The fact that membrane-bound heparan sulfate proteoglycans (HSPGs) regulate the activity of heparin-binding growth factor has been known for many years, but only during the last decade has the existence of two major families of membrane-bound HSPGs been established. One of these families, the glypicans, plays a critical a role in developmental morphogenesis. The interest in the study of glypicans has increased in the last few years as a result of the discovery that glypican-3 (GPC3) is mutated in an overgrowth and dysmorphic syndrome. The finding that GPC3 regulates body size was particularly surprising and suggested that this glypican may interact with signaling pathways that have not been traditionally associated with membrane-bound HSPGs. Here we will describe our current knowledge on the structure and function of glypicans. The involvement of these molecules in human pathologies will be discussed, including the different hypotheses with regard to the molecular basis of glypican function.
The glypican family
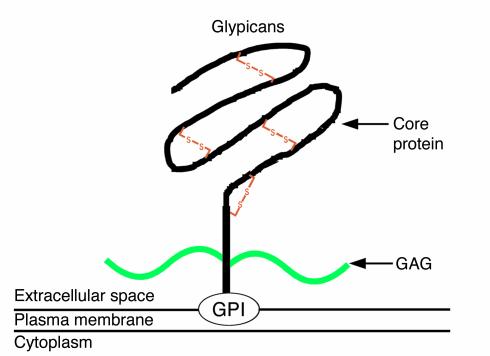
Glypicans are a family of HSPGs that are linked to the cell surface by a glycosylphosphatidylinositol (GPI) anchor (1) (Figure 1). Six members of this family (GPC1 to GPC6) have been identified in mammals (2–9), and two in Drosophila (10, 11). The size of the core protein of glypicans is similar (60–70 kDa), and, as expected, they all display an N-terminal secretory signal peptide, and a hydrophobic domain required for the addition of the GPI anchor at the C-terminus. The degree of amino acid homology among most glypicans is moderate (8), but the position of 14 cysteine residues is conserved, suggesting that the three-dimensional structure of glypicans is very similar. Another characteristic that is shared by all glypicans is the location of the insertion sites for the heparan sulfate chains, which seems to be restricted to the last 50 amino acids in the C-terminus, placing the chains close to the cell membrane (8).
Figure 1.
Schematic diagram of glypicans. GAG, glycosaminoglycan chains; GPI, glycosylphosphatidylinositol anchor.
In general, glypicans are expressed predominantly during development. Expression levels change in a stage- and tissue-specific manner, suggesting that glypicans are involved in the regulation of morphogenesis (7, 12–15).
Drosophila glypicans
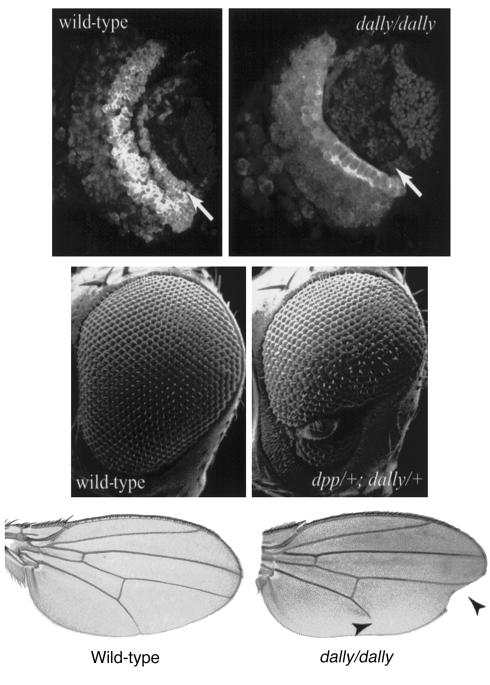
Two glypican-encoding genes are found in Drosophila, and a combination of genetic and molecular studies have revealed important features of their function during development. The best-characterized of the two Drosophila glypicans is division abnormally delayed (dally), a gene discovered by virtue of its effects on cell division patterning in the visual system (10). As its name implies, dally mutants show a failure of cell cycle progression for specific sets of dividing cells in the eye and developing brain. dally also plays a critical role in a number of other patterning events affecting wing, antenna, genitalia, and eye development (Figure 2).
