Abstract
Maize ribosome-inactivating protein is classified as a class III or an atypical RNA N-glycosidase. It is synthesized as an inactive precursor with a 25-amino acid internal inactivation region, which is removed in the active form. As the first structural example of this class of proteins, crystals of the precursor and the active form were diffracted to 2.4 and 2.5 Å, respectively. The two proteins are similar, with main chain root mean square deviation (RMSD) of 0.519. In the precursor, the inactivation region is found on the protein surface and consists of a flexible loop followed by a long α-helix. This region diminished both the interaction with ribosome and cytotoxicity, but not cellular uptake. Like bacterial ribosome-inactivating proteins, maize ribosome-inactivating protein does not have a back-up glutamate in the active site, which helps the protein to retain some activity if the catalytic glutamate is mutated. The structure reveals that the active site is too small to accommodate two glutamate residues. Our structure suggests that maize ribosome-inactivating protein may represent an intermediate product in the evolution of ribosome-inactivating proteins.
INTRODUCTION
Ribosome-inactivating proteins (RIPs) are N-glycosidases which cleave the N-glycosidic bond of adenine-4324 in eukaryotic 28S rRNA or adenine-2660 in Escherichia coli 23S rRNA (1,2). This adenine is located in a highly conserved GAGA hairpin within the α-sarcin/ricin loop. Removal of the specific adenine hinders the elongation factor 1-dependent binding of aminoacyl-tRNA and GTP-dependent binding of elongation factor 2 to the ribosome. Thus, protein synthesis is arrested at the elongation step (2,3). RIPs get access to the ribosome by firstly interacting with ribosomal proteins; for example, trichosanthin (TCS) binds to the acidic ribosomal P proteins (4,5), ricin A chain (RTA) binds to L9 and L10e (6) and pokeweed antiviral protein (PAP) binds to L3 (7–9).
RIPs are important biomedicine because they are highly cytotoxic towards human cancer cells, including lymphoma and myeloma. RTA conjugated to monoclonal antibodies anti-CD25 and anti-CD30 is being used to treat Hodgkin's lymphomas (10). Saporin is coupled to major histocompatibility complex (MHC) class I tetramers to kill antigen-specific CD8(+) T cells, which are important effector cells responsible for tissue destruction in several autoimmune and allograft-related diseases (11). TCS is used to induce midterm abortion, treat ectopic pregnancies and hydatidiform moles, reset menstruation and expel retained placenta (12). TCS and PAP have also been shown to possess anti-HIV activities (13).
Based on the number of subunits, RIPs are grouped into two classes. Type I RIPs such as TCS and saporin consist of a single polypeptide chain, with molecular weight around 30 kDa. They are actively uptaken by the alpha-2 macroglobulin receptor (α-2-MR) (14,15), which is widely distributed in different cell types such as macrophages, hepatocytes and follicular cells of the ovary (16). Type II RIPs such as ricin and abrin consist of two polypeptide chains linked by a disulphide bridge. Chain A is the catalytic subunit sharing high structural homology to type I RIPs, while chain B facilitates the intracellular delivery of chain A by interacting with carbohydrates on the cell surface (2). Both type I and II RIPs are basic proteins, with pI greater than 8. Maize RIP is an unusual RIP, which is either classified as a type III RIP (3) or considered as an atypical type I RIP (1). It is synthesized as a 34 kDa acidic inactive precursor in endosperm, with proper folding and a pI of around 6 (17). Its expression is controlled by the Opaque-2 regulatory locus (18). During germination, this precursor is converted to a two-chain active form by the elimination of 16 aa at the N-terminal region (residues 1–16), 25 aa at the acidic central region (residues 163–189) and 14 aa at the C-terminus (residues 287–300) to generate a two subunit basic protein of 248 aa (19). The two subunits of 16.5 and 8.5 kDa are tightly associated without any covalent linkage.
Among the sequences to be removed during the activation of maize RIP, the 25 aa internal region (known as internal inactivation region) is the most crucial, as removal of this region increases the activity by at least 600-fold, whereas removal of the N- or C-terminal region only increases the activity by 6- or 5-fold, respectively (17). Deletion of the internal amino acid residues represents a novel mechanism of enzyme activation in plants and resembles processing of certain hormones, such as insulin (3).
It is hypothesized that maize RIP can directly inhibit pathogens by inactivating their ribosomes and causing cell death (3). The active form is therefore a promising anti-insect and anti-fungal agent. This form has been overexpressed in transgenic rice, wheat and tobacco, for increasing the resistance to plant insects such as larvae of the cigarette beetle (Lasioderma serricorne), the tobacco hornworm (Manduca sexta) and the corn earworm (Helicoverpa zea) (20). Co-expression of the active form and a rice basic chitinase gene in transgenic rice has also led to increased resistance to sheath blight (Rhizoctonia solani) (21).
To further understand how maize RIP functions, in particular the relationship of the internal inactivation region to the rest of the protein, and to provide the first structural example of a type III RIP, we set forth to reveal the structures of the active (MOD) and inactive forms (Pro-RIP) of maize RIP and analyse the active site pocket.
MATERIALS AND METHODS
Preparation of protein samples
Maize [Δ1-16, Δ287-300]-Pro-RIP (Pro-RIP-WT in short) and [Δ1-16, Δ163-164, Δ167-189, Δ287-300]-Pro-RIP (MOD-WT in short) were obtained from Prof. R.S. Boston. The numbering of the aa residues is made according to Ref. (19). [Δ1-5]-Pro-RIP-WT (Pro-RIP), [Δ1-5]-MOD-WT (MOD), [E207A]-MOD, [E207AV238E]-MOD, [E207DV238E]-MOD and [V238E]-MOD were generated by polymerase chain reaction mutagenesis using overlapping primers and KOD DNA polymerase (Novagen). A methionine residue was added to the N-terminus as start codon. All concerned DNA were cloned into pET3a expression vector and sequenced to ensure that no secondary mutation had occurred. Proteins were overexpressed in E. coli strain C41 (DE3) (Novagen) in M9 medium (6 g/l Na2HPO4, 3 g/l KH2PO4, 1 g/l NH4Cl, 4 g/l glucose, 0.5 g/l NaCl, 100 µg/l ampicillin, 2 mM MgSO4, 0.1 mM CaCl2). Bacterial cells were grown in 37°C until OD 600 reached 0.4–0.6 and 0.4 mM IPTG was added to induce protein expression at 25°C. The cells were harvested after overnight culture by centrifugation at 4°C. Cell pellet was resuspended and sonicated in 20 mM phosphate buffer, pH 7.0 (buffer A). Cell lysate was collected by centrifugation at 4°C and loaded onto a HiTrap CM-FF column (Amersham) pre-equilibrated with buffer A, and eluted using a gradient of 0–0.5 M NaCl in buffer A. Fractions containing the target protein were pooled and dialyzed against buffer A, and loaded to a HiTrap SP column (Amersham) pre-equilibrated with buffer A. The protein was eluted using a gradient of 0–0.5 M NaCl in buffer A. Target fractions were pooled and concentrated to 5 ml for further purification by Superdex 75 gel filtration column (Amersham), which was pre-equilibrated with 20 mM Tris–HCl, 0.1 M NaCl, pH 7.0. Purified protein was concentrated and stored at −80°C.
