Abstract
Translation of most eukaryotic mRNAs involves the synergistic action between the 5′ cap structure and the 3′ poly(A) tail at the initiation step. The poly(A) tail has also been shown to stimulate translation of picornavirus internal ribosome entry sites (IRES)-directed translation. These effects have been attributed principally to interactions between eIF4G and poly(A)-binding protein (PABP) but also to the participation of PABP in other steps during translation initiation. As the rabbit reticulocyte lysate (RRL) does not recapitulate this cap/poly(A) synergy, several systems based on cellular cell-free extracts have been developed to study the effects of poly(A) tail in vitro but they generally exhibit low translational efficiency. Here, we describe that the non-nuclease-treated RRL (untreated RRL) is able to recapitulate the effects of poly(A) tail on translation in vitro. In this system, translation of a capped/polyadenylated RNA was specifically inhibited by either Paip2 or poly(rA), whereas translation directed by HCV IRES remained unaffected. Moreover, cleavage of eIF4G by FMDV L protease strongly stimulated translation directed by the EMCV IRES, thus recapitulating the competitive advantage that the proteolytic processing of eIF4G confers to IRES-driven RNAs.
INTRODUCTION
Translation initiation is a highly ordered process that involves the concerted action of several polypeptides called eukaryotic initiation factors (eIFs) and other accessory proteins which facilitate the recruitment of the ribosome onto the mRNA molecule (1). In eukaryotes, virtually all nuclear-encoded mRNAs possess an m7GpppN (where N is any nucleotide) cap structure at their 5′ terminus and a poly(A) tail (50–300 nt) at the 3′ end. These structures have been shown to act synergistically to promote translation initiation in yeast, mammals and plants in vivo (2).
The 5′ cap moiety is recognized and bound by eIF4F which is composed by the cap-binding protein eIF4E, the ATP-dependent RNA helicase eIF4A and the scaffold protein eIF4G (3). In mammals, the interaction between the 40S ribosomal subunit-associated initiation factor eIF3 and eIF4G bridges the ribosome to the 5′ end of the mRNA. A growing number of viral and cellular RNAs use an alternative cap-independent mechanism that allows the recruitment of the ribosome internally by interaction of RNA structures located in the 5′-UTR and called internal ribosome entry sites (IRES) (4,5). The mechanism of IRES-dependent translation does not require the cap-binding protein eIF4E and allows efficient protein synthesis under conditions where cap-dependent translation is either repressed or shut off (6–8).
Genetic and biochemical studies have shown that eIF4G interacts with the poly(A)-binding protein (PABP) in yeast, plants and mammals promoting a pseudocircularization of the mRNA (9–11). The circularization of a capped and polyadenylated transcript could even be visualized by high-resolution microscopy using purified yeast eIF4E, eIF4G and PABP (12). PABP is a highly conserved protein among species which covers the length of the poly(A) tail on the mRNA. It contains four RNA-recognition motifs (RRMs 1–4) and a C-terminal domain (CTD) that is responsible for many protein–protein interactions including translation factors eIF4G, eIF4B and eRF3 (13). The interaction between eIF4G and PABP stimulates translation initiation and several possible mechanisms have been proposed to explain this effect, they include: (i) ribosome recycling promoted by pseudocircularization of the mRNA, (ii) an increase in the association of 60S ribosomal subunits and (iii) a higher affinity of the eIF4F holoenzyme for the cap structure. The recent characterization of the PABP-interacting proteins, Paip1 and Paip2 (which stimulates and represses poly(A)-dependent translation, respectively) confirm that protein synthesis can be regulated by 5′ to 3′ interactions of the mRNA (14,15).
Translation in the conventional nuclease-treated rabbit reticulocyte lysate (RRL) and other nuclease-treated cell-free in vitro systems fail to recreate the selective advantage conferred by addition of the poly(A) tail to the mRNA (16–18). As a consequence, several in vitro systems that recapitulate a high level of competitiveness have been developed to study the cap/poly(A) synergy in vitro (11,16,19–21). All these cell-free systems were very successful in mimicking the competitive cellular environment and have been instrumental to improve our knowledge about the role of the cap/poly(A) tail interaction in initiation on both cap- and IRES-dependent translation (22–24). However, these in vitro systems are somehow very tedious to make and they exhibit relatively poor translational efficiency. Recently, an in vitro system based on a nuclease-treated RRL partially depleted from its ribosomes by ultracentrifugation has been engineered (25). Although the latter recapitulates well the cap/poly(A) synergy and is relatively simple to make, it exhibited very poor translational efficiencies compared to the parental reticulocyte lysate probably due to the depletion of some rate-limiting ribosome-associated factors.
Here, we show that the commercially available untreated RRL that contains endogenous mRNAs (globin and lipoxygenase principally) is able to recreate very faithfully the cap/poly(A) tail synergy in vitro. Interestingly, this system also recapitulates the selective advantage displayed by the EMCV IRES under conditions of cap-dependent translation inhibition ex vivo that is not appreciated in the nuclease-treated RRL. In summary, the results presented here place the untreated RRL as a very good and reliable in vitro system to recreate a ‘near to physiological translational environment’ that can be easily used as a tool for functional studies in translation.
