Abstract
Prostaglandins are anticancer agents known to inhibit tumor cell proliferation both in vitro and in vivo by affecting the mRNA stability. Here we report that a MAR-binding protein SMAR1 is a target of Prostaglandin A2 (PGA2) induced growth arrest. We identify a regulatory mechanism leading to stabilization of SMAR1 transcript. Our results show that a minor stem and loop structure present in the 5′ UTR of SMAR1 (ϕ1-UTR) is critical for nucleoprotein complex formation that leads to SMAR1 stabilization in response to PGA2. This results in an increased SMAR1 transcript and altered protein levels, that in turn causes downregulation of Cyclin D1 gene, essential for G1/S phase transition. We also provide evidence for the presence of a variant 5′ UTR SMAR1 (ϕ17-UTR) in breast cancer-derived cell lines. This form lacks the minor stem and loop structure required for mRNA stabilization in response to PGA2. As a consequence of this, there is a low level of endogenous tumor suppressor protein SMAR1 in breast cancer-derived cell lines. Our studies provide a mechanistic insight into the regulation of tumor suppressor protein SMAR1 by a cancer therapeutic PGA2, that leads to repression of Cyclin D1 gene.
INTRODUCTION
Most of the cellular transformation processes often involve changes in the gene expression levels rather than changes in the sequence itself. The multiple layers of control include a closely knit transcriptional program and the associated translation of the product. Posttranscriptional regulation (includes mRNA stability and translation) is one such mechanism whereby the organisms control the flow of genetic information into the proteome (1). Within the continuum of posttranscriptional regulation, mRNA stability is considered a major effector of gene regulation (2). For example, the levels of several cell cycle regulatory molecules like Cyclin D1, p21, p27 and cdks are known to oscillate from a high in dividing cells to a low in quiescent cells. Most of these oscillations are due to transcriptional control and regulated protein stability (3). The manifestation of cancer is a consequence of the loss of control of one or more of these regulations that leads to a loss of gene function. Anticancer drugs like flavopiridol and cyclopentenone prostaglandins (PGs) are potent inhibitors of cell cycle in various cell lines (4–6). These compounds cause growth arrest in cultured cells and exhibit antitumor activity in vivo primarily by affecting cell cycle regulatory molecules. Cyclopentenone PGA1 and flavopiridol are shown to affect the transcriptional rate of Cyclin D1 and levels of cdk4 (7,8). PGA2-mediated growth inhibition is attributed to the downregulation of Cyclin D1 by reduction in the mRNA transcript. Further, microarray-based studies have led to the identification of mRNA species whose transcription and/or steady-state levels are regulated by cyclopentenone PGs (9–11). A number of reports have also documented the role of some RNA-binding proteins such as ELAV family proteins or TIA-1 and TIAR that bind to the 5′ or 3′ UTR of cell cycle regulatory molecules and lead to an increased/decreased mRNA expression upon PGA2 treatment (12,13).
Recently, we have reported that SMAR1 (Scaffold/Matrix attachment region-binding protein 1) is a tumor suppressor MAR-binding protein that downregulates Cyclin D1 expression by recruiting HDAC1-mSin3A co-repressor complex at Cyclin D1 promoter locus (14). SMAR1 is documented to be a tumor suppressor protein that interacts with p53 (15) and retards B16F-10-induced melanoma in mouse model (16). It also acts as a transcriptional repressor for MARβ-mediated transcription from Eβ locus, governing V(D)J recombination in T cells. Transgenic mice overexpressing SMAR1 exhibits lymphoid organomegaly, associated with higher infiltration of lymphoid cells (17). Further, a drastic downregulation of SMAR1 in higher grades of breast cancer and cancer-derived cell lines like MCF-7, HBL-100, ZR 75.3 and ZR 75.1 has also been observed. (14,18).
In the present study, we looked for a possible reason for downregulation of SMAR1 in breast cancer-derived cell lines and a therapeutic agent to restore SMAR1 levels. We find that the stability of SMAR1 mRNA in MCF-7 cells is low and treatment of this cell line with antitumor agent PGA2 increases the stability of the transcript. The increase in SMAR1 mRNA stability is attributed to a 18 bp stem and loop structure of SMAR1 5′ UTR (ϕ1-UTR) that serves as a cognition site for binding of regulatory proteins, essential for conferring transcript stability. Interestingly, we identify a variant of SMAR1 5′ UTR (ϕ17-UTR) that lacks the stem and loop structure of ϕ1, leading to an altered stability. Moreover, we find that the expression of ϕ17-UTR, correlated to an elevated Cyclin D1 level marks the malignant nature of these breast cancer-derived cell lines. Additionally, the involvement of SMAR1 in PGA2-mediated growth arrest underscores the importance of regulation of such tumor suppressor genes by anticancer agents like PGA2.
MATERIALS AND METHODS
Cell culture and transient transfections
MCF-7 cells were maintained in DMEM supplemented with 10% fetal calf serum (Invitrogen) in presence of 5% CO2 at 37°C. Cells were seeded at a density of 1 × 106 per 35 mm dish and cultured for 24 h before transfection. MCF-7 cells were treated with PGA2 at various concentrations in fresh complete medium. Cells were harvested 24 h after PGA2 treatment. Actinomycin D (Sigma) was used at a concentration of 4 μg/ml and incubated for indicated time points.
Semi-quantitative RT–PCR
Seven micrograms of total RNA was subjected to reverse transcription in 20 μl reaction mixture containing 1× random hexanucleotide mix, 1 mM dNTPs, RNase inhibitor and MuMLV reverse transcriptase (Invitrogen). PCR reactions were carried out in a 25 μl reaction mixture using 1 μl cDNA as template. Gene-specific primers used are listed in Table 1.
Table 1.
Different sets of primers and oligos used for assays
| Figure | Primer/Oligo | Product |
|---|---|---|
| 1A and B, 2 C, D, 5 B, C | For-AGACAAACACCACGAGAAT Rev- CGGAGTTCAGGGTGATGAGTGTGAC | SMAR1 (Predominantly ϕ1 form) |
| 1C, 5 B, C | For- CTTCCTCTCCAAAATGCCAG Rev- AGAGATGGAAGGGGGAAAGA | Cyclin D1 |
| 1, 2, 5 | For- TACCACTGGCATCGTGATGGACT Rev- TTTCTGCATCCTGTCGGCAAT | β-actin |
| 2 E, G | For-CGGCACGAGACAAACACCA Rev-TGCAATCTGAACCACATCCGC | Composite UTR |
| 2 F | For-AGACAAACACCACGAGAAT Rev -TTCATCATCATCTCGTCACGA | ϕ1 |
| 2 F | For-CAGCAGCCGCAGCCCACAC Rev -TTCATCATCATCTCGTCACGA | ϕ17 |
| 2 A, B | Amplicon from For-AGACAAACACCACGAGAAT Rev- CGGAGTTCAGGGTGATGAGTGTGAC cloned in PGEM-T Easy | SMAR1 (recognizes ϕ1 form predominantly) |
| 2I | Amplicon from For-AGACAAACACCACGAGAAT Rev -TTCATCATCATCTCGTCACGA cloned in PGEM-T Easy | ϕ1 UTR |
| 2I | Amplicon from For-CAGCAGCCGCAGCCCACAC Rev –TTCATCATCATCTCGTCACGA cloned in TOPO-TA | ϕ17 UTR |
| 4 D–G | CGAAATTAACCCTCACTAAAGGGAACCCACGGTCGACAGAAACC | SL1 |
| 4G | CGAAATTAACCCTCACTAAAGGGAACCACACGGTCGACAGTGACC | Mutant 1 |
| 4 G | CGAAATTAACCCTCACTAAAGGGAACCACATAATCGACAGGGACC | Mutant 2 |
| 5 F, G | For-TGAAAATGAAAGAAGATGCAGTCG, Rev-GAAACTTGCACAGGGGTTGT | Probe II Cyclin D1 promoter |
Real Time RT–PCR
The quantitative measurements of RNA transcripts was performed by icycler iQ thermal cycler system (BioRad) using double-stranded DNA-specific flurophore SYBR Green. In a 25 μl PCR reaction, 1 μl of cDNA was amplified using 1× iQ SYBR Green Supermix (BioRad) containing 0.4 mM dNTP mix, 1.5 mM MgCl2, 50 pmol of forward and reverse primer mix, SYBR Green I, 0.5 U iTaq DNA polymerase. Resolution of the product of interest from nonspecific amplification was achieved by melt curve analysis. Quantitation was done with three different sets of cDNA samples. Sigma plot was used for statistical analysis and plotting graphs.
