Abstract
mRNA poly(A) tails affect translation, mRNA export and mRNA stability, with translation initiation involving a direct interaction between eIF4G and the poly(A)-binding protein Pab1. The latter factor contains four RNA recognition motifs followed by a C-terminal region composed of a linker and a PABC domain. We show here that yeast mutants lacking the C-terminal domains of Pab1 display specific synthetic interactions with mutants in the 5′-3′ mRNA decay pathway. Moreover, these mutations impair mRNA decay in vivo without significantly affecting mRNA export or translation. Inhibition of mRNA decay occurs through slowed deadenylation. In vitro analyses demonstrate that removal of the Pab1 linker domain directly interferes with the ability of the Pop2–Ccr4 complex to deadenylate the Pab1-bound poly(A). Binding assays demonstrate that this results from a modulation of poly(A) packaging by the Pab1 linker region. Overall, our results demonstrate a direct involvement of Pab1 in mRNA decay and reveal the modular nature of this factor, with different domains affecting various cellular processes. These data suggest new models involving the modulation of poly(A) packaging by Pab1 to control mRNA decay.
INTRODUCTION
Nuclearly encoded mRNAs of eukaryotes contain two constant distinctive features: a cap at their 5′ end and a poly(A) sequence at their 3′ end (except for histone mRNAs that are not polyadenylated in some species). These characteristic modifications provide cells with specific marks demonstrating the integrity of the corresponding mRNAs. Both the 5′ cap and poly(A) tails have been implicated in several processes including mRNA processing, mRNA nuclear export, translation and mRNA stability (1–3). Through these actions, the mRNA cap and poly(A) tail enhance strongly gene expression and contribute to its regulation. This situation contrasts significantly with the one observed in prokaryotes where RNA polyadenylation has been shown to activate mRNA turnover (4). Interestingly, a similar mechanism targeting aberrant transcripts or processing intermediates towards decay through the addition of a poly(A) tail, has recently also been observed in the nucleus of eukaryotes, thereby expanding the function of eukaryotic polyadenylation (5–8). Eukaryotic mRNA caps and poly(A) tails protect the flanking mRNA body from exonuclease attacks. This is particularly true for the 5′ cap, which can only be removed by a dedicated decapping enzyme (1). However, these modifications mediate their effects mostly through interactions with specific binding factors. Henceforth, nuclear and cytoplasmic cap binding factors have been identified and shown to participate in RNA processing and transport and to play a key role in translation initiation (3,9). Similarly, nuclear and cytoplasmic poly(A)-binding proteins (PABPs) have been described. The abundant cytoplasmic PABP was shown to contribute to translation and mRNA stability (2). It is thus not surprising that genetic deletion of the gene encoding the yeast PABP, PAB1, leads to cell death. Sequence analysis demonstrated that Pab1 and its homologues are composed of four RNA recognition motifs (RRM), followed by a linker region (L) and a C-terminal PABC domain (C) (10). Interestingly, the essential portion of Pab1 resides in the RRM region (11). Structural analyses have demonstrated that the RRM domains directly and specifically contact the associated RNA molecule (12). The RRM region also interacts with the eIF4G translation initiation factor (13,14). Thus, its essential nature may be explained by a role in translation initiation. The structures of the C-terminal yeast and human PABC domains have also been determined (10,15). This domain participates in protein–protein interactions (16) including the association with the translation termination factor eRF3 in yeast as well as ataxin-2 and some PABP-interacting proteins (PAIP) in higher eukaryotes. The eRF3-PABP interaction was proposed to be involved in translation termination (17), but no biological evidence supporting this model has been reported.
Pab1 has also been implicated in the control of mRNA stability (2,18). RNA degradation contributes to the fine-tuning of cellular transcript levels as well as to the elimination of accidentally damaged or aberrant mRNAs that could impact on the cell function. These degradation processes are mediated by a variety of enzymes and mechanisms (19). Two conserved pathways have been implicated in the degradation of functional mRNAs. Both pathways are initiated by the removal of the mRNA poly(A) tail and this deadenylation step is most often rate limiting for the decay process. Deadenylated mRNA bodies get degraded either in the 5′-3′ direction by the Xrn1 exonuclease after removal of the 5′ cap by the Dcp1–Dcp2 decapping complex or in the 3′-5′ direction by a multi-subunit exonuclease complex named exosome. These various exonucleases and decapping enzymes are also involved in the degradation of faulty mRNAs containing a premature stop codon, lacking a stop codon or inducing ribosome stalling. However, a main difference with the degradation of functional mRNAs is that the decay of these messages usually bypasses the initial deadenylation step, at least in yeast.
As deadenylation is rate limiting for the decay of functional mRNAs, it is a main target for the regulation of this process (20). Poly(A) degradation is carried out by deadenylases that show an exonucleolytic activity with a strong preference for poly(A) sequences. In yeast, the major cytoplasmic deadenylase is constituted by the Pop2–Ccr4 complex (21,22). This complex is conserved in human and Drosophila (23,24). Interestingly, both subunits of this complex display nuclease activity (21,25). Moreover, the heterodimeric Pop2–Ccr4 deadenylase forms a variety of larger complexes by associating with Not and Caf factors, which also affect deadenylation of some mRNAs (26). A second deadenylase identified in yeast is composed of the Pan2 and Pan3 subunits (27). However, its inactivation in yeast results neither in a strong growth phenotype nor in a strong block of deadenylation. It was suggested that the PAN deadenylase was primarily involved in the initial shortening of poly(A) tails of newly made mRNA (27), a possibility supported by the analysis of its mammalian homologue (28). Finally, a third deadenylase was identified in vertebrate cells, PARN (29). This protein, which shows a limited evolutionary conservation and is mainly located in the nucleoplasm, was not found to affect mRNA turnover in cultured mammalian cells (28) but affected deadenylation during meiotic maturation in Xenopus. In addition to deadenylases, some factors affecting the deadenylation process have been identified. These include RNA-binding proteins such as the members of the Smaug and PUF protein families that control mRNA decay through deadenylation and directly affect the recruitment of the Pop2–Ccr4 deadenylase (30–33). Similarly, the eRF3 translation termination factor was proposed to affect the deadenylation process (34).
