371/journal.pone.0000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnard FM, Maxwell A. Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser(83) and Asp(87) Antimicrob. gents Chemother. 2001;45:1994–2000. doi: 10.1128/AAC.45.7.1994-2000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiasa H. The Glu-84 of the ParC subunit plays critical roles in both topoisomerase IV-quinolone and topoisomerase IV-DNA interactions. Biochemistry. 2002;41:11779–11785. doi: 10.1021/bi026352v. [DOI] [PubMed] [Google Scholar]
- 40.Hockings SC, Maxwell A. Identification of four GyrA residues involved in the DNA breakage-reunion reaction of DNA gyrase. J. Mol. Biol. 2002;318:351–359. doi: 10.1016/S0022-2836(02)00048-7. [DOI] [PubMed] [Google Scholar]
- 41.Bouchillon SK, Hoban DJ, Johnson JL, Johnson BM, Butler DL, Saunders KA, Miller LA, Poupard JA. In vitro activity of gemifloxacin and contemporary oral antimicrobial agents against 27247 Gram-positive and Gram-negative aerobic isolates: a global surveillance study. Int. J. Antimicrob. Agents. 2004;23:181–196. doi: 10.1016/j.ijantimicag.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Kosowska-Shick K, Credito K, Pankuch GA, Lin G, Bozdogan B, McGhee P, Dewasse B, Choi DR, Ryu JM, et al. Antipneumococcal activity of DW-224a, a new quinolone, compared to those of eight other agents. Antimicrob. Agents Chemother. 2006;50:2064–2071. doi: 10.1128/AAC.00153-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jain A, Rajeswari MR. Preferential binding of quinolones to DNA with alternating G, C/A, T sequences: a spectroscopic study. J. Biomol. Struct. Dyn. 2002;20:291–299. doi: 10.1080/07391102.2002.10506844. [DOI] [PubMed] [Google Scholar]
- 44.Hays FA, Teegarden A, Jones ZJ, Harms M, Raup D, Watson J, Cavaliere E, Ho PS. How sequence defines structure: a crystallographic map of DNA structure and conformation. Proc. Natl Acad. Sci. USA. 2005;102:7157–7162. doi: 10.1073/pnas.0409455102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trantirek L, Stefl R, Vorlickova M, Koca J, Sklenar V, Kypr J. An A-type double helix of DNA having B-type puckering of the deoxyribose rings. J. Mol. Biol. 2000;297:907–922. doi: 10.1006/jmbi.2000.3592. [DOI] [PubMed] [Google Scholar]
- 46.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc. Natl Acad. Sci. USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haring D, Kypr J. Escherichia coli genome is composed of two distinct types of nucleotide sequences. Biochem. Biophys. Res. Commun. 2000;272:571–575. doi: 10.1006/bbrc.2000.2825. [DOI] [PubMed] [Google Scholar]