Cloning of ϕ1 and ϕ17 UTR
The ϕ1 and ϕ17 UTRs were PCR amplified using specific primers from ϕ1 and ϕ17 templates, described previously (19). The PCR products were then cloned in and PGEM-T Easy for ϕ1 and TOPO-TA vector for ϕ17. Plasmids isolated from positive colonies were digested with EcoR1 and cloned in pEGFP vector.
RNase protection assay (RPA)
To determine the half-life of SMAR1, Actinomycin D at a final concentration of 4 μg/ml was added to MCF-7 cells. For stability assays, PGA2 (70 μM) and Actinomycin D were added simultaneously to the culture media. Cells were harvested at different time points post-treatment, washed in 1× PBS and pelleted at 1000 r.p.m. Total RNA was isolated using Tri-Reagent (Sigma) and used for protection assay. Antisense RNA probes were made by linearizing ϕ1 and ϕ17 SMAR1 templates using Nco1 or Pst1, respectively and performing in vitro transcription in the presence of SP6 or T7 RNA polymerase, respectively. In a 25 μl transcription reaction containing 1 μg of linearized DNA fragment, 1× transcription buffer (Stratagene), 0.4 mM each of ATP, CTP, GTP, 0.25 mM UTP and 50 μCi [α32p] UTP (BRIT), 40 U of RNasin and 20 U of T7/SP6 RNA polymerase (Stratagene) were used to label RNA. Labeled RNA transcripts were then purified using probe quant G-50 columns (Amersham Pharmacia). Samples were heated at 90°C for 3 min before hybridization. For the hybridization reaction, 10 μg of total RNA was incubated with labeled probe (∼105 c.p.m.) in hybridization buffer at 37°C for 16 h. The reaction mixtures were then diluted using 300 μl of digestion buffer. Single-stranded RNA was then digested using RNaseT1 (15 μg/ml) and RNaseA (1 μg/ml) for 2 h at 30°C. After phenol/chloroform extraction and ethanol precipitation, samples were analyzed on 6–10% urea gel. Band intensities were quantified using phosphorimager (BioRad).
RNA electrophoretic mobility shift assays (EMSAs)
PBK-CMV ϕ1 and ϕ17 were linearized with Xho1 and in vitro transcribed with T3 polymerase to yield the respective 5′ UTR probes. The wild-type and mutant UTR minor stem and loop sequences were synthesized as forward and reverse complementary oligonuceotides (Genomechanix), containing T3 polymerase site. They were then annealed with respective pairs and in vitro transcibed with T3 polymerase to yield the probes. The sequences are:
SL1-Fwd: CGAAATTAACCCTCACTAAAGGGAACCCACGGTCGACAGAAACC,
Mutant 1-Fwd: CGAAATTAACCCTCACTAAAGGGAACCACACGGTCGACAGTGACC,
Mutant 2-Fwd: CGAAATTAACCCTCACTAAAGGGAACCACATAATCGACAGGGACC and their reverse complementary. EMSAs were performed in a 10 μl binding reaction containing 10 μg of MCF-7 cell extract, either untreated or treated with PGA2 and 12 fmol of transcript, 10 mM Tris (pH 7.4), 15 mM KCl, 5 mM MgCl2 and 10% glycerol. For competition assays, cold competitors were mixed with the hot probe 10 min prior to incubation with lysate. All reactions were carried out on ice for 20 min and then run on 8% native gel, resolved, dried and processed for autoradiography.
Immunoblotting and antibodies
Cells were scraped in 1× PBS, collected at various time points and lyzed using DIGNAM buffer. Equal amount of proteins were taken for immunoblotting. Following SDS–PAGE, the resolved proteins were transferred to PVDF membrane (Amersham). Blocking was carried out with 5% BSA in Tris-buffered saline containing 0.1% Tween-20 (TBST). The membranes were probed with primary and respective secondary antibody. Proteins were detected using ECL plus chemiluminescence substrate (Amersham). Antibodies used are Cyclin D1 and Actin (Santa Cruz). SMAR1 polyclonal antibody was raised in house as described earlier (14). Mouse secondary HRP and rabbit secondary HRP were purchased from BioRad.
Immunostaining
MCF-7 cells were seeded on cover slips and after 24 h, the cells were treated with indicated amount of PGA2. Posttreatment, cells were washed with ice-cold 1× PBS and the cells were fixed with 2% PFA for 15 min at room temperature. After subsequent quenching and blocking in PBS containing 10% FCS, SMAR1 antibody was added onto the cover slips and incubated for an hour. After three washes with cold PBS, cells were incubated with secondary rabbit FITC in dark for 45 min. Cells were then washed three times with ice-cold PBS and mounted using Anti-fade (DABCO) from Sigma. The mounted slides were then visualized using confocal laser microscope (LSM-510, Zeiss, Thornwood, NY).
Luciferase reporter assay
For luciferase assays, 1 μg of Cyclin D1 luciferase promoter construct (CD1) was cotransfected with ϕ1 or ϕ17 SMAR1 with an internal GFP control in MCF-7 cells. Luciferase assays were performed 24 h posttransfection or PGA2 treatment. For SMAR1 knockdown experiments, 100 nM siRNA was cotransfected with Cyclin D1 promoter construct. Luciferase activity was measured using Luclite substrate (Perkin Elmer, USA) and assay performed using Top-Count luminometer (Packard Life sciences, USA). Equal amounts of protein were used for luciferase assays and relative light units plotted for luciferase activity.
Chromatin Immunoprecipitation (ChIP) analysis
ChIP assay kit (Upstate Biotechnology) was performed following manufacturer's instructions. 1 × 106 cells were plated per 30 mm dish and treated with PGA2 or vehicle. After treatment, DNA–protein complexes were fixed with 1% formaldehyde at 37°C for 10 min at various time points. ChIP assays were carried out using anti-SMAR1, anti-HDAC1, anti-H3K9, anti-H4K10, RNA pol II, H3 pSer10 and H3K9 methyl antibodies (Cell Signaling). Input DNA, rabbit IgG (r-IgG), and mouse IgG (m-IgG) pulled DNA served as controls for all the experiments. Immunoprecipitated DNA was then subjected to 28 cycles of PCR using primers for probe II described earlier (14).
Cell cycle analysis
MCF-7 cells were plated at a density of 1 × 105 on a 35 mm dish. After 6 h of serum starvation, synchronized cells were transfected with SMAR1 siRNA, ϕ1 or ϕ17 SMAR1 plasmid constructs using Lipofectamine 2000 (Invitrogen). Twelve hours posttransfection, cells were treated with 30 and 70 μM PGA2 in case of siRNA transfection and cells were harvested 36 h posttransfection. After trypsinization, cells were collected and washed in PBS and pelleted at 1000 r.p.m. The resulting pellet was then re-suspended in 0.2 ml 1× PBS containing 2% fetal bovine serum and fixed in 0.8 ml 70% ethanol. Cells were kept in the fixative for 2 h and then centrifuged for 5 min at 1000 r.p.m. Cells were then treated with RNase A (75 U/ml) and re-suspended in 0.5 ml PBS containing 50 mg/ml propidium iodide (PI) and 0.1% Triton X-100. This was incubated at room temperature for 30 min and subjected to flow cytometry. PI-stained cells were analyzed for cell cycle profiles by FACS Vantage (Becton Dickinson) using Cell Quest.