Purification of ribosomes from rat liver
Rat liver tissue of 250 g was used for ribosome purification (22). In brief, the liver tissue was homogenized in ice-cold homogenization buffer [50 mM Tris–HCl (pH 7.6), 25 mM KCl, 5 mM MgCl2, 0.25 M sucrose] and was centrifuged at 13 000g for 30 min. The supernatant was filtered through glass wool and the filtrate was centrifuged at 145 000 g for 2 h. The pellet was resuspended in a buffer containing 35 mM Tris–HCl (pH 7.8), 25 mM KCl, 10 mM MgCl2, 0.15 M sucrose and 6 mM 2-mercaptoethanol. One-tenth of the volume of a freshly prepared solution of 10% sodium deoxycholate was added to release the ribosomes from the microsomal membrane. The resultant suspension was layered over an equal volume of 0.3 M sucrose pad in buffer B [35 mM Tris–HCl (pH 7.8), 600 mM KCl, 10 mM MgCl2, 6 mM 2-mercaptoethanol] and then centrifuged at 176 000 g for 90 min. The ribosomal pellet was rinsed and resuspended in buffer C [50 mM Tris–HCl (pH 7.8), 50 mM KCl, 5 mM MgCl2, 10 mM KH2CO3, 0.25 M sucrose, 6 mM 2-mercaptoethanol] to 7.5 mg/ml.
Crystallization, data collection and data analysis
Crystals of Pro-RIP and MOD were grown by mixing equal volume of protein solution and buffer in sitting drop at 16°C. The buffer used for Pro-RIP was 2 M ammonium sulphate, 0.25 M Tris–HCl and 0.1 M sodium acetate tri-hydrate, pH 3.5. The buffer for MOD was 0.2 M sodium acetate tri-hydrate, 0.1 M Tris–HCl pH 8.5 and 30% PEG 4000. MOD-Adenine crystal complex was obtained by soaking MOD crystals in the mother liquid containing saturated adenine for 72 h. Diffraction data of Pro-RIP and MOD were collected at 110 K using an in-house Rigaku MicroMax 007 X-ray generator and the synchrotron 6B beamline at Pohang Accelerator Laboratory, Pohang, Korea. Diffraction data were processed by MOSFLM. Phase determination was carried out by molecular replacement using MolRep of the CCP4 program suite. Structure of ricin A chain (PDB ID : 1RTC) with the side chains first eliminated by the Align4MR program (23) was used as the search model for Pro-RIP. For MOD, the refined Pro-RIP with the internal fragment deleted was used as the search model. The starting model was subject to rigid refinement using CNS and model building was carried out by XtalView. Further refinement was also achieved by CNS. Adenine and water molecules were added after the rest of the structure was well refined. Water molecules with Sigma values less than 1.0 in the 2Fo−Fc map were excluded. The stereochemical quality of the model was assessed by PROCHECK (24).
Ribosome-inactivating activity assay
The ribosome-inactivating activity assay was carried out using a rabbit reticulocyte lysate in vitro protein synthesis system, with L-[3, 4, 5-3H (N)]-leucine as label (NEN Life Science Products). In brief, 0.1 pM to 10 μM proteins and 5 nCi [3H]-leucine were incubated with the translation system in triplicate at 30°C for 30 min. Newly synthesized protein was precipitated with 25% trichloroacetic acid and captured by filtration through glass microfibre filters (Whatman).
Cytotoxicity assay of MOD and Pro-RIP
MTT assay was performed on human choriocarcinoma JAR cell line. In brief, JAR cells were treated with Pro-RIP or MOD for 72 h followed by incubation with MTT (5 mg/ml) for 4 h. DMSO was then added and OD595 values were measured using a colorimetric microplate reader (Model 3550, Biorad). The result was presented in terms of percentage to control as mean ± SEM. Two-way ANOVA with Bonferroni test as the post hoc test for multiple comparisons was used to compare cell viability after treatment with Pro-RIP and MOD.
Flow cytometry assay
JAR cells were seeded in a six-well plate (3 × 106 cells/well). Pro-RIP and MOD were labelled by green fluorescence dye F1640 (Roche) and incubated with the cells for 4 h. The cells were harvested and analysed by flow cytometry (BD biosciences). Uptake of the Pro-RIP and MOD was presented as mean ± SEM of percentage to control. One-way ANOVA with Dunnett's Multiple Comparison Test was used to assess the significance of protein uptake as compared to the dye control.
Pull-down assay
Pull-down assay was carried out to find the interaction between maize RIP and purified rat ribosomes. In brief, Pro-RIP and MOD columns were prepared by immobilizing purified Pro-RIP or MOD to a 1 ml NHS-column (Amersham). Then purified ribosomes were loaded onto the columns. Unbound protein was washed away by 30 ml PBS (1 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, 2.7 mM KCl, 0.005% Tween 20, pH 7.4), followed by elution buffer (1 M NaCl in PBS, pH 8.0) to elute the bound proteins. The eluted fractions were analysed by 15% SDS–PAGE. Control experiment was carried out by loading the ribosomes onto uncoupled NHS-Sepharose.