The location of Pab1 on poly(A) tails suggests that it may influence deadenylation and mRNA stability. Consistently, it appears to stimulate the activity of PAN and to inhibit the activity of purified Ccr4 in vitro (26,27). Moreover, Pab1 was proposed to participate in the recognition of premature stop codon in the non-sense-mediated decay (NMD) pathway (35). The linker and PABC domains of Pab1 were also shown to interact with the Pan3 subunit of the PAN deadenylase, suggesting a direct role in deadenylation control (36). Finally, tethering experiments have suggested that only the recruitment of Pab1 at the 3′ terminus of the associated mRNA, but not its binding to poly(A) tails, is required for mRNA stabilization; possibly not by affecting deadenylation but rather through slowed decapping (37).
We have investigated whether specific domains of Pab1 are specifically implicated in the control of mRNA decay in vivo. We show that the deletion of the carboxy-terminal region of Pab1 stabilizes reporter mRNAs in vivo by impairing deadenylation, without affecting mRNA export or translation. This conclusion is also supported by the observation that a deletion of the linker region of Pab1 increases its ability to inhibit the Pop2–Ccr4 deadenylase in vitro. These results indicate that Pab1 is directly involved in the mRNA decay process and reveal the modular nature of this protein with specific regions involved in various cellular functions. They further suggest that the carboxy-terminal domains of Pab1 may be implicated in the control of mRNA stability by regulating the poly(A) packaging, and thus the deadenylation through interactions with trans-acting factors.
MATERIAL AND METHODS
Yeast strains and growth conditions
Yeast cells were grown in standard media at 30°C, except otherwise indicated. All the strains derive from BMA64 (detailed genotypes given in Supplementary Table 1).
Strains BSY1522 (PAB1), BSY1537 (pab1ΔC), BSY1552 (pab1ΔLΔC) were generated by homologous recombination. Briefly, a DNA fragment including a stop codon followed by the TRP1Kl cassette was amplified by PCR (see Supplementary Table 2 for oligonucleotides) from plasmid pBS1479 (38) and inserted in the genome. Stop codons were inserted after amino acids 577, 490 or 405 respectively. To generate the strain BSY1600 (pab1ΔL) a DNA fragment containing a deletion of PAB1 between codons 406 and 489 and the TRP1Kl cassette was generated from BSY1522 genomic DNA by overlapping PCR and integrated in the genome. Strain BSY1677 (pab1ΔLΔC dcp1-2) was constructed by crossing BSY1624 (dcp1-2) with BSY1552, followed by tetrad dissection.
Strain BSY1461 used to overexpress the Pop2ΔN–Ccr4ΔN complex was constructed using a promoter substitution strategy (Dziembowski et al., in preparation). Briefly, cassettes containing a selectable marker and a strong promoter were amplified and successively integrated upstream of nucleotide 439 of the POP2 ORF and of nucleotide 330 of the CCR4 ORF. The cassette inserted upstream of the POP2 coding sequence contained in addition an N-terminal TAP tag for protein purification. All deletions and insertions were verified by analytical PCR and DNA sequencing.
Plasmid construction
Standard cloning procedures were used. Plasmid descriptions are presented in the Supplementary Data. The reporter plasmids pSG-LD5, pRP611 and the expression plasmid pET24-His6x-GST-TEV-ABD have been described (39–42).
Analysis of in vivo produced RNA
Reporter plasmids were introduced in yeast by LiCl-mediated transformation (43). RNA chase experiments were essentially performed as described previously (21). In the case of tetO-driven reporters, liquid cultures were grown at 30°C in SD medium lacking histidine to an OD of 08–1.0, concentrated 10 times in fresh medium and returned to 30°C. Reporter transcription was inhibited by adding 50 μg/ml doxycycline (44). Aliquots of 1 ml were taken at the indicated time points, pellets frozen in liquid nitrogen and stored at −20°C. Yeast total RNA was obtained by hot acid phenol extraction. Northern blot analyses were carried out following either formaldehyde–agarose or polyacrylamide gel electrophoresis. Reporter RNAs containing oligo(G) insertions were detected using a radiolabeled poly(C) probe. Signals were measured and quantified using PhosphorImager (GE Health Care). The mRNA half-lives were calculated using the best fit to an exponential decay. SCR1 was detected with a specific probe when used as loading control.
To assay the deadenylation rates the PGK1pG mRNA from the transcriptional chase experiments was digested with RNase H (45). As a control, a second sample from the 0 time point was treated in addition with a synthetic oligo(dT) to reveal the position of the fully deadenylated species. To estimate the deadenylation rate, signals emanating from species harboring different size of poly(A) (from fully adenylated to completely deadenylated) were measured using PhosphorImager (GE Health Care) by dividing the area in 100 cells of identical size using a column grid. These data were used to calculate, by integration, weighted averages of the poly(A) size at each time point. Those were then used to derive an apparent deadenylation rate.
To analyze the CYH2 mRNA and pre-mRNA levels, total RNA obtained by hot phenol extraction from exponentially growing cultures from cells expressing PAB1 or PAB1ΔLΔC and the Δupf1 control were analyzed by northern blot.
In vitro RNA analysis
A 201 nucleotide long synthetic RNA ending by 48A residues was produced by in vitro transcription in presence of α32-UTP from plasmid pBS2900 linearized with BsmBI. For in vitro RNA binding, 2 nM synthetic polyadenylated RNA was incubated with increasing concentrations of the Pab1 proteins for 10 min at 30°C in Tris pH 7.4 10 mM, 20 mM NaCl, Mg acetate 1 mM, DTT 2 mM, NP-40 0.02% in a final volume of 5 μl and kept on ice. A control-unrelated recombinant protein was used to balance the total protein concentration in each reaction. A 1.25 μl of glycerol 50%, Bromophenol blue 0.025%, TBE 1× was added and complexes resolved on 5% non-denaturing polyacrylamide gel (19 : 1 ratio) in TBE 0.5×. Migration was performed at 4°C at 10 V/cm before autoradiography. For in vitro RNA dissociation assays, 0.4 nM synthetic polyadenylated RNA was incubated with 0.8 nM of the different Pab1 proteins for 10 min at 30°C before the addition of a 140-fold excess of competitor poly(A) (Pharmacia Biotech). As a control, the synthetic RNA and the excess of competitor poly(A) were premixed before the addition of Pab1. Protein–RNA complexes were resolved by non-denaturing PAGE as described above.