RESULTS
PGA2 induces SMAR1 expression
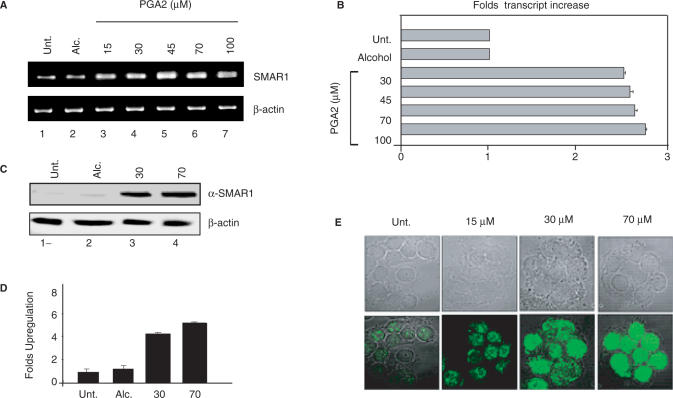
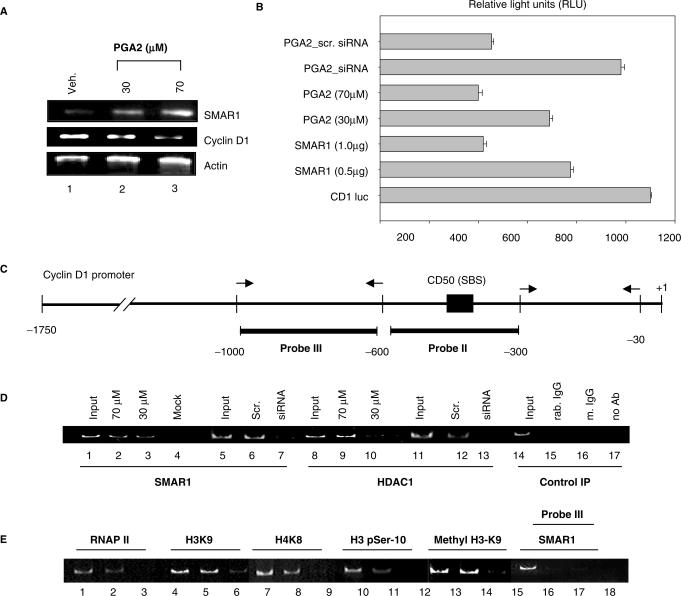
The negative regulation of Cyclin D1 by SMAR1 and PGA2 are well documented, though the mechanisms reported are different (8,14). This prompted us to check the interdependence of these pathways and look for a possible regulation of SMAR1 by PGA2. To evaluate this study, we employed MCF-7 cell line derived from mammary epithelia that is characterized by low invasive potential and induced Cyclin D1 level correlated to reduced SMAR1 expression (14). MCF-7 cells were treated either with vehicle (ethanol) or PGA2 (0–100 μM). Total RNA was isolated 24 h posttreatment and the cDNA obtained was subjected to RT–PCR and Real time RT–PCR analysis. SMAR1 transcript levels increased upto 45 μM PGA2 treatment, after which a steady-state level of the transcript was observed (Figure 1A). Quantitation by Real time RT–PCR showed that SMAR1 transcript increased 1.8- to 2-fold at 30–100 μM PGA2 (Figure 1B). The transcript profile was normalized using β-actin. PGA2-induced SMAR1 protein levels were checked using western blot analysis using 30 and 70 μM PGA2. At these concentrations, the protein expression increased by 3.8- and 4.5-fold respectively, while the protein levels were very low in untreated and vehicle-treated cells (Figure 1C and D). The steady increase in the protein amounts were then visualized using confocal microscopy. MCF-7 cells were treated with PGA2 (15, 30 and 70 μM) and the protein levels were monitored using SMAR1 antibody. A steady increase in the amount of SMAR1 was observed from 15 to 70 μM PGA2 concentrations (Figure 1E).
Figure 1.
PGA2 induces SMAR1 expression. (A) MCF-7 cells were treated with vehicle (alcohol) and various concentrations of PGA2. Total RNA was obtained from aforementioned samples and used for reverse transcription. SMAR1 and β-actin transcript levels were studied using 1 μl of cDNA in RT–PCR (B) SMAR1 transcript was quantified from the above obtained cDNA by real-time–PCR analysis. Bar graphs represent the fold changes in the transcript after PGA2 treatment. (C) Western blot analysis of SMAR1 and β-actin was performed from PGA2-treated or vehicle-treated MCF-7 cell extracts as indicated. (D) Densitometry analysis for protein expression of SMAR1 reveals that protein levels increase by 3.8- and 4.5-fold upon 30 and 70 μM PGA2 treatments. (E) Immunostaining of MCF-7 cells using anti-SMAR1 antibody was performed upon PGA2 treatment at 15, 30 and 70 μM concentrations. All the results represented indicate the average of three independent experiments.
PGA2 stabilizes SMAR1 mRNA
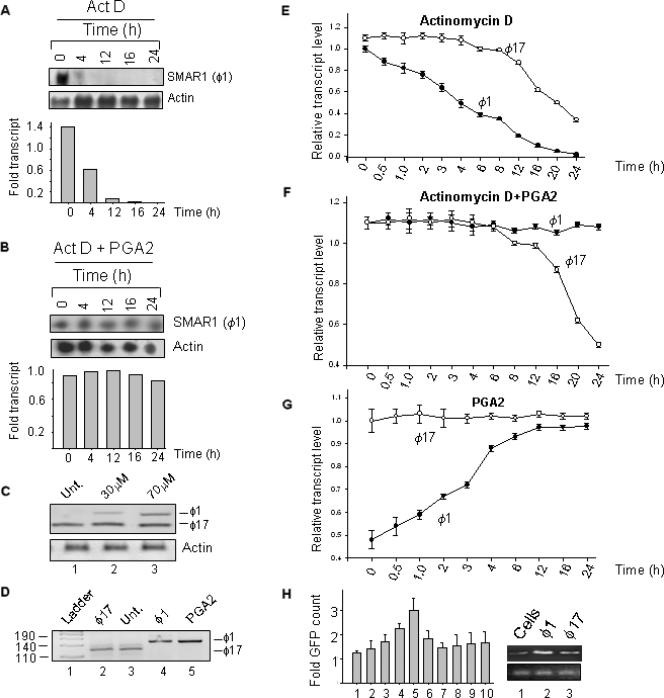
It is well known that most of the reported prostaglandin-mediated effects occur through alteration in mRNA half-life. Results from the previous section show that PGA2 treatment increased SMAR1 transcript by 2-fold but the protein level by 4-fold, indicating a possible involvement of mRNA stability. To address this issue, half-life of endogenous SMAR1 transcript was verified by blocking cellular transcription using Actinomycin D (4 μg/ml) and cells were collected at indicated time points (Figure 2A). Total RNA was isolated and subjected to RPA as described in ‘Materials and Methods’ section. As shown in Figure 2A upper panel, SMAR1 transcript level decreased after 4 h in Actinomycin D-treated samples. Interestingly, upon addition of Actinomycin D and PGA2, we observe that the levels of SMAR1 transcript remained steady till 24 h (Figure 2B, upper panel). This indicates that PGA2 stabilizes SMAR1 mRNA and maintains the steady-state levels of the transcript. The mRNA available at the given time points were normalized to the actin transcript and quantification represented as bar graph (Figure 2A and B).
Figure 2.