Binding kinetic assay using surface plasmon resonance
BIAcore 3000 surface plasmon resonance biosensor (Pharmacia Biosensor AB) was used to measure the kinetic parameters of the interaction. Pro-RIP or MOD (1 nM) in 10 mM sodium acetate, pH 5.0 was covalently linked to the dextran on the surface of CM5 sensor chip via primary amino groups using the Amine Coupling Kit (Pharmacia) at a flow rate of 5 µl/min, 25°C. A range of 0–240 nM of ribosomes in PBS were injected at a flow rate of 5 µl/min, 25°C, onto the RIP immobilized sensor chip surface. The binding surface was regenerated by 2 M NaCl between sample injections. Control experiment was carried out similarly on uncoupled sensor chip surface.
Molecular modelling of maize RIP–ribosome complex
The coordinates of yeast ribosome were derived from the cryo-EM structure (PDB ID: 1S1H and 1S1I). The adenine ring of A-2697 (analogous to A-4324 in rat 28S rRNA) was positioned manually to the adenine-binding site of MOD. Pro-RIP was then superimposed to MOD. The model was energy minimized using the program CNS.
RESULTS
Protein purification and crystallization
Pro-RIP and MOD were purified, with purities greater than 95% and the usual yield was 50 mg/l culture. Variant [E207AV238E]-MOD had the lowest yield but still reached 20 mg/l culture. Crystals of Pro-RIP, MOD and MOD-adenine complex were obtained. The cell parameters and data collection statistics are shown in Table 1.
Table 1.
Data collection and refinement statistics
| Pro-RIP | MOD | MOD–adenine | |
|---|---|---|---|
| Data collection | |||
| Resolution range (Å) | 30.0–2.38 | 20.0–2.5 | 44.9–2.8 |
| Space group | P2(1) | P3(2) | P3(2) |
| Unit cell a, b, c (Å) | 39.23, 75.62, 97.15 | 115.19, 115.19, 45.00 | 115.91, 115.91, 44.90 |
| α, β, γ (deg) | 90, 93.725, 90 | 90, 90, 120 | 90, 90, 120 |
| Total number of observation | 55 918 | 51 970 | 31 580 |
| Total number of unique | 19 282 | 21 654 | 16 621 |
| Completeness (%) | 84.1 (70.6)a | 93.9 (90.1) | 99.9 (99.9) |
| Rmerge | 0.118 (0.342) | 0.031 (0.153) | 0.073 (0.338) |
| <I>/σ (I) | 7.1 (1.6) | 22.2 (4.1) | 11.2 (1.8) |
| Multiplicity | 2.9 (2.1) | 2.4 (2.0) | 2.0 (1.9) |
| Refinement statistics | |||
| Rwork/Rfree | 0.2114/0.2299 | 0.2188/0.2635 | 0.2319/0.2934 |
| RMSD bonds/angles (Å/deg) | 0.010026/1.80921 | 0.007964/1.45764 | 0.009768/1.59545 |
| Overall Mean B (Å2) | 30.746 | 47.899 | 62.895 |
| Chain A Mean B (Å2) | 30.618 | 34.758 | 56.537 |
| Ramachandran plot (%) | |||
| Most favoured regions [A,B,L] | 89.4 | 87 | 87.1 |
| Additional allowed regions [a,b,l,p] | 10.6 | 13 | 12.3 |
| Generously allowed regions [∼a,∼b,∼l,∼p] | 0 | 0 | 0.8 |
| Disallowed regions [XX] | 0 | 0 | 0 |
aHighest resolution shell is shown in parenthesis.
The structures of Pro-RIP (PDB ID: 2PQG), MOD (PDB ID: 2PQI) and MOD–adenine (PDB ID: 2PQJ) were resolved to 2.4, 2.5 and 2.8 Å, respectively. Phase determination was carried out by molecular replacement. The refinement statistics are shown in Table 1. Crystals of MOD belong to space group P3(2), with three molecules per asymmetric unit. After refinement, the final Rwork and Rfree values were 0.2188 and 0.2635, respectively. The structure shows a large N-terminal domain (aa 21–230, aa 21 is an added methionine) and a small C-terminal domain (aa 231–283). The former consists of five α-helices and five-stranded mixed β-sheets; the latter is made up of four α-helices. M21-F27 in MOD is flexible. It exists as a β-strand in chain C but shows no secondary structure in chains A and B (Figure 1). For the MOD crystals soaked in saturated adenine solution, electron density consistent to an adenine molecule was found in the active site of the protein. The space group remained P3(2) and no major structural change was observed. The final Rwork and Rfree values were 0.2319 and 0.2934, respectively.
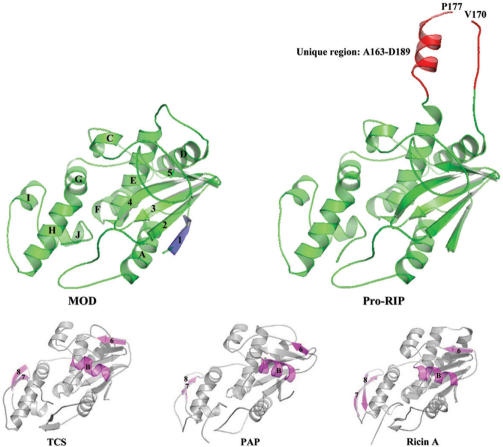
Figure 1.
Structures of Pro-RIP and MOD and their comparison to TCS, PAP and RTA. The overall structures of Pro-RIP and MOD, except the unique inactivating region Ala163–Asp189 in Pro-RIP (red), are similar. Pro-RIP and MOD have a large N-terminal and a small C-terminal domain. In the MOD structure, β-strand 1 (blue) is present in chain C but not chain A or B. The α-helix B and β-strand 6 (purple) found in the large domain of other RIPs are absent in Pro-RIP and MOD. The anti-parallel β-strands 7 and 8 in the small domain of other RIPs (purple) are replaced by a short α-helix I.