For the deadenylation protection analysis, 2 nM synthetic polyadenylated RNA was incubated for 10 min with 20 nM of the Pab1 proteins and transferred on ice as described for the binding assays. This concentration of Pab1 was empirically determined to give a partial protection for the wild-type factor. A 20 nM of purified Pop2–Ccr4 was added and deadenylation reactions carried on at 30°C. Samples of 5 μl were taken at the indicated time points, extracted with phenol–chloroform–isoamylalcohol and precipitated. Products resuspended in loading buffer (Xylene cyanol 0.05%, Bromophenol blue 0.05%, EDTA 10 mM, SDS 0.1% in formamide) were loaded on a 6% denaturing polyacrylamide gel (19 : 1 ratio), 7M urea, in TBE 0.5×. Electrophoresis was performed at room temperature at 20 W before autoradiography.
Analysis of RNA and protein production in vivo
The lacZ reporter induction and β-galactosidase measurements were performed as described previously (46). In parallel, 10 ml samples were taken to extract total RNA using hot acid phenol. A 1 μg of total RNA was used for reverse-transcription that was performed as recommended (Fermentas) using oligonucleotide OBS1846 (Supplementary Table 2). Reverse-transcription products were used for quantitative-PCR analysis on a LightCycler LC480 apparatus (Roche) following the supplier's recommendations using oligonucleotides OBS1847 and OBS1848. The average signal from duplicate PCR experiments was used to derive the relative levels of lacZ mRNA present in the original samples.
Western blot
Western blot analysis was performed using standard procedures with polyclonal antibodies against Pab1 and Stm1 (F. Wyers).
Protein expression and purification
Recombinant proteins expressed from plasmids pET22-PAB1, pBS2841, pBS2928 and pBS2926 and pBS2010 were purified by affinity chromatography (47). The Pop2ΔN–Ccr4ΔN complex was purified from strain BSY1461 following a variation of the TAP method and gel-filtration chromatography (48). Peak fractions were dialyzed against storage buffer (PBS 1×, 20% glycerol) and stored at −80°C. Complex composition and absence of contaminants were assessed by SDS-PAGE and Coomassie blue staining.
RESULTS
Phenotypes of carboxy-terminal Pab1 deletion mutants and genetic interaction with DCP1 and the Pop2–Ccr4 complex
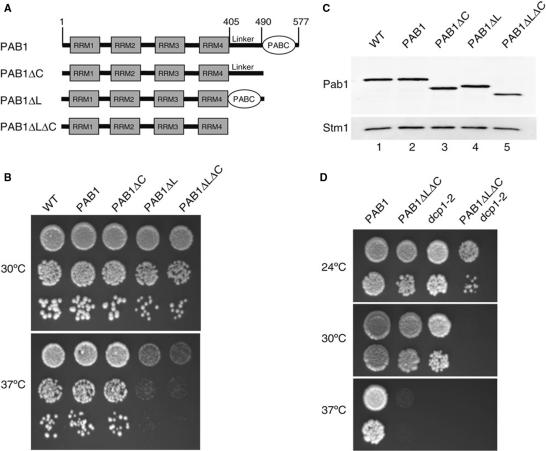
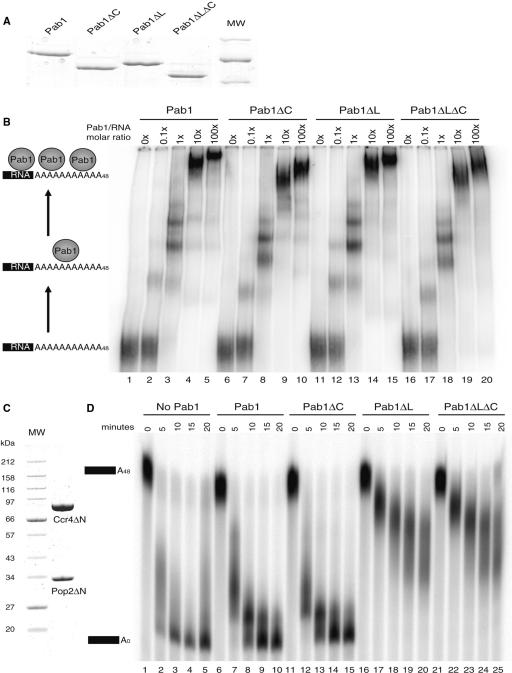
The N-terminal region of Pab1 encompassing the four RRM sequences has been implicated in translation initiation and nucleocytoplasmic shuttling and shown to be sufficient for yeast viability (11,13,14,49). Thus, we decided to investigate the in vivo role of the C-terminal region of yeast Pab1. Phylogenetic conservation and structural data indicate that it can be subdivided into two domains, a linker region (L) and a C-terminal PABC domain (C) (Figure 1A). Thus, we generated mutant strains lacking either the PABC domain (represented as ΔC), the linker region (ΔL) or both (ΔLΔC) at the genomic PAB1 locus (Figure 1A). Consistent with previous analyses (11), these mutants are viable, however, deletion of the linker alone or in combination with the PABC domain generated a temperature-sensitive phenotype (Figure 1B) while the PABC deletion did not induce a significant growth phenotype. The growth defects of the ΔL and ΔLΔC mutants could result either from a specific function of these regions or indirectly from an altered Pab1 level. Indeed, C-terminal deletions might have destabilized the protein. However, western blot analysis of Pab1 levels demonstrated comparable accumulation of this factor in the various genetic backgrounds (Figure 1C). These results reveal an important role of the Pab1 C-terminal region for its activity. To establish a potential link between these domains and mRNA decay, we next combined the ΔLΔC deletion with mutations in various mRNA decay factors. Construction of double mutants carrying the ΔLΔC mutation and a pop2 (or ccr4) deletion by crosses and dissection revealed a synthetic lethal phenotype (data not shown). Moreover, when combined with a thermosensitive mutation of the yeast decapping factor Dcp1 the ΔLΔC mutant generated a synthetic phenotype since it was, unlike the single mutants, unable to grow at 30°C (Figure 1D). In contrast, the ΔLΔC mutant did not exacerbate the thermosensitive phenotype of a dis3 mutant (data not shown, Dis3 is the catalytic subunit of the exosome (50)). These observations globally supported a role for the Pab1 C-terminal region in mRNA decay and suggested further a preponderant involvement in the main 5′-3′ mRNA decay pathway.
Figure 1.