PGA2 stabilizes SMAR1 mRNA. (A) Equal number of MCF-7 cells were seeded in 35 mm dishes and 24 h later treated with 4 μg/ml Actinomycin D. Cells were collected after indicated time points and total RNA isolated using Tri reagent. In case of PGA2 treatment, 70 μM PGA2 was added to cells 12 h prior to Actinomycin D treatment. Stabilization of SMAR1 transcript by PGA2 was verified using RPA. Antisense probe from Nco1 digested SMAR1 fragment from PGEM-T Easy was used and protection assays were performed as described in ‘Materials and Methods’ section. Upper panel shows that the half-life of SMAR1 in MCF-7 cells is about 4 h. (B) Upper panel shows protection of SMAR1 mRNA over the indicated time intervals upon Actinomycin D and PGA2 treatment. The lower panels show actin mRNA levels used as control. The bar graphs both in A and B represents the quantification of transcripts normalized to actin. (C) RT–PCR analysis of cDNA from untreated and PGA2-treated MCF-7 cells using composite UTR primers reveals the amplification of a lower size fragment in cells (lane1) while in treated sample, the upper band becomes prominent with increasing PGA2 concentration (lanes 2 and 3). (D) Amplification of specific UTRs of cDNA from control and PGA2-treated cells reveals that ϕ17 is present in untreated cells (lane 3) and ϕ1 in PGA2-treated samples (lane 5) as checked by amplification with template control (lanes 2 and 4). The pUC mix marker (Fermentas) in lane 1 was used to identify the molecular size of the fragments. (E) Quantitation of transcript derived from ϕ1 and ϕ17 after Actinomycin D treatment reveals that the half-life of ϕ1 is between 4 and 6 h and that of ϕ17 between 14 and 16 h. The transcripts were normalized to β-actin control. (F) Real-time–PCR analysis of ϕ1 and ϕ17 transcript upon PGA2 and Actinomycin D treatment reveals that ϕ1 form remains constant while the ϕ17 form does not respond to PGA2 treatment and there is a decline in the transcript after 16 h time point. The transcripts were normalized to β-actin control. (G) Real-time–PCR analysis of ϕ1 and ϕ17 transcripts in PGA2-treated MCF-7 cells reveals that the ϕ1 form increases 1.5–2-fold between 6 and 16-h interval and thereafter remains steady till 24-h time point while ϕ17 amount remains almost unchanged after PGA2 treatment. (H) The responsiveness of the two UTRs to PGA2 was verified using GFP reporter assay. One microgram of each of the UTR-GFP vector was transiently transfected in separate sets of experiments, cells collected 24 h posttransfection and verified for the GFP counts after addition of PGA2 (30 and 70 μM). There was a 1.8–2-fold increase in the GFP count in the ϕ1 UTR (bars 3–5) compared to the ϕ17 UTR (bars 7–9) that did not show any significant increase in GFP count. Lanes 6 and 10 represent vehicle controls with each of the UTR plasmids. EGFP vector and ethanol were used as controls (lanes 1 and 2). Transfection of plasmids was verified using SMAR1 RT–PCR (right panel). The bars are a depiction of three sets of experiments and error bars denote the mean SD.
SMAR1 has two 5′ UTR variants in MCF-7 cells
The stability and translation of most of the transcripts are associated with the untranslated regions, both at 5′ and 3′ UTRs. Primers were designed to amplify the sequence from the start of exon1 and a part of exon 2 that was expected to yield an intact SMAR1 5′ UTR of 142 bp (18). The amplimer of 5′ UTR from the cDNA of MCF-7 cells untreated and treated with PGA2 were checked on a 10% polyacrylamide gel. Interestingly, untreated cells showed a single amplimer (Figure 2C, lane 1) while in case of PGA2 treatment (30 and 70 μM), there was an additional amplimer (Figure 2C, lanes 2 and 3). The fast migrating amplimer (lower band) was common to both untreated and PGA2 treated cells, while there was an induction of the slower migrating amplimer (upper band) specifically in PGA2-treated cells. Thus, the upper and the lower bands represent two variants of 5′ UTR. Earlier, we have reported the identification of three clones of SMAR1 from mouse thymocyte cDNA library (19). Out of these, two clones shared the same open reading frame and same translational reading frame of downstream sequence, but differed only by 18 bases in 5′ UTR. The two clones were denoted as ϕ1 (containing full-length 5′ UTR) and ϕ17 (containing variant 5′ UTR lacking 18 bases). When checked using ϕ1 and ϕ17 template controls (Figure 2D, lanes 2 and 4), we find that the size of upper band obtained upon PGA2 treatment corresponded to the ϕ1 form (the upper band in Figure 2C, lanes 2 and 3; Figure 2D, lane 5) and the lower band in untreated and PGA2-treated cells corresponds to ϕ17 form (the lower band in Figure 2C, lanes 1–3, Figure 2D, lane3). This was further verified by cloning the corresponding PCR products in PGEMT-Easy vector and DNA sequencing. RT–PCR analysis of cDNA from MCF-7 cells (untreated or treated with 70 μM PGA2, lanes 1 and 2, respectively) with primers specific for ϕ1-UTR and ϕ17-UTR forms was performed. The results revealed that upon PGA2 treatment there was no apparent change in the ϕ17 form but there was an increase in the ϕ1 form of SMAR1 (Supplementary Figure S1A). The map of SMAR1 gene and the primers used are depicted in Supplementary Figure S1B.
The responsiveness of 5′ UTR to PGA2 is essential for SMAR1 mRNA stability
Next we checked if the responsiveness of SMAR1 UTR to PGA2 contributed to the stability of SMAR1 transcript. The antisense UTR transcript specific to each form was hybridized with the total RNA from MCF-7 cells untreated or treated with PGA2, as described in ‘Materials and Methods’ section. Protected bands revealed that the ϕ17 form was predominant in untreated cells, unlike the ϕ1 form (almost 3–4-times lower than ϕ17, verified by densitometry). An increase in protection of ϕ1 transcript was observed upon PGA2 treatment while ϕ17 form remained almost unchanged (Supplementary Figure S1C). To validate the transcript profiles of ϕ1-UTR and ϕ17-UTR containing SMAR1, cDNA obtained was subjected to Real time RT–PCR analysis using primers specific to the UTRs and melt curve analysis performed. The transcript profile obtained after Actinomycin D treatment showed that the half-life of ϕ1 was 4–6 h while that of ϕ17 varied between 14–16 h (Figure 2E). Though the transcript level of ϕ1 remained stable after PGA2 and Actinomycin D treatment, ϕ17 transcript levels started to decline from 12 h, as verified by melt curve analysis (Figure 2F). Quantification of transcripts revealed that ϕ1-SMAR1 level increased 2-fold at 6 h time point of PGA2 treatment and remained steady till 24 h, while there was a negligible change in ϕ17-SMAR1 level (Figure 2G). These results point out at the early response of ϕ1-UTR to PGA2 treatment that stabilizes the transcript till 24 h. To further assess the response of the two UTRs to PGA2, we performed reporter assays, where the UTRs from both forms were cloned upstream of gfp gene in pEGFP vector. In case of ϕ1-UTR transfection, we observed a 2–3-fold increase in GFP expression upon PGA2 treatment compared to the untreated cells (Figure 2H, lanes 3–5). ϕ17-UTR transfection however showed only a minor change in the basal expression of GFP, showing that ϕ1 and not ϕ17 is responsive to PGA2 (Figure 2H, lanes 7–9). Thus, we demonstrate that in MCF-7 cells, ϕ1 form of SMAR1 and not ϕ17 form responds to PGA2.
ϕ1-UTR stem and loop structure is critical for PGA2-induced complex formation
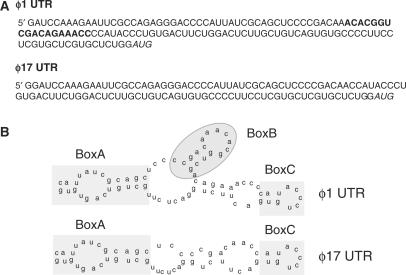
The natural tendency of RNA is to form highly stable secondary and tertiary structures and the alteration in these structures represent a well-known regulatory mechanism for many cellular processes (20). To determine the differences in the secondary structure that could implicate a functional significance, we analyzed the ϕ1 and ϕ17 UTR sequences as shown in Figure 3A. Prediction of the secondary structures of the two UTRs was done using M-fold secondary structure prediction software (21). After energy minimizations, we observed that ϕ17-UTR lacks a small stem and loop structure, pertaining to those missing 18 bases (Figure 3B).
Figure 3.
The ϕ1 form hosts an extra stem and loop (A) The sequence of ϕ1 and ϕ17 reveals that the 5′ UTRs vary by 18 bases. The missing bases are indicated in bold and the transcription start site indicated in italics. (B) Secondary structure analysis of the UTRs by M-fold prediction software indicates the presence of an extra stem and loop structure, marked by a shaded box indicated as BoxB. BoxA and BoxC denote the structures that are common in both the RNA.