The crystals of Pro-RIP belong to space group P2(1) with two monomers per asymmetric unit. The final Rwork and Rfree values were 0.2114 and 0.2299, respectively. The overall structures of Pro-RIP and MOD are very similar, except the presence of a unique internal inactivating region Ala163–Asp189 in Pro-RIP (Figure 1), which is rich in acidic residues. Clear electron densities were observed in flexible loop regions Ala163–Val170 and Pro177–Ala179, and a long α-helix in Ala180–Ala188. When aligned to type I and type II RIPs, maize RIP showed some significant structural differences (Figure 1). There are no α-helix B and β-strand 6 (purple) in the large domain and the anti-parallel β-strands 7 and 8 in the small domain of other RIPs (purple) are replaced by a short α-helix.
Construction of MOD variants and ribosome-inactivating activity assay
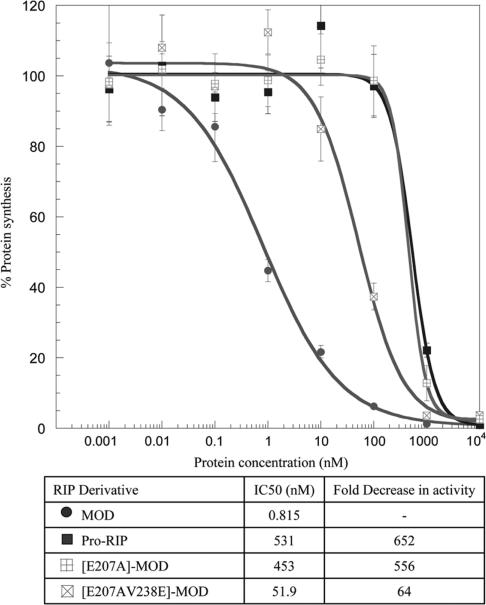
[E207A]-MOD, [E207AV238E]-MOD, [E207DV238E]-MOD and [V238E]-MOD were constructed and expressed in E. coli. [E207A]-MOD and [E207AV238E]-MOD were soluble and purified. On the other hand, [E207DV238E]-MOD and [V238E]-MOD formed inclusion bodies. The purified proteins were assayed for ribosome-inactivating activity. Percentage inhibition of protein synthesis could be fitted to sigmoidal curves (Figure 2), indicating concentration-dependent inhibition. MOD exhibited the highest protein-synthesis inhibition activity whereas Pro-RIP had the least. Compared to the wild-type MOD, the activity of [E207A]-MOD decreased by about 556-fold. Variant [E207AV238E]-MOD was 9-fold more active than [E207A]-MOD.
Figure 2.
Inhibition of in vitro protein synthesis by maize RIP variants. The potency of maize RIP variants to inhibit in vitro protein synthesis was measured by adding 1 pM to 10 µM of protein samples to nuclease-untreated rabbit reticulocyte lysate as described in Materials and Methods section. IC50, the concentration of protein required to achieve 50% inhibition, was determined by fitting the data to a four-parameter logistic equation (n = 5).
Cytotoxicity and cellular uptake of MOD and Pro-RIP
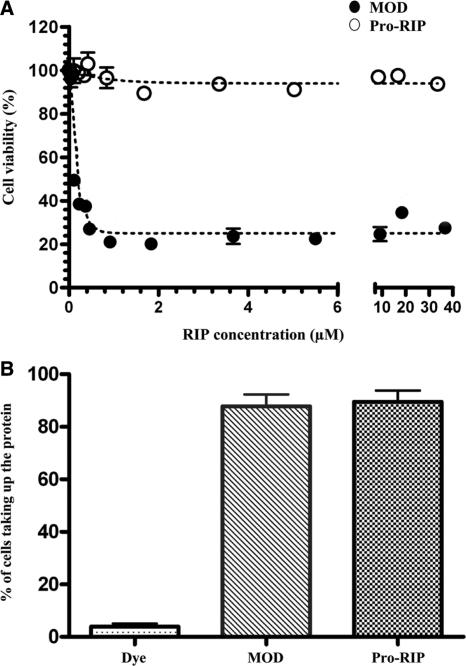
The cytotoxicities of Pro-RIP and MOD to choriocarcinoma JAR cells were evaluated by MTT assay. MOD was found to be more effective in reducing the viability of JAR (IC50 = 0.37 µM) when compared to Pro-RIP (IC50 > 40 µM) (Figure 3A).
Figure 3.
(A) Cytotoxicity assays of Pro-RIP and MOD to JAR cells. Cytotoxicity is expressed as percentage to control in terms of mean ± SEM. The difference in cell viability under the same concentration of Pro-RIP and MOD is statistically significant (P < 0.05, n = 5). (B) Uptake of Pro-RIP and MOD by JAR cells as measured by flow cytometry. Proteins were labelled by green fluorescence dye F1640 (Roche) and incubated with JAR cells for 4 h. Uptake of the two proteins was significant as compared to the control experiment where only the dye was incubated with the cells (P < 0.05, n = 3).
Uptake of Pro-RIP and MOD by JAR cells was analysed by flow cytometry. JAR cells were incubated with protein labelled fluorescent dye F1640 (Roche) for 4 h. It was found that both Pro-RIP and MOD entered JAR cells with similar efficiency (Figure 3B).
Protein–protein interaction and molecular modelling of maize RIP–ribosome complex
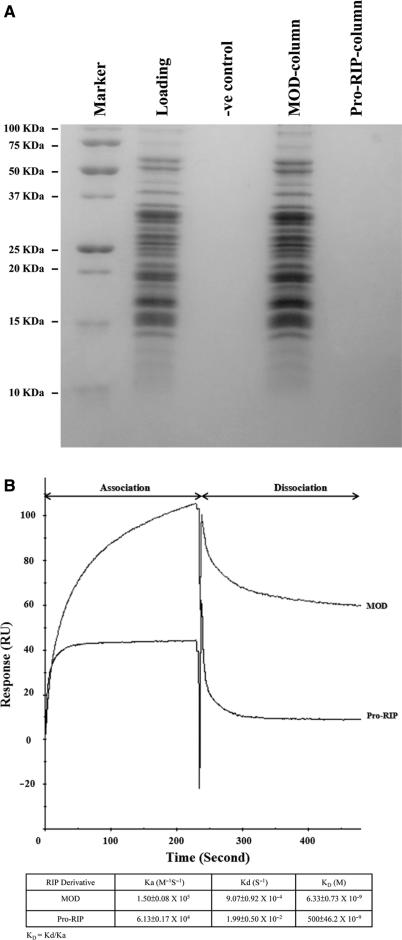
Pull-down assays were carried out to study the interaction between the two forms of maize RIP and purified rat ribosomes. MOD and Pro-RIP were coupled to NHS Hi-Trap columns, with efficiencies of 85–90%. Purified rat ribosomes were loaded onto the two RIP-coupled NHS columns, which were then washed by PBS. The bound ribosomes were eluted and analysed by SDS–PAGE. Our results showed that only MOD, but not Pro-RIP interacts with ribosomes (Figure 4A). Rat ribosomes did not interact with uncoupled NHS column.