Pab1 Carboxy-terminal deletion mutants and their phenotypes. (A) Structural organization of Pab1 and structure of the various deletion mutants used in this study. The various domains present in PABP (RRM, RNA recognition motif; PABC, PAB C-terminal domain) are depicted and the residues corresponding to the Pab1 truncation end points indicated. (B) Growth phenotype of the various Pab1 mutants at 30°C and 37°C. Dilutions of exponentially growing liquid cultures of the indicated strains were deposited on YPDA plates and their growth was scored after 2 days of incubation at the indicated temperatures. (C) Steady-state levels of the various Pab1 versions. Results of a western blotting experiment using total proteins from the indicated strains are presented. Pab1, and the loading control, Stm1, were detected with polyclonal sera. (D) Synthetic phenotype of a C-terminal Pab1 deletion combined with a dcp1 mutation. The assay is identical to the one presented in panel B except that growth was scored after 2.5 days.
Carboxy-terminal deletions of Pab1 stabilize reporter mRNAs
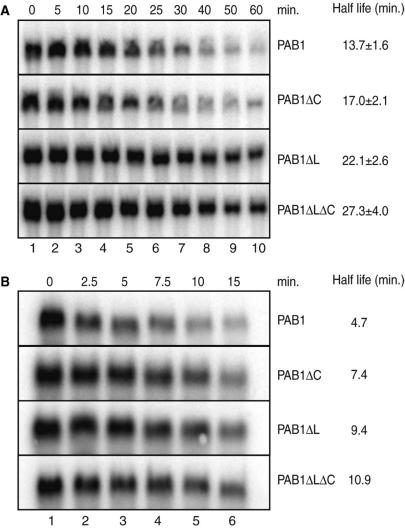
To assess more directly the involvement of the carboxy-terminal region of Pab1 in mRNA decay, we performed transcriptional chase experiments. Wild-type and mutant strains lacking this region were transformed with plasmids encoding mRNA reporters under the control of a tetO-regulated promoter. Following growth in conditions allowing constitutive reporter expression, mRNA production was rapidly repressed by the addition of doxycycline (44). Northern blot analysis of RNA extracted at various time points after transcription repression allowed the quantification of the reporter half-lives in the different backgrounds. Importantly, these assays were performed at permissive temperature to avoid indirect effects. These experiments revealed that the half-life of the PGK1pG reporter mRNA were modestly affected in the pab1ΔC strain (half-life of about 17 min compared to roughly 14 min in the wild-type strain). In contrast, the PGK1pG mRNA half-life was significantly increased to ∼22 min in the pab1ΔL mutant, and to over 27 min in the pab1ΔLΔC strain (Figure 2A).
Figure 2.
C-terminal truncation of Pab1 stabilizes the PGK1pG and MFA2pG mRNA reporters. The results of transcriptional chase experiments assayed by northern blotting are depicted. (A) Results obtained with the tetO-PGK1pG reporter pBS2813 in strains expressing PAB1, PAB1ΔC, PAB1ΔL and PAB1ΔLΔC. Indicated half-lives are the mean (and SD) of four different experiments. (B) Results obtained with the tetO-MFA2pG reporter pBS2762. Numbers on the top represent the time points after the transcription repression, in minutes.
To test whether the stabilization observed was general, we performed a similar experiment using the short-lived MFA2pG mRNA reporter (Figure 2B). We observed again a stabilization of this reporter in the mutant strains. Importantly, the effects were proportionally similar to those of the previous analysis, with the linker deletion alone having a stronger effect than the deletion of the PABC domain alone but a weaker effect than the complete deletion. These analyses demonstrate that the carboxy-terminal region of Pab1, in particular the linker region, affects mRNA decay and indicate that this effect is not specific to a given mRNA.
Carboxy-terminal deletions of Pab1 do not affect mRNA export and translation
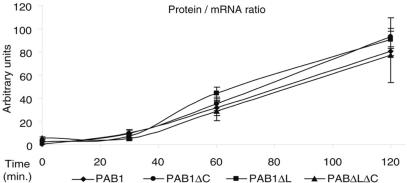
Pab1 has been linked to mRNA translation through its interaction with the factors eIF4G and eRF3 (17,51) and has also been involved in mRNA nucleocytoplasmic transport (49,52). As these cellular processes may indirectly affect mRNA stability, we tested whether deletions of the carboxy-terminal domains of Pab1 affected translation or mRNA export. To that purpose, we introduced a LacZ gene under the control of a GAL-inducible promoter in the various strains and followed the reporter mRNA level and translation at various time points after transcription induction. The mRNA levels were assayed by quantitative RT-PCR while translation was revealed by assaying β-galactosidase production. We observed that the ratios of protein produced to the level of the cognate mRNA were comparable for all strains at each time point (Figure 3). This result suggests that there is neither a detectable delay in mRNA export to the cytoplasm nor a significant nuclear accumulation of mRNA in the mutant cells compared to the wild-type. Furthermore, the similar protein-to-mRNA ratios indicate that the translation rate per mRNA molecule is also equivalent in the various strains. While we cannot exclude minor effect(s) of Pab1 C-terminal truncation on nucleocytoplasmic mRNA transport and/or translation, these data support a direct effect of the Pab1mutants on mRNA decay.
Figure 3.
The Pab1 C-terminal truncation does not affect mRNA export and translation. The ratio of the β-galactosidase protein to its mRNA was assayed at various time points after induction of the lacZ reporter in strains expressing PAB1, PAB1ΔC, PAB1ΔL or PAB1ΔLΔC. β-galactosidase production was assayed enzymatically and lacZ mRNA levels monitored by quantitative RT-PCR. The ratios of protein-to-mRNA levels, plotted in arbitrary units, are the mean ± SD from two independent experiments.