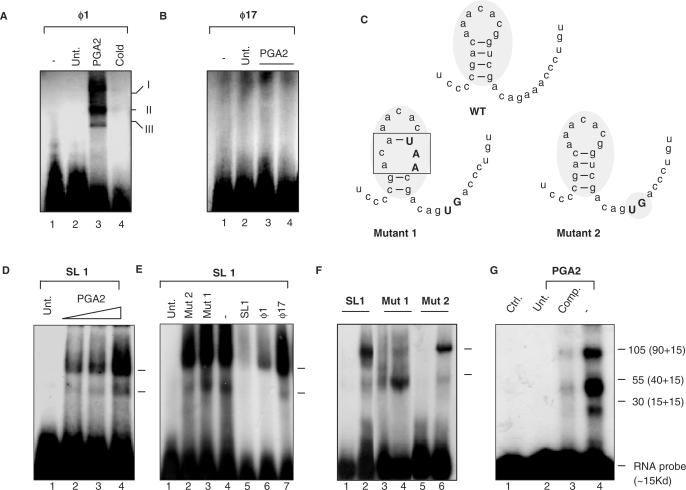
The major factor that controls mRNA turnover is the association of RNA-binding proteins with sequences forming stable secondary structures. We checked for the association of such proteins with SMAR1 UTR, in the presence and absence of PGA2. RNA EMSA were performed to check for putative nucleoprotein complex formation on ϕ1-UTR. Labeled RNA obtained by transcription of the respective 5′ UTR templates (ϕ1 and ϕ17) were used to perform EMSAs. The binding reactions were performed as described in ‘Materials and Methods’ section. The mixture was then run on 6% native gel and subjected to autoradiography. We could detect three specific nucleoprotein complexes on ϕ1-UTR, in the PGA2-treated lane alone compared to the untreated cell lysate (Figure 4A, lanes 2 and 3). Upon addition of 50-fold molar excess self-cold competitor, we find a complete abolishment of all three shifted complexes, revealing the specificity of the complex formation (Figure 4A, lane 4). ϕ17-UTR however failed to form nucleoprotein complex, indicating that the missing 18 bases holds the key determinant to bind to certain factors that in turn govern the stability and translation of SMAR1 (Figure 4B). To validate this, RNA was transcribed from the ϕ1 template encoding the stem and loop structure (named SL1 hereafter). Using this transcript as probe, EMSAs were performed as described earlier. SL1 also showed nucleoprotein complex formation, comparable to the ϕ1-UTR upon PGA2 treatment (Figure 4D). Further, competition experiments with 10-fold molar excess cold competitors showed the specificity of the complex (Figure 4E). To identify the importance of the secondary structure imparted by the sequence, binding assays using two mutants were performed. Mutant 2 had a minor modification of the primary sequence but the secondary structure remained largely unperturbed (where nucleotides outside boxB were modified) while in Mutant 1, the stem–loop structure pertaining to boxB was predicted to be partly disrupted (Figure 4C). When compared to SL1 (Figure 4F, lane 2) both Mutant 1 and Mutant 2 showed a reduced complex formation, although to varying degrees. Mutant 1, where the core stem structure was disrupted, lacked the specific complex formation compared to Mutant 2 (Figure 4F, lanes 4 and 6, respectively). Thus, we emphasize that the secondary structure imparted by these 18 bases to SMAR1–UTR is critical for the complex formation. Further, to identify the number and putative molecular weights of the components of specific nucleoprotein complex, UV cross-linking experiments were performed as described in ‘Materials and Methods’ section. The binding mixture employing SL1 from the intact UTR as probe was subjected to UV cross-linking, resolved on 10% SDS–PAGE gel followed by autoradiography. After subtraction of the molecular weight of the RNA probe (∼15 kDa), we find the involvement of three specific proteins of ∼90, 40 and 15 kDa in forming the ribonucleoprotein complex (Figure 4G). Though the identity of these factors remains unknown, we believe that these factors that bind to the stem and loop structure of UTR upon PGA2 treatment could be the key in stabilizing the SMAR1 mRNA, resulting in altered protein levels.
Figure 4.
SMAR1 UTR binds to complexes in a PGA2-dependent manner. EMSAs for different UTRs were performed as described in Materials and Methods section. (A) PGA2 induces ribonucleoprotein complex formation (indicated as I, II and III) on SMAR1 5′ UTR probe. PGA2 treatment (70 μM) induces RNP complex formation on the 5′ UTR of ϕ1 (lane 3). Ten-fold molar excess cold self-competitor demonstrates the specificity of the complex (lane 4) (B) The ϕ17 5′ UTR of SMAR1 does not show any complex formation under the same conditions (lane 1: free probe, lane 2: cells only, lanes 3 and 4: 30 and 70 μM PGA2 treatment). (C) Depiction of the secondary structure of the wild-type and the mutant 18-mer. The upper case fonts indicate the mutated residues and the shaded box represents the change in the secondary structure. (D) The synthetic stem–loop structure of ϕ1 UTR (SL1) shows complex formation upon PGA2 treatment in a dose-dependent manner. (E) Competition studies using specific and nonspecific competitors on SL1 to show the specificity of complex formation upon PGA2 treatment. (lane 1: free probe, lane 2: Mutant 2, lane 3: Mutant 1, lane 4: no competitor, lane 5: SL1, lane 6: ϕ1, lane 7: ϕ17). (F) SL1 and Mutant 1 and 2 were used as probe as EMSAs performed upon PGA2 treatment. Mutant 1 showed a drastic reduction in the nucleoprotein complex formation (lanes 3: untreated cell lysate, lane 4: PGA2 treated cell extract) while Mutant 2 (lane 5: untreated cell lysate, lane 6: PGA2-treated cell lysate) showed a similar albeit a lesser complex formation to SL1 (lane 1: untreated cell lysate, lane 2: PGA2-treated cell extract), indicating the importance of the intact secondary structure for complex formation. (G) Samples were prepared as mentioned in ‘Materials and Methods’ section and subjected to UV cross-linking. After incubation, samples were size fractionated using SDS–PAGE. Three predominant species of ∼105, 55 and 30 kDa were resolved and observed (lane 4). Specific cold competitor (lane 3) and probe alone (lane 1) were then used to validate the complex formation. The actual molecular weights was obtained by deduction of 15 kDa (the molecular weight of ∼45 nt RNA) from these and the correct sizes identified to be ∼90, 30 and 15 kDa.
The responsiveness of 5′ UTR to PGA2 is essential for Cyclin D1 downregulation
Since SMAR1 is known to possess transcriptional regulatory functions, the implication of the responsiveness of 5′ UTR to PGA2 in vivo was verified. The inverse correlation of Cyclin D1 downregulation and SMAR1 induction was first verified upon treatment with 30 and 70 μM PGA2 (Figure 5A). The results so far suggest that PGA2 treatment stabilizes ϕ1 mRNA, the product of which results in Cyclin D1 repression. To confirm this, we performed luciferase reporter assays where Cyclin D1 regulation by ϕ1 and PGA2 were compared. Cotransfection of 0.5 and 1 μg of ϕ1 SMAR1 along with Cyclin D1 promoter construct (CD1-luc) revealed that ϕ1 downregulated Cyclin D1 by 1.5- and 2.5-fold, comparable to PGA2-treated samples. Moreover, PGA2 treatment after knockdown of SMAR1 using the specific siRNA showed inefficient downregulation of Cyclin D1 (Figure 5B). This reveals the importance of SMAR1 produced from ϕ1 transcript in PGA2-mediated repression of Cyclin D1. As discussed earlier, the recruitment of SMAR1 and the associated corepressor complex to Cyclin D1 promoter is well documented, and hence we verified if SMAR1 occupied Cyclin D1 promoter in response to PGA2 (Figure 5C). We observed the interaction of HDAC1 and SMAR1 upon PGA2 treatment (Supplementary Figure S2A and B). Chromatin immunoprecipitation experiments using MCF-7 cells either untreated or treated with 70 μM PGA2 were performed. Amplification of probe II region in SMAR1 and HDAC1 immunoprecipitated from PGA2 treated cells (30 and 70 μM) showed the recruitment of SMAR1 corepressor complex on Cyclin D1 promoter (Figure 5D, lanes 2 and 3, 9 and 10,). siRNA-treated, SMAR1-pulled chromatin was used to verify the specificity of the recruitment (Figure 5D, lanes 6 and 7, 12 and 13). These results are consistent with the direct recruitment of HDAC1 and SMAR1 on Cyclin D1 promoter that is responsible for the observed repressive effects. Upon over expression, SMAR1 is shown to deacetylate the histones at H3K9 and H4K8 loci that is accompanied by dephosphorylation of H3-phospho-ser 10 at Cyclin D1 promoter locus. Therefore, we checked if these modifications persist upon PGA2 treatment or if any additional histone modifications are involved in repression of Cyclin D1. We observed a decrease in acetylation at H3K9 (Figure 5E, lanes 5 and 6), H4K8 (Figure 5E, lanes 8 and 9) and phosphorylation of H3p-ser 10 (Figure 5E, lanes 11 and 12) in PGA2-treated cells compared to mock-treated cells. In addition to this, H3K9 monomethylation status that is well documented to have a role in transcriptional activation was investigated. As shown in Figure 5E, lanes 14 and 15, PGA2 treatment decreased the methylation of probe II region compared to mock treatment. To establish the repression of transcription brought about by SMAR1, the recruitment of RNAP II on Cyclin D1 promoter locus was studied. Decreased amplification of probe II region in RNAP II-pulled samples compared to mock treated suggests decreased transcription from Cyclin D1 promoter locus (Figure 5E, lanes 2 and 3). Nonspecific probe III was used as negative control (lanes 17 and 18). Mouse and rabbit IgG served as negative controls (Figure 5D, lanes 15 and 17).