MATERIALS AND METHODS
DNA constructions
The pGEM-Renilla vector was recently described and used as template to generate polyadenylated and non-polyadenylated transcripts by digestion with EcoRI or SmaI, respectively (26). The 80 nt region containing the T7 promoter was deleted by PvuII/BamHI digestion and the resulting vector was used to insert the different 5′-UTRs. The human β-globin 5′-UTR with the authentic initiation codon was obtained by hybridizing two synthetic oligodeoxyribonucleotides (Eurogentec) and cloned into the double-digested vector (EcoRV and BamHI restriction sites and T7 promoter were added with the oligos) generating the pGlobin-renilla vector. The EMCV and HCV IRES were obtained by PCR using the pXL EMCV (27) and the pDC HCV vectors (28), respectively as templates. Both PCR products were EcoRV/BamHI-digested and cloned in the double-digested vector (T7 promoter and restriction sites were added by PCR) generating pEMCV-renilla and pHCV-renilla vectors, respectively. In the case of pHCV-renilla, as the HCV IRES contains two SmaI sites, the SmaI site at the end of the renilla coding region was changed to an XhoI site by site-directed mutagenesis (QuickChange® XL, Stratagene).
In vitro transcription
The four possible combinations, capped/polyadenylated (+/+), capped/non-polyadenylated (+/−), uncapped/polyadenylated (−/+) and uncapped/non-polyadenylated (−/−) were obtained by in vitro transcription using the linear template of pRenilla vector that was linearized either at the SmaI (non-polyadenylated RNAs) or at the EcoRI site (polyadenylated RNAs) (Figure 1A). It should be noted that non-polyadenylated RNA do not contain any 3′-UTR after the luciferase coding region. Uncapped RNAs were obtained by using 2 µg of linear DNA template, 20 U of T7 RNA polymerase (Promega Co., Madison, WI, USA), 40 U of RNAsin (Promega Co, Madison, WI, USA), 10 mM of each ribonucleotide triphosphate, 30 mM DTT in transcription buffer [40 mM Tris–HCl (pH 7.9), 6 mM MgCl2, 2 mM spermidine and 10 mM NaCl]. For capped RNAs, the rGTP concentration was reduced to 0.48 mM and the m7GpppG cap analogue (Invitrogen, Co) was added to a final concentration of 1.92 mM as previously described (29). The transcription reaction was carried out at 37°C for 1.5 h and the RNAs were precipitated with LiCl at 2.5 M final concentration. The integrity of the RNAs was checked by electrophoresis on non-denaturating agarose gels and their concentration was quantified by spectrophotometry at 260 nm using Nanodrop (NanoDrop Technologies, Wilmington, Delaware, USA).
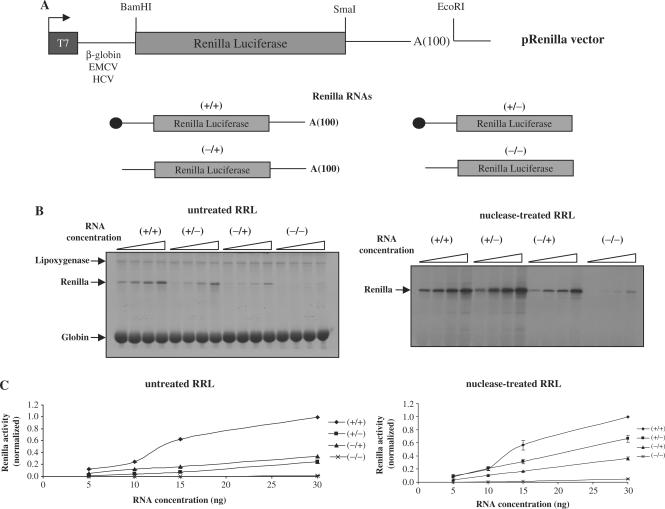
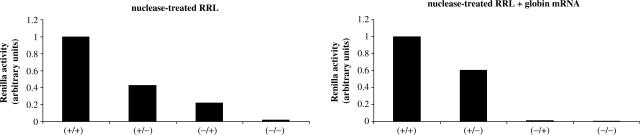
Figure 1.