Carboxy-terminal deletion of Pab1 does not affect an NMD substrate
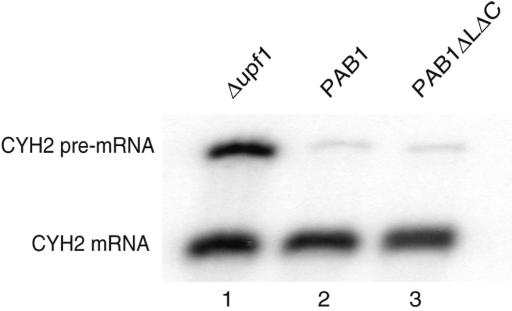
To test if the deletion of the Pab1 C-terminal region affected every mRNA decay event, we analyzed whether it affected mRNA decay by the NMD pathway. For this purpose, we assayed the steady-state level of the CYH2 pre-mRNA (53). Northern blot analysis demonstrated that, unlike in an NMD defective Δupf1 strain, the CYH2 pre-mRNA did not accumulate significantly in the PAB1ΔLΔC background (Figure 4). To definitively rule out a mild role of the Pab1C-terminal region in NMD, we also followed the decay of a GAL-driven PGK1pG reporter containing a premature termination codon (PGK1NSpG) (42) in a chase assay. Again, no difference was detected between cells expressing the full-length Pab1 or the ΔLΔC truncated version (data not shown). Altogether, these results demonstrate that the Pab1 C-terminal region does not affect all mRNA degradation pathways. Furthermore, the observation of an active NMD pathway (process that is translation-dependent) provides an independent evidence that translation is not dramatically altered in the Pab1 truncation mutant.
Figure 4.
CYH2 pre-mRNA does not accumulate in the PAB1ΔLΔC background. Total RNA extracted from exponentially growing cultures of the control Δupf1 (lane 1), wild-type (lane 2) or PAB1ΔLΔC mutant (lane 3) strains were analyzed by northern blotting to reveal both the CYH2 mRNA and pre-mRNA, with the latter being an NMD substrate and the former providing an internal loading control (53).
Carboxy-terminal deletion of Pab1 slows down reporter mRNA deadenylation and produces extended Poly(A) tails
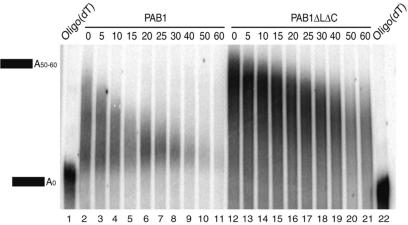
The specific effect of the Pab1 C-terminal truncation on the decay of reporters degraded by pathways targeting functional mRNAs and the lack of effect on substrates containing a premature stop codon were consistent with an effect on deadenylation, as the latter bypasses the deadenylation step in yeast (54). Moreover, the direct interaction of Pab1 with the poly(A) tail, its reported stimulation of Pan2 (36) and inhibition of Ccr4 (26) also suggested that deadenylation could be affected by the Pab1 C-terminal truncation. To test this possibility, we analyzed the length of the poly(A) tail of the PGK1pG reporter mRNA during a transcriptional chase experiment. To improve the resolution, RNAs were digested by RNaseH using an oligonucleotide complementary to the mRNA body and fractionated on denaturing polyacrylamide gels before northern blotting. This analysis demonstrated that the PGK1pG mRNA extracted from the Pab1ΔLΔC background (Figure 5, lanes 12–21) presented longer poly(A) tails compared to the mRNA extracted from cells expressing full-length PAB1 (Figure 5, lanes 2–11). Moreover, the size of these mRNAs decreased more slowly in the mutant strain indicating a reduced deadenylation rate. Quantification of this effect revealed that deadenylation was slowed down ∼2.5-fold by the removal of the C-terminal region of Pab1. This value agrees relatively well with the 2.0–2.3-fold longer half-life of the reporter mRNAs (Figure 2). These data indicate that mRNA stabilization results, at least in part, from an impaired deadenylation.
Figure 5.
Deletion of the Pab1 C-terminal region impairs deadenylation. Evolution of poly(A) tail length of the PGK1pG reporter mRNA in chase experiments was assayed by northern blotting after RNase H-mediated release of the reporter 3′ UTR region and fractionation on high-resolution polyacrylamide gels. A comparison of the wild-type strain (PAB1, lanes 2–11) and of the strain expressing PAB1ΔLΔC (lanes 12–21) is depicted. Numbers on the top represent the time points after the transcription repression, in minutes. In lanes 1 and 22, RNA samples from the 0 time points were digested by RNase H in the presence of oligo(dT) in addition to the PGK1 specific oligonucleotide, thus revealing the migration position of fully deadenylated RNA.
Removal of the Pab1 linker domain impedes Pop2–Ccr4 mediated deadenylation in vitro
In Saccharomyces cerevisiae, deadenylation is mainly carried out by the Pop2–Ccr4 complex (21,22). As the C-terminally truncated Pab1 slowed down deadenylation in vivo, we tested whether these mutations could directly impact on the Pop2–Ccr4 activity in vitro. For this purpose, we expressed in Escherichia coli and purified 6His-tagged recombinant Pab1 versions carrying C-terminal truncations equivalent to those analyzed in vivo (Figure 6A). We verified, using a gel retardation assay with a radioactively labeled polyadenylated RNA substrate, that the recombinant proteins were functional by testing their in vitro binding capacity (Figure 6B). Consistent with previous results that attributed the Pab1 RNA-binding activity to its RRM region (11,55), wild-type Pab1, Pab1ΔC, Pab1ΔL and Pab1ΔLΔC formed similar complexes of lower mobility in a concentration-dependent manner, indicating that the mutant proteins were still functional in vitro (Figure 6B, lanes 2–20). Interestingly, these results are coherent with the observation that the mutant strains are viable.
Figure 6.
Deletion of the Pab1 linker domain increases Pop2–Ccr4 deadenylase inhibition. (A) Coomassie blue-stained gel showing the protein profile of the purified 6His-tagged recombinant Pab1 and deletion mutant thereof used for the binding assay and poly(A) protection experiment. (B) Recombinant Pab1 (lanes 1–5), Pab1ΔC (lanes 6–10), Pab1ΔL (lanes 11–15) and Pab1ΔLΔC (lanes 16–20) are active as assayed by poly(A) binding in a gel shift assay. Values above the lanes indicate the relative Pab1/RNA molar ratio. (C) Coomassie blue-stained gel showing the protein profile of the Pop2–Ccr4 deadenylase fraction used in the protection assay. (D) Inhibition of Pop2–Ccr4 mediated deadenylation by full-length and C-terminally truncated Pab1. The panel shows products of time course deadenylation reactions performed in the absence of added Pab1 (lanes 1–5), or in the presence of Pab1 (lanes 6–10) or truncated variants (Pab1ΔC: lanes 11–15, Pab1ΔL: lanes 16–20, Pab1ΔLΔC: lanes 21–25) after fractionation on denaturing polyacrylamide gels. The protein concentration used was selected to give a partial inhibition for the wild-type factor. Samples were then taken at the indicated time points. Fully adenylated (A48) and deadenylated (A0) species are indicated.