Figure 4.
Interaction of maize RIP and eukaryotic ribosomes in vitro. (A) 100 nM of purified rat ribosomes were loaded onto the indicated columns. After washing away the unbound ribosomes, the bound ribosome was eluted and analysed by 15% SDS–PAGE. Loading denotes ribosomes before interaction. −ve control denotes ribosomes loaded onto uncoupled NHS column. (B) 60 nM of purified rat ribosomes were loaded onto the MOD-coupled or Pro-RIP-coupled CM5 sensor chip. The kinetic rates and dissociation constants of the interaction between maize RIP and rat ribosomes were analysed by BIAcore 3000 biosensor.
A BIAcore 3000 surface plasmon resonance biosensor (Pharmacia Biosensor AB) was used to study the kinetic parameters of the interaction. 5100 RU of Pro-RIP and 5200 RU of MOD were coupled onto the surface of CM5 sensor chips via primary amide groups. When 60 nM of purified rat ribosomes at 5 µl/min were loaded onto the RIP-coupled CM5 sensor chips, both association and dissociation responses of MOD were higher than those of Pro-RIP. Within a 240 s window, the association signal rapidly reached 105 RU with a slow dissociation ended at 60 RU. On the contrary, Pro-RIP associated slowly to 40 RU and dissociated sharply to 10 RU (Figure 4B). Control experiment was carried out similarly on uncoupled sensor chip surface. To determine the binding affinity (KD) of the ribosomes to MOD and Pro-RIP, ribosomes of different concentrations were allowed to interact with the maize RIP-immobilized sensor chip surface. The KD values of MOD and Pro-RIP were 6.33 ± 0.73 and 500 ± 46.20 nM, respectively. The 80-fold decrease of binding affinity of Pro-RIP was due to the slower association rate and faster dissociation rate.
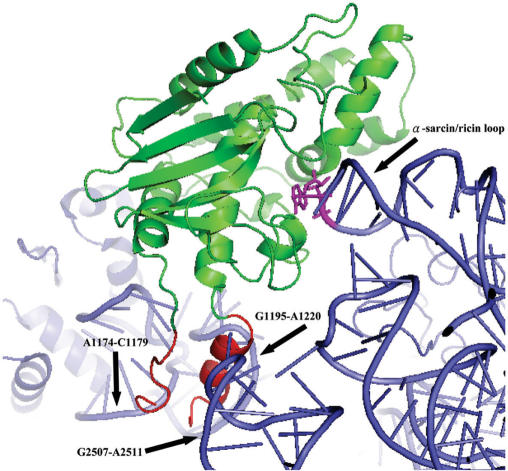
In silico docking of Pro-RIP to yeast ribosome showed that if the adenine ring of A-2697 (analogous to A-4324 of rat 28S rRNA) is placed properly into the active site pocket, the internal inactivation region would clash with A1174-C1179, G1195-A1220 and G2507-A2511 of the 25S rRNA (Figure 5). Therefore, this region of Pro-RIP probably sterically hinders the interaction of the protein with the ribosome.
Figure 5.
Docking of Pro-RIP to yeast ribosome. The adenine ring of A-2697 in the α-sarcin/ricin loop (analogous to A-4324 in rat 28S rRNA) is shown in pink. The internal inactivating region is coloured in red and the rest of Pro-RIP is coloured in green. As indicated, the internal inactivating region of Pro-RIP clashes into at least three sites of the 25S rRNA.
DISCUSSION
Maize RIP represents a unique class of RIP. It has a prominent internal acidic region spanning Ala163–Asp189, which is absent in type I and type II RIPs. Previously, it was predicted that the internal acidic region would alter the arrangement of key residues in the active-site cleft or disrupt protein folding, thus affecting the catalytic activity (25). To shed light on its biochemical function, we set out to elucidate the structures of Pro-RIP and MOD.
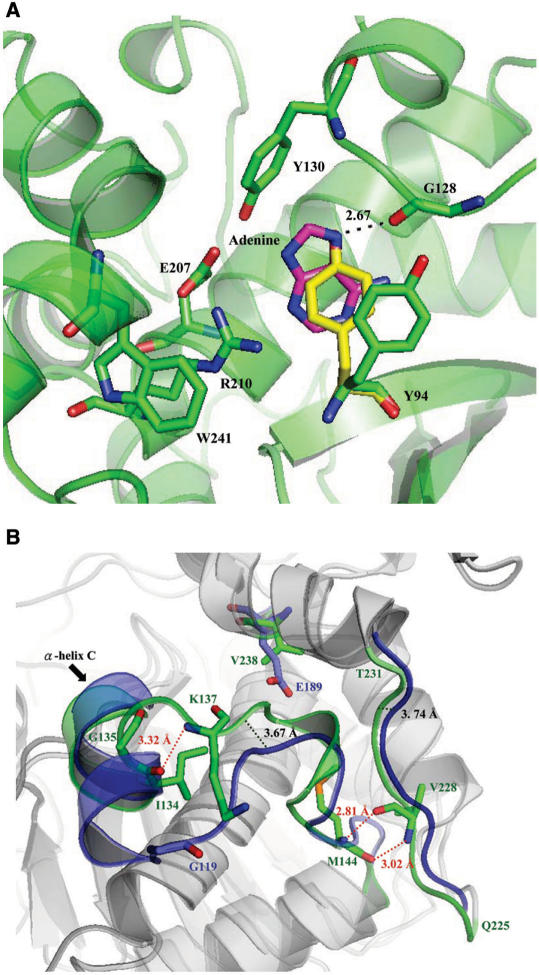
Secondary structure analysis of MOD-WT and Pro-RIP-WT by nnPredict program (26) showed that the N-terminal 5 aa is a flexible loop and crystallization of MOD and Pro-RIP was only achieved by deleting these 5 aa (data not shown). The overall structures of Pro-RIP and MOD are very similar, except the presence of the internal inactivation region Ala163–Asp189 in Pro-RIP (Figure 1). Each protein has two domains, consisting of five α-helices and five-stranded mixed β-sheet in the large N-terminal domain. The small C-terminal domain is composed of four α-helices, with a bend between helices G and H. This helical bend is conserved among RIPs. The conserved active site residues Tyr94, Tyr130, Glu207, Arg210 and Trp241 are located at the cleft between the N-terminal and C-terminal domains. The tyrosine rings of Tyr94 and Tyr130 in MOD are facing each other, which may facilitate the insertion of A-4324 adenine ring of 28S RNA. In Pro-RIP, the tyrosine ring of Tyr94 assumes a different conformation. It flips towards the adenine-binding site such that it becomes perpendicular to the ring of Tyr130, with its hydroxyl group hydrogen-bonded to the carboxyl group of Gly128 (Figure 6A). RIPs in Family Poaceae have more tryptophan residues than those in other families. In maize RIP, there are a total of six tryptophan residues of which only Trp241 in the active site is conserved. The only two cysteine residues (Cys51 and Cys206) of maize RIP are 14.37 Å apart, without disulphide-bond linkage.