(A) Schematic diagram, not to scale, of the constructs used in this study. The cap structure in the RNAs is indicated by a black circle, the 100 nt length poly(A) tail is indicated by A(100). (B) Capped/polyadenylated (+/+), capped/non-polyadenylated (+/−), uncapped/polyadenylated (−/+) or uncapped/non-polyadenylated (−/−) Globin-renilla RNAs were translated at 0.05, 0.1, 0.15 or 0.3 mg/ml in the untreated RRL (left panel) or the nuclease-treated lysate (right panel) as described in Materials and Methods section. Translational products were analysed by 15% SDS–PAGE and autoradiography. Synthesis of endogenous globin and lipoxygenase together with exogenous renilla products are indicated by arrows on the figure. Densitometric quantification of renilla synthesis at 0.1 mg/ml from Globin-renilla RNA translation in the untreated and nuclease-treated lysates has been plotted at the bottom of each panel. (C) Capped/polyadenylated (+/+), capped/non-polyadenylated (+/−), uncapped/polyadenylated (−/+) or uncapped/non-polyadenylated (−/−) Globin-renilla RNAs were translated at 0.05, 0.1, 0.15 or 0.3 mg/ml in the untreated RRL (left panel) or the nuclease-treated lysate (right panel) as described in Materials and Methods section. Renilla luciferase activity was then determined and normalized as described in Materials and Methods section. The results presented are representative of three independent experiments are expressed as means ± SD.
Capped and polyadenylated ‘natural’ globin mRNAs were purchased from Gibco (Gibco BRL).
Preparation of the untreated RRL and in vitro translation reactions
The untreated RRL was supplemented as previously described [(30,31), Morley, S., personal communication]. Briefly, 1 ml of untreated RRL (Promega Co., Madison, WI, USA) was supplemented with 25 µM haemin (Fluka), 25 µg/ml creatine phosphokinase (Sigma-Aldrich Co.), 5 mg/ml creatine phosphate (Fluka), 50 µg/ml of bovine liver tRNAs (Sigma-Aldrich Co.) and 3 mM of d-glucose (Sigma-Aldrich Co.). In vitro transcribed RNAs were translated in 10 µl of the supplemented untreated RRL or the Flexi® Rabbit Reticulocyte System 50% (v/v) each (Promega Co., Madison, WI, USA) in the presence of KCl (75 mM), MgCl2 (0.5 mM), 20 µM of amino acids mix minus methionine and 0.6 µCi of [35S]-methionine (GE Healthcare Life Sciences) for 30 min at 30°C. Reactions were stopped with 2 × SDS-loading buffer and the products were resolved by 15% SDS–PAGE. Gels were dried and subjected to autoradiography using Biomax films (Eastman Kodak Co.). Densitometric analyses were performed by Phospho Imaging with a Fuji FLA-5100 imaging system (Fujifilm Life Sciences) using the ImageReader FLA-5000 software (Fujifilm Life Sciences).
Renilla luciferase activity
Renilla activity from translation reactions carried out without [35S]-methionine was measured in a Veritas™ Luminometer (Turner Biosystems) using the Renilla Luciferase Assay System (Promega, Madison Co).
Recombinant GST-Paip2
BL21(DE3)CodonPlus-RP bacteria (Stratagene) were transformed with pGEX–Paip2 (a kind gift from Dr Dalla Venezia). Bacterial culture was grown at 30°C to a density of A600 = 0.4, then induced with IPTG at a final concentration of 1 mM and shaken for additional 3 h at 20°C. The cells were lysed by freeze/thaw cycles, lysozyme addition and sonication in buffer A (50 mM MOPS–KOH pH 7.2, 50 mM NaCl, 10 mM EDTA, 2 mM DTT and 10% glycerol). The GST-Paip2 proteins were purified from the soluble fraction using Glutathione Sepharose 4B (GE Healthcare Life Sciences). Proteins were washed with buffer B (50 mM MOPS–KOH pH 7.2, 100 mM NaCl, 10 mM EDTA, 2 mM DTT, 10% glycerol and 1% Triton-X100) and eluted with buffer A + 5 mM reduced glutathione (Sigma-Aldrich Co). The fractions were analysed by Coomassie blue staining. The appropriate fraction was dialysed against buffer C (20 mM HEPES–KOH pH 7.5, 100 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT and 10% glycerol). The recombinant GST-Paip2 proteins were analysed for size and integrity by SDS gel electrophoresis, followed by staining with Coomassie blue. Protein concentration was determined with the Biorad Protein Assay (Bio-Rad Laboratories, Inc.).
GST-Paip2 and poly(rA) competition
Translation mixtures were pre-incubated either with different concentrations of recombinant GST-Paip2 or free poly(rA) (GE Healthcare Life Sciences) for 5 min at 30°C prior to addition of [35S]-methionine and RNAs.
FMDV L protease
pMM-1 vector that codes for FMDV L protease under the T7 promoter (32) was linearized with XbaI and transcribed as indicated for uncapped RNAs. The Flexi® RRL system 50% (v/v) (Promega Co, Madison, WI, USA) was programmed with 2 µg of MM-1 RNA and treated as previously described (33).