To reconstitute a deadenylation system in vitro, we further prepared a highly pure active core deadenylase. For this purpose, we overexpressed Pop2–Ccr4 in yeast cells (Figure 6C). N-terminally truncated versions of these proteins were used because of the higher yield and lower level of contaminants. Importantly, these truncated proteins lack only non-conserved regions, are fully functional in vivo and display an in vitro deadenylation activity identical to the full-length proteins (Figure 6D, lanes 1–5 and data not shown). Addition of full-length recombinant Pab1 to the in vitro reactions before incubation with the Pop2–Ccr4 deadenylase slightly slowed down the deadenylation reaction (lanes 6–10), consistently with the previously reported inhibitory effect of this factor on Ccr4 activity (26). Addition of the Pab1ΔC protein had an effect similar to the addition of the wild-type factor (lanes 11–15). Interestingly, the Pab1ΔL and Pab1ΔLΔC proteins inhibited more strongly the Pop2–Ccr4-mediated deadenylation than the wild-type Pab1 factor (lanes 16–25). Overall, the in vitro results are parallel to those obtained in vivo with the linker region having a stronger effect on deadenylation than the PABC domain. The increased Pop2–Ccr4 inhibition by the truncated Pab1 version is unanticipated and indicates that the inhibitory function of the full-length protein may be further increased.
Removal of the Pab1 linker domain modifies its dissociation from Poly(A) tails
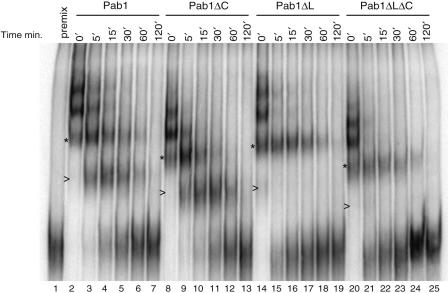
Two models can be proposed to explain the effect of the Pab1 linker deletion on Pop2–Ccr4 mediated deadenylation. On the one hand, Pab1 could interact directly with Pop2–Ccr4, thereby modulating their activity. On the other hand, Pab1 could affect deadenylation by controlling poly(A) packaging, and thus substrate access by Pop2–Ccr4. The former possibility appears unlikely because Pab1 has not been reported as a Ccr4 or Pop2 interacting partner and we have not been able to detect Pab1 above background level in Pop2-TAP purifications or in co-immunoprecipitation assays (data not shown). Thus, we tested whether the deletion of the C-terminal domains of Pab1 was affecting its interaction with poly(A). As a sensitive measure of Pab1 affinity for poly(A), we assayed its dissociation from a polyadenylated RNA. Complexes were formed by incubating a radioactively labeled polyadenylated substrate with an excess of Pab1. At time zero, a 140-fold excess of unlabeled poly(A) was added, preventing the detection of complexes resulting from the reassociation of Pab1 with RNA once it has dissociated from the labeled substrate (Figure 7, lane 1). The dissociation of Pab1 was monitored by analyzing the complexes remaining on native gels at various time-points after unlabeled poly(A) addition (Figure 7, lanes 2–25). This revealed that the dissociation of wild-type Pab1 occurred in a stepwise manner (Figure 7, lanes 2–7). Taking into account the number of adenines covered by the yeast Pab1 [25 nt/protein monomer, (11)] and the size of the poly(A) of the substrate RNA (48 adenines), a maximum of two Pab1 molecules are expected to bind to the substrate poly(A) tail while additional subunits could bind non-specifically to the 153 residue-long mRNA body. The Pab1ΔC protein presented a dissociation pattern similar to the wild-type factor (lanes 8–13). Remarkably, for the Pab1ΔL and Pab1ΔLΔC proteins a different dissociation pattern was observed (lanes 14–19 and 20–25). In these cases, a rapid initial dissociation of Pab1 bound non-specifically to the mRNA body was followed by the slow release of the two remaining Pab1 molecules in a single step. (Note that only trace amount of the complex containing a single Pab1 molecule could be detected, see Figure 7, lane 21 and compare with Figure 6B. The same RNA and electrophoresis conditions were used in these two experiments and the relative mobility of the complexes are identical) The absence of the band corresponding to a single bound Pab1 molecule when the Pab1ΔL or Pab1ΔLΔC versions are used suggests an altered dissociation from the poly(A) tails of the versions lacking the linker region. These data support the idea that mRNA stabilization induced by Pab1 truncations results from a different interaction of truncated Pab1 with poly(A). This different mode of binding may cause a different packaging of poly(A) tails and/or a slower cooperative release of Pab1, thus affecting deadenylase access.
Figure 7.
The absence of the linker domain affects Pab1 dissociation from Poly(A) tails. Dissociation from bound RNA of Pab1 (lanes 2–7), Pab1ΔC (lanes 8–13), Pab1ΔL (lanes 14–19) and Pab1ΔLΔC (lanes 20–25) analyzed in a gel shift assay. Dissociation was assayed in time-course reaction after the addition of excess unlabeled poly(A). To demonstrate that sufficient unlabeled competitor was present a control reaction was performed by mixing the synthetic RNA and the excess of competitor poly(A) before the addition of Pab1 (lane 1). Values above lanes 2–25 indicate the time in minutes after the addition of an excess of competitor Poly(A) to the reactions. (>) and (*)indicate the position of migration of complexes containing 1 or 2 molecules of Pab1 bound to the polyadenylated RNA for the wild-type factor and each mutant form, respectively (compare also with Figure 6B).
DISCUSSION
We have analyzed the biological function of the C-terminal region of the poly(A) binding protein. Our data reveal the specific involvement of Pab1 C-terminal domains in the mRNA decay process and uncover the modular nature of the Pab1 factor. Importantly, Pab1 C-terminal domains appear to modulate specifically its interaction with poly(A) and thus the deadenylation step during the mRNA decay.