Figure 6.
(A) Active site pockets of Pro-RIP and MOD-adenine complex. The structure of Pro-RIP was aligned to that of the MOD-adenine complex. The adenine is coloured in pink and with Tyr94 of MOD and Pro-RIP in green and yellow, respectively. In Pro-RIP, the ring of Tyr94 has flipped towards the adenine-binding site and forms hydrogen bond with Gly128. (B) Comparison of the size of the active site pocket in MOD (green) and TCS (blue). As indicated by black dotted lines, Ile134-Met144 and Gln225-Thr231 in MOD are found to bend towards the active site. As a result, the space surrounding Val238 is too small for the side chain of a glutamate residue. Hydrogen bonds are indicated by red dotted lines.
The overall root mean square deviation (RMSD) among the structures of TCS, PAP, RTA and SO6 is 0.755. However, the value increases to 1.516 when MOD is included, suggesting that maize RIP is more structurally distinct. Compared to other RIPs, the α-helix B and β-strand 6 in the large domain are missing in MOD, and the anti-parallel β-strands 7 and 8 in the small domain are replaced by a short α-helix (Figure 1). These structural differences, nevertheless, do not significantly diminish the ability of MOD to inhibit protein synthesis in vitro, implying that the variable regions are not crucial for the enzymatic activity. In Pro-RIP, the internal inactivating region Ala163–Asp189 is very rich in acidic residues. Our structure shows that the internal inactivating region consists of a flexible loop (Ala163–Ala179) and a long α-helix (Ala180–Ala188) (Figure 1). Since this fragment is located on the protein surface and is 15.5 Å away from the active site, the conformation of the protein and the active-site cleft should not be affected.
At the C-terminal region, the hydrogen bonds Val280 N to Pro60 O and Val280 O to Leu62 N are conserved among RIPs. In TCS, the corresponding hydrogen bonds are Leu240 N to Pro35 O and Leu 240 O to Leu37 N. Deletion of these hydrogen bonds has been shown to disrupt the folding of TCS. Therefore, the C-terminal region in TCS can only be deleted up to Leu240 for an active variant (27). It will be of interest to investigate if the hydrogen bonds orchestrated by Val280 also play a role in the folding of Pro-RIP and MOD.
Our biochemical studies showed that the presence of the internal inactivation region in Pro-RIP affects the cytotoxicity but not the uptake to JAR cells (Figure 3A and B). Pull-down assay indicated that MOD, but not Pro-RIP, interacts with rat ribosomes (Figure 4A). Surface plasmon resonance analysis also showed that the binding affinity (KD) of Pro-RIP is about 80-fold higher than that of MOD (Figure 4B). These indicated that the internal inactivation region of Pro-RIP might obstruct the protein to dock onto the ribosome. Indeed, in silico docking of Pro-RIP to yeast ribosome showed that the internal inactivation region clashes with multiple sites of the 25S rRNA (Figure 5). It has been reported that the inhibitory activity of Pro-RIP on maize ribosomes is lower than that of MOD (28). Therefore, the internal inactivation region may serve the purpose of preventing the maize RIP from attacking its cognate ribosomes in vivo.
In TCS and saporin, the ribosome-binding sites are located between the anti-parallel β-sheets 7 and 8 in the C-terminal domain (5,29). Since the corresponding region in Pro-RIP is replaced by a short alpha-helix, and the internal inactivation region is located on the surface of helices D and E in the N-terminal domain, we predict that the ribosome interaction site on maize RIP may be different from that of TCS and saporin.
In the active sites of many RIPs of dicotyledonous plants, there are two glutamate residues. The carboxyl group of one of the glutamate residues stabilizes the oxocarbenium ion-like transition state. The other glutamate residue serves as a backup when the catalytic glutamate is mutated (30,31). Interestingly, in the active site pocket of Pro-RIP and MOD, only one glutamate residue (Glu207) is found. The residue corresponds to the backup glutamate that has turned into valine (Val238). This phenomenon is also observed in other known RIPs of the Family Poaceae, such as rice RIP (GenBank: BAB85659) and JIP60 (GenBank: AAB33361). This raises the puzzle of the absence of the backup glutamate in these RIPs. We found that the ribosome-inactivating activity of [E207A]-MOD decreased by about 556-fold (Figure 2), confirming the importance of this residue. Variants [V238E]-MOD and [E207D-V238E]-MOD were expressed as inclusion bodies, showing the existence of two long side chains in the active site disrupts the folding and/or structure of the protein. On the other hand, the variant [E207AV238E]-MOD was soluble and regained partial ribosome-inactivating activity, indicating that a glutamate residue may be placed in position 238 as a backup for improving the activity.