RESULTS
The untreated RRL as a model system for cap/poly(A) synergy studies in vitro
The untreated RRL from a commercial origin (Promega) was supplemented with haemin, creatine phosphokinase, creatine phosphate and tRNAs as it was described in the original publication of Pelham and Jackson (31). For all experiments described in this article, we have used RNA constructs that contain the renilla luciferase gene (pRenilla vector) driven by either the human β-globin 5′-UTR, the EMCV IRES or the HCV IRES as stated (Figure 1A). The untreated RRL contains two endogenous mRNAs, lipoxygenase and globin, which are actively translated in this system to produce proteins that run at 68 and 15 kDa, respectively (Figure 1B, left panel). Translation of the Globin-renilla RNA in the untreated lysate system revealed that the presence of both the cap and the poly(A) tail on the RNA (+/+ on the figure) conferred a real advantage for translation (Figure 1B). Addition of the cap and the poly(A) tail together strongly stimulated translation of Globin-renilla RNA (more than 4-fold) at 10 ng of RNA (Figure 1B, left panel) indicating that the untreated RRL was able to recapitulate the synergistic effect of the cap structure and the poly(A) tail in vitro. At the same RNA concentrations, the effects of the cap and the poly(A) tail on Globin-renilla RNA translation resulted in a modest increase (<20%) in the nuclease-treated reticulocyte lysate (Figure 1B, right panel) as it was previously described (17). Moreover, the synergistic effects observed in the untreated RRL were RNA concentration-sensitive since at high RNA concentration (over 30 ng of RNA) no synergy was observed (data not shown). Interestingly, translation of the (−/+) and (−/−) Globin-renilla RNA, was very weak demonstrating the importance of the cap and poly(A) structures for efficient translation in competitive conditions. Quantification of the luciferase activity from three independent experiments has been carried out and the results are presented as graphs (Figure 1C). It should be noted that the variations in translational efficiencies did not reflect any significant differences in RNA stability in the treated and untreated lysates (data not shown).
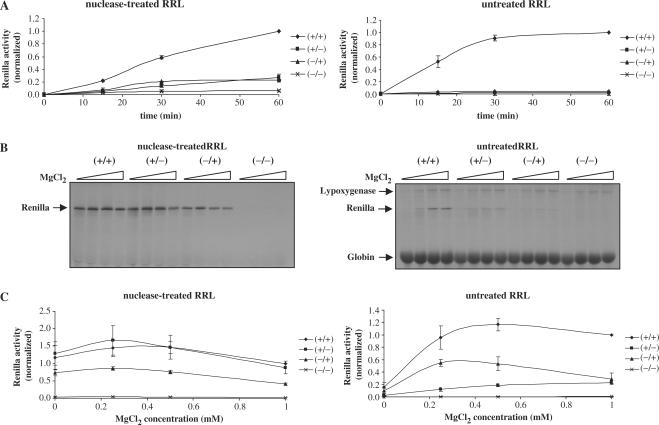
The next step was to investigate the role of the cap and the poly(A) tail in kinetics studies. For this, a low concentration (10 ng) of Globin-renilla RNAs was translated in both reticulocyte lysate systems and protein production was analysed at 0, 15, 30 and 60 min by analysis of the renilla luciferase activity (Figure 2A). It is interesting to note that the kinetics of translation of the capped and polyadenylated transcript in the competitive system is much higher than that of the other RNAs (compare +/+ with all others). It is noteworthy that such a difference between the (+/+) and all other transcripts can be observed right from the early time points (Figure 2A, compare left and right panel).
Figure 2.
(A) Different combinations of Globin-renilla RNAs (+/+), (+/−), (−/+) or (−/−) were translated at 0.1 mg/ml for 0, 15, 30 or 60 min in the nuclease-treated RRL (left panel) or untreated RRL (right panel) as indicated on the figure. Renilla luciferase activity was then determined and normalized as described in Materials and Methods section.(B) Globin-renilla RNAs (+/+), (+/−), (−/+) or (−/−) at 0.1 mg/ml were translated in both nuclease-treated RRL (left panel) or untreated RRL (right panel) in the presence of 0, 0.25, 0.5 or 1 mM added MgCl2. The resulting translation products were resolved by 15% SDS–PAGE and autoradiography and the position of the protein products are indicated by arrows on the figure. (C) Globin-renilla RNAs (+/+), (+/−), (−/+) or (−/−) at 0.1 mg/ml were translated in both nuclease-treated RRL (left panel) or untreated RRL (right panel) in the presence of 0, 0.25, 0.5 or 1 mM added MgCl2. Renilla luciferase activity was then determined and normalized as described in Materials and Methods section. The results presented are representative of three independent experiments are expressed as means ± SD.
Another important matter was to study the effects of salt concentration changes in both nuclease-treated and untreated systems as it had been described to be an important parameter for translational accuracy and efficiency in vitro (31,34). Whereas preferentially similar values for the optimal KCl concentration were obtained for (+/+) and (+/−) RNAs in both systems (data not shown), a different profile was observed when changes in the MgCl2 concentration were imposed. At high Mg2+ concentration, an inhibitory effect was observed for all RNAs tested in the nuclease-treated lysate system (Figure 2B and C). However, the situation is completely different in the competitive system with a net increase in translation for the (+/+) RNA construct at elevated level of exogenously added Mg2+. Once again, the cap and poly(A) addition have a synergistic effect at all Mg2+ tested (Figure 2B and C for the quantification of the luciferase activity). Taken together, these results show that the untreated RRL system recapitulates faithfully the cap/poly(A) synergy together with a stimulatory response to kinetics and salt concentration changes, thus mimicking physiological conditions encountered within cells.