Previous analyses had already established the existence of several functional features in the poly(A) binding protein. Thus, the N-terminal RRM region has been implicated in poly(A) binding (11,55), interaction with the eIF4G translation initiation factor (56) and association with nucleocytoplasmic transport factors Xpo1 and Kap108 (52). In fact, only a short segment of this region, overlapping RRM4, is essential to provide Pab1 function (11). These pleiotropic functions of the poly(A) binding protein could suggest that the effect of its C-terminal truncation on mRNA stability reported here could be indirect. This is particularly true in the case of translation as its inhibition (e.g. through the use of inhibitors or mutants) strongly affects mRNA stability (57) and it has been proposed that mRNA stabilization by Pab1 requires ongoing translation (37). However, our data are inconsistent with a role for translation in the mRNA stabilization promoted by Pab1 C-terminal truncation. Indeed, we did not detect a significant effect of these mutants on translation, and in vivo mRNA stabilization by truncated Pab1 could be reproduced in vitro using purified deadenylase, thus bypassing the need for any translation event. Similarly, the C-terminal deletions did not prevent nuclear mRNA export, consistent with the fact that this region has not been implicated in Pab1 nucleocytoplasmic shuttling. Altogether, it is thus unlikely that the mRNA stabilization that we observe arises from an indirect effect of Pab1 on another cellular process. Therefore, contrasting with previous conclusions (37), our in vivo and in vitro data demonstrate that Pab1 is directly implicated in mRNA decay. Moreover, our results reveal a specific involvement of the Pab1 C-terminus in this process.
Which are the targets of the C-terminal region of Pab1 that affect mRNA decay? Our data demonstrate that Pab1 mutants impair mRNA deadenylation. This effect could be mediated in vivo by a Pab1 partner associating directly with its C-terminal region such as Pbp1, eRF3 and/or Pan3. Pbp1 binds to the Pab1 linker region but has been shown to control proper poly(A) tail synthesis rather than decay (58). The eRF3 interacts with Pab1 through its C-terminal PABC domain. However, the function of this conserved interaction in mRNA decay remains unclear (17). The eRF3 has been reported to affect mRNA decay by altering deadenylation (34) but there is no evidence supporting that this occurs through an interaction with Pab1. Moreover, we could not duplicate these original results (data not shown), possibly owing to strain differences. Because Pab1 stimulates Pan2 and interacts directly with Pan3 through its C-terminal region (36), an alternative interpretation could be that removal of the corresponding Pab1 domain inhibits deadenylation by preventing Pan2 action. Several observations argue against this interpretation. First, Pan2 is only responsible for a minor deadenylase activity in yeast implicated in the initial step of mRNA deadenylation and its deletion does not stabilize the reporters analyzed here as strongly as Pab1 truncations [data not shown, (22,28,59)]. Moreover, Pab1 truncation stabilized mRNAs even in a PAN2 deletion background (data not shown), arguing that these processes occur through independent pathways. Finally, our observation that the deletion of the Pab1 C-terminal region enhances its inhibitory activity towards purified Pop2–Ccr4 deadenylase in vitro suggests that Pab1 effects occur directly through this enzyme. This interpretation is consistent with the observation that Pop2–Ccr4 is the major yeast deadenylase, and thus essential for the normal decay of the reporters we assayed (22).
Two possible mechanisms explaining how Pab1 affects Pop2–Ccr4 could be envisaged. A first possibility was that the Pab1 C-terminal region interacted directly or indirectly with Pop2–Ccr4, stimulating its activity, e.g. by increasing its local concentration (45). However, this situation is unlikely as a physical interaction between Pab1 and the Pop2–Ccr4 deadenylase was neither reported nor detected. Thus, we favor the possibility that truncations of the Pab1 C-terminal domains alter poly(A) packaging and/or Pab1 release, and thus prevent an efficient access of the Pop2–Ccr4 deadenylase. This is consistent with the stronger inhibition of Pop2–Ccr4 activity by the truncated Pab1 in a reconstituted in vitro system and the evidence for an altered mode of dissociation from poly(A) for these mutants (Figure 7). Further evidence for an altered mode of binding of the truncated Pab1 can be seen upon close examination of Figure 6B. Indeed, lane 18 reveals a lower level of RNA bound by monomeric Pab1ΔLΔC compared to the wild-type Pab1 (lane 3). This argues again for increased cooperativity in binding that is consistent with the absence of monomeric complex during dissociation. Interestingly, the C-terminal region of Xenopus PABP has been implicated in its cooperative binding to poly(A) (60). A similar property was reported for the mammalian protein (61). While these results are not directly comparable to those obtained in yeast (Figure 6), they support the idea that the C-terminal domains of Pab1 affect poly(A) binding and may thus restrict substrate access. It is noteworthy, however, that assay of the LacZ reporter indicated that translation was not affected by the Pab1 truncation. As translation initiation is a critical step for gene expression, the limited growth phenotype of the Pab1 truncation mutants support also this conclusion. These observations suggest again that the truncations of Pab1 did not affect its capacity to load onto poly(A) (a step required for translation initiation) but rather the mode of poly(A) packaging or Pab1 release.
In any case, it is noteworthy that the Pab1 C-terminal truncation increases the inhibition of Pop2–Ccr4. This indicates that the repression mediated by wild-type Pab1 is not maximal and suggests that deadenylation may be regulated positively or negatively by interacting trans-acting factors. It is interesting that deletion of the linker domain produces a stronger phenotype than deletion of the PABC domain in each of the assays performed. This may suggest that the linker domain directly affects the mRNA decay process while the PABC domain may only have a regulatory function. Alternatively, the PABC domain may only affect indirectly mRNA decay by influencing (sterically) access to the linker region.