Figure 5.
Responsiveness of SMAR1 5′ UTR is essential for exerting its transcriptional modulatory function. (A) RT–PCR analysis of SMAR1, Cyclin D1, and β-actin transcript levels in PGA2-treated (lanes 2 and 3 at 30 and 70 μM PGA2) or vehicle-treated MCF-7 cells (lane 1). Actin was used as loading control (lower panel). (B) Luciferase activity from Cyclin D1 promoter was studied after transfection with the indicated plasmids, siRNA or PGA2 treatment and relative light units were plotted in the bar graph. Experiments were performed as three independent sets and error bars denote the mean deviation. From below, Bar 1 denotes the luciferase activity of CD1 promoter. Bars 2 and 3 denote the repression of Cyclin D1 by the intact UTR containing SMAR1 while bars 4 and 5 indicate down regulation of Cyclin D1 by the 30 and 70 μM PGA2. Use of SMAR1 specific siRNA and PGA2 treatment shows inefficient downregulation of Cyclin D1 luciferase activity. (C) Schematic representation of Cyclin D1 promoter and SMAR1-binding site (SBS) in probe II region. (D) Chromatin immunoprecipitation was performed as described in ‘Material and Methods’ section. Recruitment of SMAR1 (lanes 2 and 3) and HDAC1 (lanes 9 and 10) on probe II region of Cyclin D1 promoter was studied upon PGA2 treatment in MCF-7 cells using antirabbit SMAR1 and antimouse HDAC1 antibody. Endogenous SMAR1 was depleted using SMAR1 siRNA and studied for recruitment of SMAR1 (lane 7) and HDAC1 (lane13) in MCF-7 cells treated with PGA2 (70 μM). Scrambled siRNA (scr) was used as negative control that did not alter SMAR1 or HDAC1 recruitment (lanes 6 and 12, respectively). Chromatin immunoprecipitation using mouse IgG, rabbit IgG or no antibody are used as negative control (lanes 15–17). Reverse cross-linked sheared genomic DNA is used as positive control (input) for amplification (lanes 1, 5, 8, 11 and 14). Transfection of empty vector was performed as mock control (lane 4). (E) To study the status of histone modifications at Cyclin D1 promoter upon PGA2 treatment, ChIP assays were performed using antibodies against RNA pol II, acetyl H3K9, acetyl H4K8, H3 p-ser10 and monomethyl H3K9 and in MCF-7 cells untreated (lanes 2, 5, 8, 11 and 14) or treated with 70 μM PGA2 (lanes 3, 6, 9, 12 and 15) and Probe II region of Cyclin D1 promoter was amplified from the immunoprecipitated DNA. Reverse cross-linked sheared genomic DNA is used as positive control (input) for amplification (lanes 1, 4, 7 and 10). Probe III region was used as control to verify the specificity of SMAR1 binding to the promoter region.
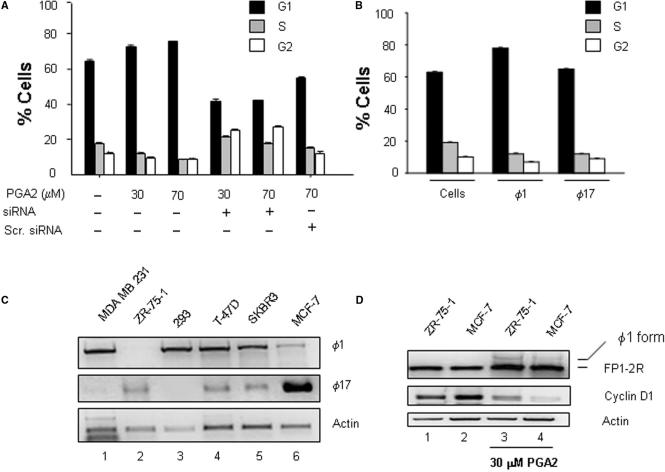
Cell cycle arrest function of PGA2 mediated by SMAR1 depends on the 5′ UTR
Previous studies by Bhuyan et al. (22) showed that PGA2 exerts growth inhibitory activity in many human cell lines. Consistent with this, we found that PGA2 treatment caused G1 phase arrest by DNA content analysis using flow cytometry. We then checked if this growth inhibitory effect of PGA2 is mediated by SMAR1. Hundred-nanomolar SMAR1 siRNA was transfected in MCF-7 cells and 16 h posttransfection, PGA2 was added to the culture media. Cell cycle analysis was done 24 h post-PGA2 treatment, after checking the knockdown of SMAR1 (data not shown). There was a marked shift of the cells from G1 to S and G2 phase, indicating that PGA2 was no longer able to arrest cells in G1 phase in the absence of SMAR1 (Figure 6A). The results so far suggest the involvement of PGA2 in stabilizing ϕ1 SMAR1 and it is also clear that SMAR1 is required for PGA2-mediated growth arrest. This prompted us to investigate the difference in growth arrest function of SMAR1, in context of the two UTRs it can host. For this, MCF-7 cells were transiently transfected with equal amounts of ϕ1 or ϕ17-SMAR1 and analyzed for cell cycle progression. DNA content analysis revealed a 1.2–2.5-fold higher number of cells in G1/S phase in case of ϕ1-SMAR1 transfection in comparison to ϕ17- SMAR1 (Figure 6B). This hints that the growth inhibitory effect of PGA2 is mediated at least partially by ϕ1-SMAR1. To check if the PGA2-mediated induction of SMAR1 specifically leads to Cyclin D1 downregulation, western blot analysis to check the status of other cyclins was performed. The results were then verified using siRNA treatment of SMAR1. The specificity of SMAR1 upregulation upon PGA2 treatment was also verified using siRNA treatment (Supplementary Figure S3A). The status of other cyclins (Cyclin A, B and associated cyclin kinases cdks 4 and 6) was unaltered (Supplementary Figure S3B). This result was corroborated using cDNA microarray results (Supplementary Figure S4).
Figure 6.
PGA2 requires SMAR1 for its growth-inhibitory function in MCF-7 cells. (A) MCF-7 cells were analyzed by flow cytometry 24 h post-PGA2 treatment in the presence or absence of SMAR1 siRNA. The treatments done are indicated in the graph. The graph represents the average percentage of cells in G1, S and G2 phase in three independent experiments and error bars denote SD. Analysis shows that in the absence of SMAR1 (knocked down by siRNA), there is a marked shift of cells from G1 to S and G2 phase. (B) MCF-7 cells transiently transfected with ϕ1 or ϕ17 were collected 24 h posttransfection and processed for DNA content analysis. The graph indicates that ϕ1 retains the cell cycle arrest function while ϕ17 is inefficient in imposing cell cycle arrest at G1/S phase. (C) RT–PCR analysis of UTR specific to ϕ1 and ϕ17 in different breast cancer-derived cell lines. Equal amount of RNA was reverse transcribed and cycling reactions performed under limiting conditions for all sets of primers as indicated. (D) RT–PCR analysis of cDNA from ZR-75-1 and MCF-7 cells untreated (lanes 1 and 2) or treated with 30 μM PGA2 (lanes 3 and 4), product amplified using FP1-2R primers to visualize the composite UTR forms.