Paip2 and free poly(rA) specifically inhibit translation of a capped/polyadenylated RNA in the untreated RRL system
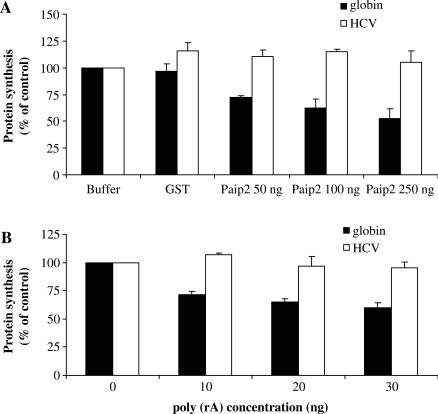
It has recently been shown that the PABP-interacting protein 2 (Paip2) was able to down regulate poly(A)-dependent translation by a two-step mechanism: it initially disrupts the PABP–poly(A) tail interactions (15) and it directly competes with eIF4G for binding to PABP (35). Therefore, the effect of recombinant GST-Paip2 addition has been studied on the translation of Globin-renilla (+/+) RNA in the untreated lysate system (Figure 3A). The data show that translation of exogenous capped/polyadenylated was specifically inhibited by addition of recombinant GST-Paip2 in a dose-dependent manner (reaching ∼50% of inhibition at the highest concentration tested). These effects were not due to the presence of the GST fragment in the fusion protein since the GST alone did not influence translation of this RNA. As expected, no effect could be observed on translation driven by the HCV IRES, which has the ability to initiate protein synthesis independently from eIF4G, PABP and the poly(A) tail (compare globin and HCV in Figure 3A).
Figure 3.
(A) Capped and polyadenylated Globin-renilla RNAs (+/+) and uncapped and non-polyadenylated HCV-renilla RNAs (−/−) were translated in the untreated reticulocyte lysate that had been pre-incubated with 50, 100 or 250 ng of GST-Paip2 recombinant protein or (B) 10, 20 or 30 ng of free poly(rA). Protein production was determined by densitometric quantification of the renilla protein product and is expressed as percent of the control (no inhibitor added, set to 100%). Data were obtained from densitometric analyses of at least three independent experiments and are expressed as means ± SD.
As additional evidence, we have also added free poly(rA) in trans to the untreated reticulocyte system. It has been shown that a free poly(rA) oligoribonucleotide has the ability to specifically inhibit cap/poly(A) tail-dependent translation by competing for PABP binding (17,36,37). Therefore, translation of Globin-renilla (+/+) RNA was studied under these experimental conditions (Figure 3B). Addition of increasing concentrations of the poly(A) polymer resulted in the inhibition of translation of Globin-renilla (+/+) RNA albeit to a lesser extent than that obtained with Paip2. Actually, addition of free poly(rA) inhibited as much as 40% of translation compared to the control situation but we failed to observe a clear dose-dependent effect. This result is somehow similar to that obtained by Munroe and Jacobson (∼50% of inhibition) (17). A possible explanation for this relatively low effect on cap/poly(A) translation could come from the fact that the poly(rA) competitor only disrupts the interactions between the PABP and the poly(A) tail without affecting the PABP–eIF4G interaction (35). However, this effect was clearly dependent on the presence of the poly(A) tail since no inhibition of HCV IRES-dependent translation was observed in the presence of poly(rA) (compare globin and HCV in Figure 3B). The degree of inhibition obtained with both GST-Paip2 and poly(rA) in the untreated RRL are in complete agreement with ex vivo data obtained in HeLa cells showing that the knockout of PABP by RNA interference has only a little effect (no more than a 20% of inhibition) on translation of an exogenous RNA (38). These results show that translation of exogenous Globin-renilla (+/+) RNA in the untreated RRL system is operated by a cap- and poly(A)-dependent mechanism that can be selectively inhibited by blocking the interactions between PABP and its partners.
The untreated RRL as a system to study IRES-dependent translation
Several studies have now shown that the poly(A) tail can stimulate picornavirus IRES-mediated translation to a certain extent (22–24). Recently, using a competitive in vitro system from Krebs-2 cell extracts, Svitkin and colleagues (21) have shown that the presence of competitors RNAs was required to observe an important stimulation of translation driven by the EMCV IRES following the cleavage of eIF4G by picornaviral proteases. Such a system recapitulates faithfully the situation encountered during picornaviral infection and can be explained by the fact that the low availability of eIF4E bound to eIF4F was critical to confer a strong advantage to IRES-mediated translation which does not require the cap-binding protein.