Overall, our work reveals a direct involvement of Pab1 in mRNA decay. This effect involves inhibition of the Pop2–Ccr4 deadenylase by the absence of the Pab1 C-terminal region, particularly the linker domain. Our observations confirm the pleiotropic function of the poly(A)-binding protein that appears to be a highly modular factor implicated in translation, nucleocytoplasmic transport and mRNA decay. Pab1 could thus be a central target to coordinate these processes in eukaryotic cells. Particularly, given the preponderant role of deadenylation on mRNA decay control, and the variety of known interactions involving the carboxy-terminal region of Pab1, we can easily imagine how these interactions may regulate deadenylation, and thus the mRNA decay process, e.g. by stabilizing or destabilizing Pab1 at the poly(A) tails. Further, work should allow the identification of such putative regulators and contribute to elucidate how the cells select mRNAs to be degraded in given conditions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank our group members for help and advice, in particular A. Dziembowski, C. Faux, A.L. Finoux, F. Lacroute and F. Wyers for plasmids, antibodies, protocols and support with strain construction. Plasmids carrying the tetO promoter were a generous gift from E. Herrero while the original PGK1pG and MFA2pG were kindly provided by R. Parker. A. Dziembowski, S. Camier, A.L. Finoux and F. Mauxion and one of the referees are gratefully acknowledged for critical reading of the manuscript. E.S. was supported by CNRS and ARC fellowships. This work was supported by La Ligue contre le Cancer (Équipe Labellisée 2005) and the CNRS. Funding to pay the Open Access publication charges for this article was provided by CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cougot N, van Dijk E, Babajko S, Seraphin B. ‘Cap-tabolism’. Trends. Biochem. Sci. 2004;29:436–444. doi: 10.1016/j.tibs.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Kuhn U, Wahle E. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Lewis JD, Izaurralde E. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 1997;247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- 4.Kushner SR. mRNA decay in prokaryotes and eukaryotes: different approaches to a similar problem. IUBMB Life. 2004;56:585–594. doi: 10.1080/15216540400022441. [DOI] [PubMed] [Google Scholar]
- 5.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 7.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 9.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 10.Kozlov G, Siddiqui N, Coillet-Matillon S, Trempe JF, Ekiel I, Sprules T, Gehring K. Solution structure of the orphan PABC domain from Saccharomyces cerevisiae poly(A)-binding protein. J. Biol. Chem. 2002;277:22822–22828. doi: 10.1074/jbc.M201230200. [DOI] [PubMed] [Google Scholar]
- 11.Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 13.Kessler SH, Sachs AB. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell Biol. 1998;18:51–57. doi: 10.1128/mcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozlov G, Trempe JF, Khaleghpour K, Kahvejian A, Ekiel I, Gehring K. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl Acad. Sci. USA. 2001;98:4409–4413. doi: 10.1073/pnas.071024998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozlov G, De Crescenzo G, Lim NS, Siddiqui N, Fantus D, Kahvejian A, Trempe JF, Elias D, Ekiel I, et al. Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. EMBO J. 2004;23:272–281. doi: 10.1038/sj.emboj.7600048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosson B, Couturier A, Chabelskaya S, Kiktev D, Inge-Vechtomov S, Philippe M, Zhouravleva G. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol. Cell Biol. 2002;22:3301–3315. doi: 10.1128/MCB.22.10.3301-3315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 19.Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 20.Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′–3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 21.Daugeron MC, Mauxion F, Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 23.Bianchin C, Mauxion F, Sentis S, Seraphin B, Corbo L. Conservation of the deadenylase activity of proteins of the Caf1 family in human. RNA. 2005;11:487–494. doi: 10.1261/rna.7135305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Chiang YC, Denis CL. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown CE, Sachs AB. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 29.Korner CG, Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J. Biol. Chem. 1997;272:10448–10456. doi: 10.1074/jbc.272.16.10448. [DOI] [PubMed] [Google Scholar]
- 30.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 31.Jeske M, Meyer S, Temme C, Freudenreich D, Wahle E. Rapid ATP-dependent deadenylation of nanos mRNA in a cell-free system from Drosophila embryos. J. Biol. Chem. 2006;281:25124–25133. doi: 10.1074/jbc.M604802200. [DOI] [PubMed] [Google Scholar]
- 32.Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr. Biol. 2005;15:284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 33.Goldstrohm AC, Seay DJ, Hook BA, Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- 34.Hosoda N, Kobayashi T, Uchida N, Funakoshi Y, Kikuchi Y, Hoshino S, Katada T. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 2003;278:38287–38291. doi: 10.1074/jbc.C300300200. [DOI] [PubMed] [Google Scholar]
- 35.Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 36.Mangus DA, Evans MC, Agrin NS, Smith M, Gongidi P, Jacobson A. Positive and negative regulation of poly(A) nuclease. Mol. Cell Biol. 2004;24:5521–5533. doi: 10.1128/MCB.24.12.5521-5533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coller JM, Gray NK, Wickens MP. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 1998;12:3226–3235. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 39.Ekstrom F, Stier G, Sauer UH. Crystallization of the actin-binding domain of human alpha-actinin: analysis of microcrystals of SeMet-labelled protein. Acta Crystallogr. D. Biol. Crystallogr. 2003;59:724–726. doi: 10.1107/s0907444903002063. [DOI] [PubMed] [Google Scholar]
- 40.Guarente L, Yocum RR, Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc. Natl Acad. Sci. USA. 1982;79:7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 43.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belli G, Gari E, Aldea M, Herrero E. Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast. 1998;14:1127–1138. doi: 10.1002/(SICI)1097-0061(19980915)14:12<1127::AID-YEA300>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 45.Finoux AL, Seraphin B. In vivo targeting of the yeast Pop2 deadenylase subunit to reporter transcripts induces their rapid degradation and generates new decay intermediates. J. Biol. Chem. 2006;281:25940–25947. doi: 10.1074/jbc.M600132200. [DOI] [PubMed] [Google Scholar]
- 46.Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprevote O, Seraphin B. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J. 2004;23:4847–4856. doi: 10.1038/sj.emboj.7600482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hernandez H, Dziembowski A, Taverner T, Seraphin B, Robinson CV. Subunit architecture of multimeric complexes isolated directly from cells. EMBO Rep. 2006;7:605–610. doi: 10.1038/sj.embor.7400702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunn EF, Hammell CM, Hodge CA, Cole CN. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev. 2005;19:90–103. doi: 10.1101/gad.1267005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2006 doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 51.Tarun SZ, Jr, Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl Acad. Sci. USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA. 2005;11:517–531. doi: 10.1261/rna.7291205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc. Natl Acad. Sci. USA. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muhlrad D, Parker R. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA. 1999;5:1299–1307. doi: 10.1017/s1355838299990829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burd CG, Matunis EL, Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol. Cell Biol. 1991;11:3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 57.Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 58.Mangus DA, Amrani N, Jacobson A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell Biol. 1998;18:7383–7396. doi: 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boeck R, Tarun S, Jr, Rieger M, Deardorff JA, Muller-Auer S, Sachs AB. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- 60.Kuhn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J. Mol. Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- 61.Melo EO, Dhalia R, Martins de Sa C, Standart N, de Melo Neto OP. Identification of a C-terminal poly(A)-binding protein (PABP)-PABP interaction domain: role in cooperative binding to poly (A) and efficient cap distal translational repression. J. Biol. Chem. 2003;278:46357–46368. doi: 10.1074/jbc.M307624200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.