Breast cancer-derived cell lines from mammary epithelia primarily express ϕ17 form
To find out if the mechanism of downregulation of SMAR1 by altering the mRNA stability is common to breast cancer-derived cell lines, we screened for the presence of the two forms of SMAR1 UTR in ZR75-1, SKBR-3, T47D and MDA-MB-231. Different cell lines were cultured, RNA isolated and limited cycle RT–PCR analysis was performed. RT–PCR analysis showed that ZR-75-1 failed to show the product that corresponds to ϕ1-UTR form but expresses ϕ17-UTR only (Figure 6C, lane 2 upper and middle panel). This was similar to MCF-7 cells where a very low amplification of ϕ1-UTR transcript was observed (Figure 6C, lane 6, upper and middle panel). Other cell lines like MDA-MB-231, SKBR3 and T-47D expressed the ϕ1-UTR form predominantly and ϕ17-UTR form to a lower extent (Figure 6C, lanes 1, 4 and 5, upper and middle panel). HEK-293 cells, where ϕ1 form is expressed predominantly and ϕ17-UTR is undetectable was used as control (Figure 6C, lane 3, upper and middle panels). The transcript profile we have obtained is in accordance with our previous result, where we have observed that ZR-75-1 and MCF-7 express very low amount of endogenous SMAR1 protein, whereas other cell lines showed a relatively high SMAR1 expression (14). Since the UTR profile of ZR-75-1 and MCF-7 cells was similar, we checked the response of this cell line to PGA2 treatment and compared to MCF-7 cell line. Upon 30 μM PGA2 treatment, we found a significant induction of the ϕ1-UTR form compared to the untreated cells (Figure 6D, lanes 1 and 3, upper panel). This was strikingly similar to MCF-7 cells where PGA2 treatment leads to induction of ϕ1-UTR form (Figure 6D, lanes 2 and 4, upper panel). Cyclin D1 profiles in these cell lines untreated or treated with PGA2 showed that the repression of Cyclin D1 occurred in case of PGA2-treated samples that correlate with the emergence of ϕ1-UTR form.
DISCUSSION
The primary effect of hormonal therapeutics like PGA2 has been documented through alteration of half-life of mRNA (23,24). The significant role of mRNA stability in gene expression makes it an essential component in pathways whereby tissues and organs respond to stress. Several anticancer agents like PGA2 rely on their ability to alter the mRNA stability of target cell cycle regulatory molecules, to govern the cell cycle fate. The essence of mRNA stability is the balance between regulation by destabilization determinants or translational inhibitory structures that are rendered favorable for transcription upon appropriate stimuli (25).
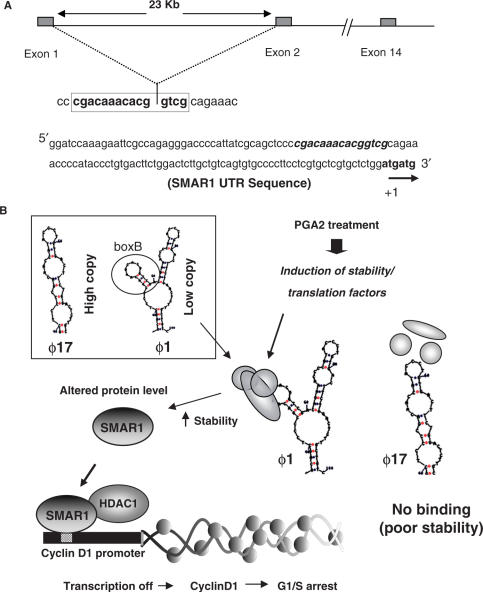
The current study stems from the fact that SMAR1 transcript is found to be drastically downregulated in higher grades of breast cancer and breast cancer-derived cell lines (14,18). Our aim was to decipher the reason for downregulation of SMAR1 in breast cancer-derived cell lines, specifically in context to MCF-7 cell line. Considering that both SMAR1 and PGA2 are documented to halt cell cycle at G1/S phase, it was an interesting possibility that PGA2 regulates SMAR1, thereby bringing about the repression of a common target gene, i.e Cyclin D1. MCF-7 cell line is characterized by low levels of SMAR1 transcript that is attributed to a half-life of 4 h, which in turn leads to a poor endogenous protein level. PGA2 treatment, however, renders the transcript stable till 24 h and elevates SMAR1 level in the cell line. Since majority of the cases involving mRNA stability rely on the ability of the UTR present on the 5′ or 3′ end of the transcript to respond to varying stimuli, the aim was to decipher the role of SMAR1 UTR in conferring stability to the transcript. Amplification of the 5′ UTR end of SMAR1 lead to the identification of a variant form of SMAR1-UTR in MCF-7 cells. This has been previously identified as ϕ17-SMAR1 and possesses the same ORF and reads out SMAR1 protein similar to the ϕ1 clone. SMAR1 transcript profile also reveals that this form is predominant in MCF-7 cells, contributing to almost 65% of the observed total transcript (data not shown). The half-life of the transcript with respect to the two UTR forms is also different, with ϕ1 having a half-life of 4–6 h while that of ϕ17 is 14–16 h. Since the secondary structure of RNA is thought to play a significant role in imparting stability and hence dictating the protein translation, we analyzed the secondary structure of both the UTRs. The ϕ1 UTR of SMAR1 is a thermodynamically stable structure and differs from its variant form (ϕ17) in hosting a minor stem–loop structure, the one formed by the missing 18 bases. Recent studies highlight the importance of binding of regulatory proteins to different structures posed by the RNA (26). Further investigations indicated that this sequence is critical for SMAR1 stabilization brought about by nucleoprotein complex formation on the stem–loop structure. The competition studies using different mutants showed that the secondary structure of the loop was important. It is thus possible that upon PGA2 treatment certain proteins that recognize this fold, bind to the stem and loop structure and regulate mRNA stability, resulting in increased SMAR1. This could be an alternate selection mechanism in tumor cells, whereby a variant UTR containing a transcript, encoding a tumor suppressor protein is retained for yet other unknown functions. Interestingly, we also find that the growth-inhibitory response of cells to PGA2 is in part due to the stabilization of SMAR1 mRNA by 5′ UTR, as the knock down of SMAR1 leads to a compensation in the growth-arrest function of PGA2. Considering that we have identified a 5′ UTR variant that is most likely a splice variant of the full-length UTR, it is tempting to hypothesize that RNA-binding factors involved in splicing regulate SMAR1 mRNA stability. This possibility cannot be ruled out as the splicing site between first and second exon junction of SMAR1 that spans 23 kb, hosts the 18 base stem–loop structure critical for SMAR1 stabilization. It is plausible that the recruitment of ribonucleoproteins like TIA-1 that aid in 5′ UTR splicing (27) is regulated by another stabilizing factor. This is essential for splicing of the 60 bases from the 3′ end of first exon to 5′ end of the second exon, to form the composite stem–loop structure (Figure 7A). In breast cancer-derived cells, low level of endogenous SMAR1 and subsequently high level of Cyclin D1 leads to cell proliferation, malignant transformation, and hence neoplasia. Under conditions that favor proliferation or lead to malignant transformation, cancer cells might have evolved such strategies to downregulate tumor suppressor proteins like SMAR1, as witnessed by low levels of the protein in breast cancer-derived cell lines. Treatment with anticancer therapeutics that lead to cell cycle arrest, reverse this effect and proteins like SMAR1 that downregulate proliferative genes are upregulated. It is also possible that SMAR1 transcript stabilized by PGA2 has a definitive link with the estrogen responsiveness of cell line, at earlier stages of transformation. In case of advanced cancer cases, however, this posttranscriptional control mechanism might be defective. Therefore, in spite of having ϕ1-SMAR1 and expressing SMAR1 protein, the existence of alternative pathways that nullify the effect of SMAR1 cannot be ruled out. Thus, our studies provide a mechanism where a therapeutic agent PGA2 used to treat malignancies, alters the mRNA stability of tumor suppressor protein SMAR1, that downregulates the proliferative gene Cyclin D1.