Compared to other RIPs, the main chains of Ile134-Met144 and Gln225-Thr231 of maize RIP exhibit some significant structural differences. With reference to TCS, these two chains have shifted 3.67 and 3.74 Å inward, respectively (Figure 6B). In maize RIP, α-helix C (Tyr130-Ile134) is shorter, due to the presence of Gly135 as an α-helix C-cap terminator (32). The hydrogen bond between Ile134 O and Lys137 N ensures an inward shift of the fragment Ile134-Met144. Moreover, the hydrogen bonds between Met144 O and Val228 N and Met144 N and Val228 O result in a tight packing of Gln225-Thr231 towards the active site. As a result, the active-site pocket becomes too small to accommodate an extra glutamate residue. Molecular modelling indicated that if Val238 is mutated to glutamate, Glu238 would clash with the side chain of Leu139 and Leu230 (Supplementary Figure 1A). This is consistent with our finding that MOD variant V238E expressed as inclusion bodies. Furthermore, maize RIP is not prepared to have a glutamate residue in position 238. In TCS, the corresponding Glu189 is stabilized by Arg122 and Gln156 (Supplementary Figure 1B). On the other hand, the latter 2 aa have become Leu139 and Val203 in maize RIP (Supplementary Figure 1C).
Interestingly, known bacterial RIPs including Shiga toxin have only one catalytic glutamate in the active site (Supplementary Figure 1D). In Shiga toxin, the position for the backup glutamate is occupied by Thr200. Previous phylogenetic analysis of representative plant and bacterial RIPs has indicated that maize RIP is more related to the latter (33). Hence, it is likely that RIPs having one catalytic glutamate presents a prototype that acquires a second glutamate residue in the active site during evolution.
In conclusion, we have solved the crystal structures of Pro-RIP and MOD, the precursor and the mature form of maize RIP. Our data reveal the structural and functional differences of the two forms and the role of the internal inactivation region for its biological activities. The structure of the active-site pocket also indicates that maize RIP may be an intermediate step of evolution from prokaryotes to higher plants.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are indebted to Prof. R.S. Boston of North Carolina State University for the clones of maize RIP. Thanks are due to Dr Siu-Hong Chan for editing the manuscript. This work was supported by a grant (CUHK 4606/06M) from the Research Grants Council of Hong Kong SAR. Work in Korea was supported by a research grant from the 21C Frontier Functional Proteomics Center. Funding to pay the Open Access publication charges for this article was provided by Department of Biochemistry, The Chinese University of Hong Kong.
Conflict of interest statement. None declared.
REFERENCES
- 1.Stirpe F, Battelli MG. Ribosome-inactivating proteins: progress and problems. Cell. Mol. Life Sci. 2006;63:1850–1866. doi: 10.1007/s00018-006-6078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartley MR, Lord JM. Cytotoxic ribosome-inactivating lectins from plants. Biochim. Biophys. Acta. 2004;1701:1–14. doi: 10.1016/j.bbapap.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Motto M, Lupotto E. The genetics and properties of cereal ribosome-inactivating proteins. Mini. Rev. Med. Chem. 2004;4:493–503. doi: 10.2174/1389557043403873. [DOI] [PubMed] [Google Scholar]
- 4.Chan SH, Hung FS, Chan DS, Shaw PC. Trichosanthin interacts with acidic ribosomal proteins P0 and P1 and mitotic checkpoint protein MAD2B. Eur. J. Biochem. 2001;268:2107–2112. doi: 10.1046/j.1432-1327.2001.02091.x. [DOI] [PubMed] [Google Scholar]
- 5.Chan DS, Chu LO, Lee KM, Too PH, Ma KW, Sze KH, Zhu G, Shaw PC, Wong KB. Interaction between trichosanthin, a ribosome-inactivating protein, and the ribosomal stalk protein P2 by chemical shift perturbation and mutagenesis analyses. Nucleic Acids Res. 2007;35:1660–1672. doi: 10.1093/nar/gkm065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vater CA, Bartle LM, Leszyk JD, Lambert JM, Goldmacher VS. Ricin A chain can be chemically cross-linked to the mammalian ribosomal proteins L9 and L10e. J. Biol. Chem. 1995;270:12933–12940. doi: 10.1074/jbc.270.21.12933. [DOI] [PubMed] [Google Scholar]
- 7.Rajamohan F, Ozer Z, Mao C, Uckun FM. Active center cleft residues of pokeweed antiviral protein mediate its high-affinity binding to the ribosomal protein L3. Biochemistry. 2001;40:9104–9114. doi: 10.1021/bi002851p. [DOI] [PubMed] [Google Scholar]
- 8.Hudak KA, Dinman JD, Tumer NE. Pokeweed antiviral protein accesses ribosomes by binding to L3. J. Biol. Chem. 1999;274:3859–3864. doi: 10.1074/jbc.274.6.3859. [DOI] [PubMed] [Google Scholar]
- 9.Di R, Tumer NE. Expression of a truncated form of ribosomal protein L3 confers resistance to pokeweed antiviral protein and the Fusarium mycotoxin deoxynivalenol. Mol. Plant Microbe Interact. 2005;18:762–770. doi: 10.1094/MPMI-18-0762. [DOI] [PubMed] [Google Scholar]
- 10.Schnell R, Borchmann P, Staak JO, Schindler J, Ghetie V, Vitetta ES, Engert A. Clinical evaluation of ricin A-chain immunotoxins in patients with Hodgkin's lymphoma. Ann. Oncol. 2003;14:729–736. doi: 10.1093/annonc/mdg209. [DOI] [PubMed] [Google Scholar]
- 11.Hess PR, Barnes C, Woolard MD, Johnson MD, Cullen JM, Collins EJ, Frelinger JA. Selective deletion of antigen-specific CD8+ T cells by MHC class I tetramers coupled to the type I ribosome-inactivating protein saporin. Blood. 2007;109:3300–3307. doi: 10.1182/blood-2006-06-028001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw PC, Lee KM, Wong KB. Recent advances in trichosanthin, a ribosome-inactivating protein with multiple pharmacological properties. Toxicon. 2005;45:683–689. doi: 10.1016/j.toxicon.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 13.Parikh BA, Tumer NE. Antiviral activity of ribosome inactivating proteins in medicine. Mini. Rev. Med. Chem. 2004;4:523–543. doi: 10.2174/1389557043403800. [DOI] [PubMed] [Google Scholar]