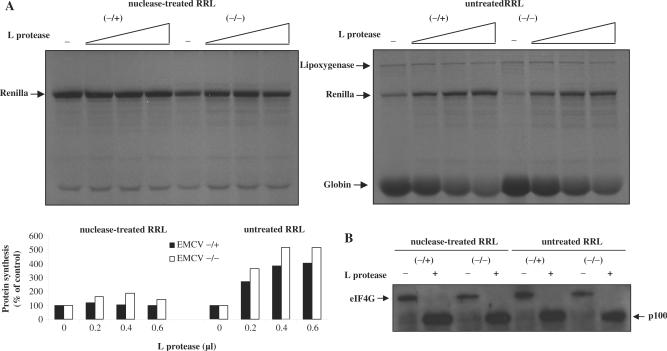
In order to analyse the ability of the untreated RRL to recreate these effects, polyadenylated and non-polyadenylated versions of an EMCV-renilla RNA (designed EMCV −/+ and EMCV −/−, respectively) were translated in parallel in the nuclease-treated (Figure 4A, left panel) and the untreated (Figure 4A, right panel) RRLs in the presence of increasing concentrations of an in vitro-synthesized FMDV L protease. As seen in the figure, translation of both EMCV RNAs was moderately stimulated by the cleavage of eIF4G in the nuclease-treated RRL. In sharp contrast with these data, a large increase of EMCV IRES-mediated translation upon addition of the L protease was observed in the untreated competitive lysate (over 5-fold enhancement, Figure 4A, right panel). Interestingly, at the same time, translation of endogenous globin mRNA was inhibited (see bottom of the gel) thus providing an internal control to monitor the extent of cap-dependent translation inhibition. In the context of our experimental conditions, treatment with 0.6 µl of L protease resulted in the cleavage of virtually all endogenous eIF4GI contained in the lysate as visualized by western blotting analysis (Figure 4B).
Figure 4.
(A) Uncapped polyadenylated (−/+) and uncapped non-polyadenylated (−/−) EMCV-renilla RNAs were translated at 0.5 mg/ml during 30 min at 30°C in both the nuclease-treated (left panel) and untreated RRLs (right panel) that were pre-incubated with 0, 0.2, 0.4 or 0.6 µl of an in vitro-synthesized FMDV L protease (see Materials and Methods section). Translation products were resolved by 15% SDS-PAGE followed by autoradiography. Densitometric analysis of the renilla protein product was performed and expressed as percentage of the control (100%, no l protease added) at the bottom of each panel. (B) At the end of the in vitro translation incubation, samples (1 µl) from the experiment described above were run on a 10% SDS–PAGE and the proteins transferred to PVDF membrane and incubated with antibodies specific to the C-terminal part of eIF4GI.
These results are in complete agreement with those obtained by Svitkin and colleagues and indicate that inhibition of cap-dependent translation by eIF4G cleavage (L protease) in the untreated RRL strongly stimulates IRES-mediated translation thus recreating the competitive advantage observed for IRES-dependent translation in vivo. It should be noted that similar results were obtained by using m7GpppG cap analogue (data not shown).
Addition of saturating concentration of globin mRNA in the nuclease-treated reticulocyte lysate
Data obtained so far show that the untreated reticulocyte lysate recapitulates cap/poly(A) synergy and the selective advantage of IRES-driven translation under conditions where cap-dependent translation was inhibited. The fact that cap/poly(A) synergy cannot be observed in the nuclease-treated RRL suggests that the presence of endogenous globin mRNAs may play a key role in the regulation of translation probably by competing for the translational apparatus. However, another possibility could be that the treatment of the reticulocyte lysate with micrococcal nuclease may also alter some components of the translational apparatus rendering them limiting for translation of exogenously added mRNAs. Thus, to address this question we have added saturating amounts of exogenous globin mRNAs (Gibco) to a nuclease-treated reticulocyte lysate programmed with luciferase mRNAs (Figure 5, right panel). An incubation in the nuclease-treated lysate without inclusion of exogenous globin mRNAs was carried out in parallel (Figure 5, left panel). From these graphs it can be observed that there is a difference in the translational pattern as the uncapped transcripts were not expressed in the system supplemented with exogenous globin (right graph). However, it is also clear that cap/poly(A) synergy was not restored by the addition of competitor RNAs although both the cap and the poly(A) provided a selective advantage. Taken together these results show that the cap/poly(A) synergy is not totally attributable to competition for the translational apparatus in this system.
Figure 5.
Different combinations of Globin-renilla RNAs (+/+), (+/−), (−/+) or (−/−) were translated at 0.1 mg/ml for 30 min at 30°C in the nuclease-treated RRL (left graph) or nuclease-treated RRL supplemented with 0.15 mg/ml of globin mRNAs (Gibco) (right graph) as indicated on the figure. Translation products were quantified by analysis of luciferase activity.
DISCUSSION
As early as in the 1980s the first reports from the Jacobson's group showed that addition of free poly(A) to mammalian translation extracts was inhibitory for capped and polyadenylated mRNAs but stimulatory for poly(A)-deficient mRNAs suggesting a possible role for the poly(A) tail in translational control (17,37). Later, the characterization of PABPs in yeast and mammals introduced a possible role for the poly(A) tail in translation (39–41). In the early 1990s, Sachs and Davis (42) showed that mutations in the yeast PAB1 gene (encoding Pabp1) resulted in an inhibition of translation and cell growth indicating that this gene was essential in yeast.