Figure 7.
Diagrammatic representation of SMAR1 regulation by PGA2. (A) The genomic organization of SMAR1 shows that the untranslated exon 1 and a part of exon 2 (23 kb apart) forms the intact 5′ UTR of SMAR1. The junction of these exons is marked by the stem and loop structure of 5′ UTR. The intact 5′UTR sequence of SMAR1. (B) In MCF-7 cells, ϕ17, a variant 5′UTR of SMAR1 is present in higher amount compared to the ϕ1 form. (E) Factors induced by PGA2 bind to the stem and loop structure of ϕ1, stabilize the mRNA and altered SMAR1 protein level. SMAR1 protein then recruits HDAC1 corepressor complex to Cyclin D1 promoter, represses transcription, and hence causes G1/S phase arrest.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors would like to thank Dr G. C. Mishra, Director, National Centre for Cell Sciences, Pune, India, for his constant support. This work is supported by grants from Department of Biotechnology, Govt. of India. L.P. and S.S. are recipients of Senior Research Fellowships from University Grants Commission and K.S. from Council for Scientific and Industrial Research, Govt. of India. Funding to pay the Open Access publication charges for this article was provided by National Centre for Cell Science, Pune, India.
Conflict of interest statement. None declared.
REFERENCES
- 1.Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- 2.Ing NH. Steroid hormones regulate gene expression posttranscriptionally by altering the stabilities of messenger RNAs. Biol. Reprod. 2005;72:1290–1296. doi: 10.1095/biolreprod.105.040014. [DOI] [PubMed] [Google Scholar]
- 3.Pagano M, Tam SW, Theodoras AM, Beer-Romero P, Del Sal G, Chau V, Yew PR, Draetta GF, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 4.Carlson B, Lahusen T, Singh S, Loaiza-Perez A, Worland PJ, Pestell R, Albanese C, Sausville EA, Senderowicz AM. Down-regulation of cyclin D1 by transcriptional repression in MCF-7 human breast carcinoma cells induced by flavopiridol. Cancer Res. 1999;59:4634–4641. [PubMed] [Google Scholar]
- 5.Hsiang CH, Straus DS. Cyclopentenone causes cell cycle arrest and represses cyclin D1 promoter activity in MCF-7 breast cancer cells. Oncogene. 2002;21:2212–2226. doi: 10.1038/sj.onc.1205293. [DOI] [PubMed] [Google Scholar]
- 6.Fukushima M, Sasaki H, Fukushima S. Prostaglandin J2 and related compounds. Mode of action in G1 arrest and preclinical results. Ann. NY Acad. Sci. 1994;744:161–165. doi: 10.1111/j.1749-6632.1994.tb52733.x. [DOI] [PubMed] [Google Scholar]
- 7.Gorospe M, Liu Y, Xu Q, Chrest FJ, Holbrook NJ. Inhibition of G1 Cyclin-dependent kinase activity during growth arrest of human breast carcinoma cells by prostaglandin A2. Mol. Cell. Biol. 1996;16:762–770. doi: 10.1128/mcb.16.3.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin S, Wang W, Wilson GM, Yang X, Brewer G, Holbrook NJ, Gorospe M. Down-regulation of cyclin D1 expression by prostaglandin A (2) is mediated by enhanced cyclin D1 mRNA turnover. Mol. Cell. Biol. 2000;20:7903–7913. doi: 10.1128/mcb.20.21.7903-7913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan J, Yang X, Wang W, Wood WH, Becker KG, Gorospe M. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc. Natl Acad. Sci. USA. 2002;99:10611–10616. doi: 10.1073/pnas.162212399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santoro MG, Amici C. Control of the growth of a human erythroleukemic cell line by prostaglandins. Adv. Prostaglandin Thromboxane Leukot. Res. 1987;17B:969–971. [PubMed] [Google Scholar]
- 11.Amici C, Sistonen L, Santoro MG, Morimoto RI. Antiproliferative prostaglandins activate heat shock transcription factor. Proc. Natl Acad. Sci. USA. 1992;89:6227–6231. doi: 10.1073/pnas.89.14.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Millard SS, Vidal A, Markus M, Koff A. A U-rich element in the 5′ untranslated region is necessary for the translation of p27 mRNA. Mol. Cell Biol. 2000;20:5947–5959. doi: 10.1128/mcb.20.16.5947-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, Wang W, Fan J, Lal A, Yang D, Cheng H, Gorospe M. Prostaglandin A2-mediated stabilization of p21 mRNA through an ERK-dependent pathway requiring the RNA-binding protein HuR. J. Biol. Chem. 2004;279:49298–49306. doi: 10.1074/jbc.M407535200. [DOI] [PubMed] [Google Scholar]
- 14.Rampalli S, Pavithra L, Bhatt A, Kundu TK, Chattopadhyay S. Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex. Mol. Cell Biol. 2005;25:8415–8429. doi: 10.1128/MCB.25.19.8415-8429.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jalota A, Singh K, Pavithra L, Jameel S, Chattopadhyay S. Tumor suppressor SMAR1 activates and stabilizes p53 through its arginine-serine-rich motif. J. Biol. Chem. 2005;280:9450–9459. doi: 10.1074/jbc.M413200200. [DOI] [PubMed] [Google Scholar]
- 16.Kaul R, Mukherjee S, Ahmed F, Bhat MK, Chhipa R, Galande S, Chattopadhyay S. Direct interaction with and activation of p53 by SMAR1 retards cell-cycle progression at G2/M phase and delays tumor growth in mice. Int. J. Cancer. 2003;103:606–615. doi: 10.1002/ijc.10881. [DOI] [PubMed] [Google Scholar]
- 17.Kaul-Ghanekar R, Majumdar S, Jalota A, Gulati N, Dubey N, Saha B, Chattopadhyay S. SMAR1 transgenice mice data. J. Biol. Chem. 2005;280:9450–9459. doi: 10.1074/jbc.M412206200. [DOI] [PubMed] [Google Scholar]
- 18.Singh K, Mogare D, Giridharagopalan RO, Gogiraju R, Pande G, Chattopadhyay S. p53 Target Gene SMAR1 is dysregulated in breast cancer: its role in cancer cell migration and invasion. PLoS ONE. 2007;2:e660. doi: 10.1371/journal.pone.0000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chattopadhyay S, Kaul R, Charest A, Housman D, Chen J. SMAR1, a novel, alternatively spliced gene product, binds the Scaffold/Matrix-associated region at the T cell receptor beta locus. Genomics. 2000;68:93–96. doi: 10.1006/geno.2000.6279. [DOI] [PubMed] [Google Scholar]
- 20.Klaff P, Riesner D, Steger G. RNA structure and the regulation of gene expression. Plant Mol. Biol. 1996;32:89–106. doi: 10.1007/BF00039379. [DOI] [PubMed] [Google Scholar]
- 21.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhuyan BK, Adams EG, Badiner GJ, Li LH, Barden K. Cell cycle effects of prostaglandins A1, A2, and D2 in human and murine melanoma cells in culture. Cancer Res. 1986;46:1688–1693. [PubMed] [Google Scholar]
- 23.Ross J. mRNA stability in mammalian cells. Microbiol. Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staton JM, Thomson AM, Leedman PJ. Hormonal regulation of mRNA stability and RNA-protein interactions in the pituitary. J. Mol. Endocrinol. 2000;25:17–34. doi: 10.1677/jme.0.0250017. [DOI] [PubMed] [Google Scholar]
- 25.Audic Y, Hartley RS. Post-transcriptional regulation in cancer. Mol. Biol. Cell. 2004;96:479–498. doi: 10.1016/j.biolcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Liebhaber SA. mRNA stability and the control of gene expression. Nucleic Acids Symp. Ser. 1997;36:29–32. [PubMed] [Google Scholar]
- 27.Guiner C, Lejeune F, Galiana D, Kister L, Breathnach R, Stevenin J, Konczak F. TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 2001;276:40638–40646. doi: 10.1074/jbc.M105642200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.