- 14.Chan WL, Shaw PC, Tam SC, Jacobsen C, Gliemann J, Nielsen MS. Trichosanthin interacts with and enters cells via LDL receptor family members. Biochem. Biophys. Res. Commun. 2000;270:453–457. doi: 10.1006/bbrc.2000.2441. [DOI] [PubMed] [Google Scholar]
- 15.Cavallaro U, Soria MR. Targeting plant toxins to the urokinase and alpha 2-macroglobulin receptors. Semin. Cancer Biol. 1995;6:269–278. doi: 10.1006/scbi.1995.0035. [DOI] [PubMed] [Google Scholar]
- 16.Zheng G, Bachinsky DR, Stamenkovic I, Strickland DK, Brown D, Andres G, McCluskey RT. Organ distribution in rats of two members of the low-density lipoprotein receptor gene family, gp330 and LRP/alpha 2MR, and the receptor-associated protein (RAP) J. Histochem. Cytochem. 1994;42:531–542. doi: 10.1177/42.4.7510321. [DOI] [PubMed] [Google Scholar]
- 17.Hey TD, Hartley M, Walsh TA. Maize ribosome-inactivating protein (b-32). Homologs in related species, effects on maize ribosomes, and modulation of activity by pro-peptide deletions. Plant Physiol. 1995;107:1323–1332. doi: 10.1104/pp.107.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bass HW, Webster C, OBrian GR, Roberts JK, Boston RS. A maize ribosome-inactivating protein is controlled by the transcriptional activator Opaque-2. Plant Cell. 1992;4:225–234. doi: 10.1105/tpc.4.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walsh TA, Morgan AE, Hey TD. Characterization and molecular cloning of a proenzyme form of a ribosome-inactivating protein from maize. Novel mechanism of proenzyme activation by proteolytic removal of a 2.8-kilodalton internal peptide segment. J. Biol. Chem. 1991;266:23422–23427. [PubMed] [Google Scholar]
- 20.Dowd PF, Holmes RA, Pinkerton TS, Johnson ET, Lagrimini LM, Boston RS. Relative activity of a tobacco hybrid expressing high levels of a tobacco anionic peroxidase and maize ribosome-inactivating protein against Helicoverpa zea and Lasioderma serricorne. J. Agric. Food Chem. 2006;54:2629–2634. doi: 10.1021/jf058180p. [DOI] [PubMed] [Google Scholar]
- 21.Kim JK, Jang IC, Wu R, Zuo WN, Boston RS, Lee YH, Ahn IP, Nahm BH. Co-expression of a modified maize ribosome-inactivating protein and a rice basic chitinase gene in transgenic rice plants confers enhanced resistance to sheath blight. Transgenic Res. 2003;12:475–484. doi: 10.1023/a:1024276127001. [DOI] [PubMed] [Google Scholar]
- 22.Spedding G. Isolation and analysis of ribosomes from prokaryotes, eukaryotes, and organelles. In: Spedding G, editor. Ribosomes and Protein Synthesis. Oxford: Oxford University Press; 1990. pp. 9–12. [Google Scholar]
- 23.Cheung YY, Allen MD, Bycroft M, Wong KB. Crystallization and preliminary crystallographic analysis of an acylphosphatase from the hyperthermophilic archaeon Pyrococcus horikoshii. Acta Crystallogr. D. 2004;60:1308–1310. doi: 10.1107/S0907444904010996. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 25.Bass HW, Krawetz JE, OBrian GR, Zinselmeier C, Habben JE, Boston RS. Maize ribosome-inactivating proteins (RIPs) with distinct expression patterns have similar requirements for proenzyme activation. J. Exp. Bot. 2004;55:2219–2233. doi: 10.1093/jxb/erh243. [DOI] [PubMed] [Google Scholar]
- 26.Ragulina LE, Makeev V, Esipova NG, Tumanian VG, Vlasov PK, Bogush VG, Debabov VG. An analysis of the secondary structure of spider spidroins I and II belonging to different species. Biofizika. 2004;49:1147–1149. [PubMed] [Google Scholar]
- 27.Chan SH, Shaw PC, Mulot SF, Xu LH, Chan WL, Tam SC, Wong KB. Engineering of a mini-trichosanthin that has lower antigenicity by deleting its C-terminal amino acid residues. Biochem. Biophys. Res. Commun. 2000;270:279–285. doi: 10.1006/bbrc.2000.2395. [DOI] [PubMed] [Google Scholar]
- 28.Krawetz JE, Boston RS. Substrate specificity of a maize ribosome-inactivating protein differs across diverse taxa. Eur. J. Biochem. 2000;267:1966–1974. doi: 10.1046/j.1432-1327.2000.01200.x. [DOI] [PubMed] [Google Scholar]
- 29.Savino C, Federici L, Ippoliti R, Lendaro E, Tsernoglou D. The crystal structure of saporin SO6 from Saponaria officinalis and its interaction with the ribosome. FEBS Lett. 2000;470:239–243. doi: 10.1016/s0014-5793(00)01325-9. [DOI] [PubMed] [Google Scholar]
- 30.Wong KB, Ke YB, Dong YC, Li XB, Guo YW, Yeung HW, Shaw PC. Structure/function relationship study of Gln156, Glu160 and Glu189 in the active site of trichosanthin. Eur. J. Biochem. 1994;221:787–791. doi: 10.1111/j.1432-1033.1994.tb18792.x. [DOI] [PubMed] [Google Scholar]
- 31.Robertus JD, Monzingo AF. The structure of ribosome inactivating proteins. Mini. Rev. Med. Chem. 2004;4:477–486. doi: 10.2174/1389557043403837. [DOI] [PubMed] [Google Scholar]
- 32.Bang D, Gribenko AV, Tereshko V, Kossiakoff AA, Kent SB, Makhatadze GI. Dissecting the energetics of protein alpha-helix C-cap termination through chemical protein synthesis. Nat. Chem. Biol. 2006;2:139–143. doi: 10.1038/nchembio766. [DOI] [PubMed] [Google Scholar]
- 33.Girbes T, Ferreras JM, Arias FJ, Stirpe F. Description, distribution, activity and phylogenetic relationship of ribosome-inactivating proteins in plants, fungi and bacteria. Mini. Rev. Med. Chem. 2004;4:461–476. doi: 10.2174/1389557043403891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.