Major breakthrough work was carried out by Gallie and coworkers who showed that the cap and poly(A) tail conferred an important translational competitive advantage to mRNAs delivered directly to the cytoplasm of yeast, plants and mammalian cells (2). They further show that the stimulatory effect added by both structures at the mRNA 5′ and 3′ termini were greater than additives and they introduced the term ‘synergy’ to the field.
The design of the first yeast-based cell-free translation system that could recapitulate the cap/poly(A) synergistic effect was very instrumental for functional in vitro translational studies (16). A condition to obtain synergistic stimulation of translation by the cap and poly(A) tail seems to require the presence of competitors RNAs since synergy was observed in non-nuclease-treated extracts or nuclease-treated extracts supplemented with an excess of exogenous RNAs (16,18,43).
Several cell-free translation systems have been successfully developed since and they all can reproduce well the ‘physiological cytoplasmic environment’ (16,18,19,21,22,24,43). However, major drawbacks from these systems come from the fact that they are usually technically difficult to prepare and that they are engineered ‘in house’ on a relatively small scale thus posing the problem of reproducibility.
To circumvent these problems, Kean and colleagues developed an in vitro system based on the conventional nuclease-treated RRL that was partially depleted from ribosomes and initiation factors by ultracentrifugation. Although, this system was able to recreate the effects of polyadenylation, its translational efficiency remained extremely low compared to the parental lysate and they require from 1 to 10 days to obtain a detectable signal on the autoradiography (23,25).
Here we describe the translational properties of the untreated reticulocyte lysate which appear to be a very good model system to faithfully recapitulate cap/poly(A) synergy. We show that exogenously added RNAs can be translated in this system and protein production can be measured by incorporation of [35S] methionine as described previously (31). This results in the labelling of the lipoxygenase at 68 kDa and globin at 15 kDa together with the protein product from the exogenously added RNA such as renilla luciferase (Figure 1B). The difference in size allows a clear distinction between these three proteins which facilitates the interpretation of the data. Moreover, endogenous globin production can be used as an internal control for cap-dependent translation when various inhibitors are added (Figure 4). The reason why the nuclease-treated RRL is unable to restitute cap/poly(A) synergy remains unknown but the experiment presented in Figure 5 suggests that it is not entirely due to the lack of competition with endogenous RNAs and may be also attributable to some effect of the micrococcal treatment itself.
In summary, the untreated RRL system exhibits very interesting properties to study in vitro translation as: (i) it recapitulates faithfully cap and poly(A) synergy (Figure 1B), (ii) it responds well to variations of divalent salt concentration (Figure 2A), (iii) it is sensitive to Paip2 and poly(rA) additions which are known inhibitors of poly(A)-dependent translation (Figure 3) and (iv) it mimics the beneficial competitive environment for IRES-driven translation that is encountered following the cleavage of eIF4G by viral proteases (Figure 4).
All these properties make the untreated reticulocyte lysate a convenient alternative in vitro method for studies on translational control.
ACKNOWLEDGEMENTS
We wish to acknowledge Dr Dalla Venezia (University of Rockfeller, Lyon) for donating the pGEX-Paip2 construct and Dr Simon Morley for donating antibodies against eIF4G. This work was supported by a grant from the ANR and an ACI jeune chercheur. R.S.R. is supported by a Conicyt/French Embassy in Chile doctoral fellowship, E.P.R. holds a grant from the MENRT and O.M. is supported by SIDACTION. Funding to pay the Open Access publication charges for this article was provided by the Agence Nationale de la Recherche (ANR).
Conflict of interest statement. None declared.
REFERENCES
- 1.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 2.Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes. Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 3.Prevot D, Darlix JL, Ohlmann T. Conducting the initiation of protein synthesis: the role of eIF4G. Biol. Cell. 2003;95:141–156. doi: 10.1016/s0248-4900(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 4.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 6.Jackson RJ, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez-Lastra M, Rivas A, Barria MI. Protein synthesis in eukaryotes: the growing biological relevance of cap-independent translation initiation. Biol. Res. 2005;38:121–146. doi: 10.4067/s0716-97602005000200003. [DOI] [PubMed] [Google Scholar]
- 8.Sarnow P. Translational control during virus infection. Virus Res. 2006;119:1. doi: 10.1016/j.virusres.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le H, Tanguay RL, Balasta ML, Wei CC, Browning KS, Metz AM, Goss DJ, Gallie DR. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J. Biol. Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- 11.Tarun SZ, Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 12.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 13.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craig AW, Haghighat A, Yu AT, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 15.Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 16.Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol. Cell. Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preiss T, Muckenthaler M, Hentze MW. Poly(A)-tail-promoted translation in yeast: implications for translational control. RNA. 1998;4:1321–1331. doi: 10.1017/s1355838298980669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gebauer F, Corona DF, Preiss T, Becker PB, Hentze MW. Translational control of dosage compensation in Drosophila by sex-lethal: cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J. 1999;18:6146–6154. doi: 10.1093/emboj/18.21.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Preiss T, Hentze MW. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 21.Svitkin YV, Herdy B, Costa-Mattioli M, Gingras AC, Raught B, Sonenberg N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell. Biol. 2005;25:10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergamini G, Preiss T, Hentze MW. Picornavirus IRESes and the poly(A) tail jointly promote cap- independent translation in a mammalian cell-free system. RNA. 2000;6:1781–1790. doi: 10.1017/s1355838200001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulous S, Malnou CE, Michel YM, Kean KM, Borman AM. Comparison of the capacity of different viral internal ribosome entry segments to direct translation initiation in poly(A)-dependent reticulocyte lysates. Nucleic Acids Res. 2003;31:722–733. doi: 10.1093/nar/gkf695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svitkin YV, Imataka H, Khaleghpour K, Kahvejian A, Liebig HD, Sonenberg N. Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA. 2001;7:1743–1752. [PMC free article] [PubMed] [Google Scholar]
- 25.Michel YM, Poncet D, Piron M, Kean KM, Borman AM. Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 2000;275:32268–32276. doi: 10.1074/jbc.M004304200. [DOI] [PubMed] [Google Scholar]
- 26.Dizin E, Gressier C, Magnard C, Ray H, Decimo D, Ohlmann T, Dalla Venezia N. BRCA1 interacts with poly(A)-binding protein: implication of BRCA1 in translation regulation. J. Biol. Chem. 2006;281:24236–24246. doi: 10.1074/jbc.M602176200. [DOI] [PubMed] [Google Scholar]
- 27.Ohlmann T, Jackson RJ. The properties of chimeric picornavirus IRESes show that discrimination between internal translation initiation sites is influenced by the identity of the IRES and not just the context of the AUG codon. RNA. 1999;5:764–778. doi: 10.1017/s1355838299982158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prevot D, Decimo D, Herbreteau CH, Roux F, Garin J, Darlix JL, Ohlmann T. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 2003;22:1909–1921. doi: 10.1093/emboj/cdg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohlmann T, Lopez-Lastra, Darlix JL. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J. Biol. Chem. 2000;275:11899–11906. doi: 10.1074/jbc.275.16.11899. [DOI] [PubMed] [Google Scholar]
- 30.Jackson RJ, Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- 31.Pelham HR, Jackson RJ. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur. J. Biochem. 1976;67:247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- 32.Medina M, Domingo E, Brangwyn JK, Belsham GJ. The two species of the foot-and-mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology. 1993;194:355–359. doi: 10.1006/viro.1993.1267. [DOI] [PubMed] [Google Scholar]
- 33.Ronfort C, De Breyne S, Sandrin V, Darlix JL, Ohlmann T. Characterization of two distinct RNA domains that regulate translation of the Drosophila gypsy retroelement. RNA. 2004;10:504–515. doi: 10.1261/rna.5185604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson RJ. Potassium salts influence the fidelity of mRNA translation initiation in rabbit reticulocyte lysates: unique features of encephalomyocarditis virus RNA translation. Biochim. Biophys. Acta. 1991;1088:345–358. doi: 10.1016/0167-4781(91)90124-5. [DOI] [PubMed] [Google Scholar]
- 35.Karim MM, Svitkin YV, Kahvejian A, De Crescenzo G, Costa-Mattioli M, Sonenberg N. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc. Natl Acad. Sci. USA. 2006;103:9494–9499. doi: 10.1073/pnas.0603701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borman AM, Michel YM, Malnou CE, Kean KM. Free poly(A) stimulates capped mRNA translation in vitro through the eIF4G-poly(A)-binding protein interaction. J. Biol. Chem. 2002;277:36818–36824. doi: 10.1074/jbc.M205065200. [DOI] [PubMed] [Google Scholar]
- 37.Jacobson A, Favreau M. Possible involvement of poly(A) in protein synthesis. Nucleic Acids Res. 1983;11:6353–6368. doi: 10.1093/nar/11.18.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida M, Yoshida K, Kozlov G, Lim NS, De Crescenzo G, Pang Z, Berlanga JJ, Kahvejian A, Gehring K, et al. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J. 2006;25:1934–1944. doi: 10.1038/sj.emboj.7601079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adam SA, Nakagawa T, Swanson MS, Woodruff TK, Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol. Cell. Biol. 1986;6:2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sachs AB, Bond MW, Kornberg RD. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986;45:827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- 41.Sachs AB, Kornberg RD. Nuclear polyadenylate-binding protein. Mol. Cell. Biol. 1985;5:1993–1996. doi: 10.1128/mcb.5.8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sachs AB, Davis RW. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 43.Tarun SZ, Jr, Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail- dependent translation. Proc. Natl Acad. Sci. USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]