Figure 2.
Three phenotypes of Dally mutants. The top two panels show third instar larval brains stained with anti–Cyclin B antibody to reveal patterns of cell division (anterior to the left, dorsal up). The wild-type displays two bands of dividing cells that generate components of the visual system. One of these two sets of dividing cells is absent in dally mutants (arrows). The middle panels show scanning electron micrographs of a wild-type and an abnormal eye of a dpp/+; dally/+ transheterozygote. Both dpp and dally mutations are completely recessive, and the eye defects found in animals heterozygous for both mutations reveal a functional link between Dally, a Drosophila glypican, and decapentaplegic (Dpp), a homolog of bone morphogenetic protein 4. The bottom row shows wing margin notching and incomplete wing vein formation (arrowheads) associated with dally mutations. These defects are likely the result of defects in Wingless (a Wnt) and Dpp signaling, respectively.
The first clues as to the molecular basis of these phenotypes came from experiments on the genetic interaction between dally and decapentaplegic (dpp). dpp encodes a protein with approximately 75% sequence identity to bone morphogenetic protein 4 (BMP4), a member of the TGF-β superfamily of secreted growth factors (16). Dpp plays an instrumental role in the control of cell division and patterning during Drosophila development, and human BMP4 can functionally replace Dpp in dorsal-ventral patterning of the Drosophila embryo (17). Genetic studies of dally and dpp showed that the eye, genital, and antennal phenotypes of dally mutants are at least in part a consequence of reductions in Dpp signaling (18). These findings were consistent with the proposal that glypicans can serve as growth factor coreceptors, regulating the activity of receptor activation by Dpp.
Biochemical characterization of Dally has shown that, like its vertebrate counterparts, Dally is GPI-linked and heparan sulfate–modified (19). Dally therefore is situated at the cell surface like the vertebrate glypicans, where it could readily influence growth factor reception. Recent structural analyses have shown that the nature of the glycosaminoglycan chains attached to proteoglycans like Dally are also highly conserved between vertebrates and Drosophila (20). The disaccharide composition of Drosophila heparan sulfate is very similar to vertebrate forms, with all the principal structural variants represented in a tissue- and developmental stage–specific manner.
A number of mutations affecting heparan sulfate biosynthesis or modification in Drosophila have a profound effect on Wingless (Wg) signaling (21). Wg is a Wnt family member, essential for a number of patterning events throughout both embryonic and postembryonic development. The demonstration that heparan sulfate biosynthesis was critical for normal Wg activity suggested that one or more proteoglycans participated in Wg-directed events. Further study of dally mutants demonstrated that the glypican encoded by this gene does indeed promote Wg activity (19, 22).
More recently, dally-like (dly), the second Drosophila glypican, has been implicated in Wg-mediated patterning of the embryo (11). RNA interference inhibition of dly produces an epidermal patterning abnormality like that found in wg mutant embryos. Manipulation of dly expression has provided some interesting clues that glypicans can regulate growth factor distributions in extracellular spaces and, in some contexts at least, block growth factor signaling (11). Ectopic expression of Dly in the developing wing disc dramatically increases the levels of Wg detectable extracellularly but, somewhat paradoxically, inhibits Wg-directed patterning at the developing wing margin. These findings suggest that high levels of glypicans may serve to prevent Wg from productively interacting with its signaling receptor, despite an increase in Wg levels at cell surfaces.
In summary, genetic studies of Drosophila have thus far implicated glypicans in signaling mediated by Wnt and TGF-β families of growth factors. Interestingly, there is selective participation of Dally in these pathways, depending on the cellular context. The basis of this selectivity is unknown. It is also important to note that so far the genetic studies in Drosophila have not implicated glypicans in the regulation of the activity of any member of the FGF family, although there is in vitro evidence indicating that glypicans can bind and modulate the activity of some members of this family (23, 24). Further analysis of dally and the other Drosophila glypican encoded by dly will likely identify additional signaling pathways that are modulated by these GPI-linked proteoglycans. A great deal of work remains to be done in order to understand how specific heparan sulfate modifications might modulate glypican function. The conservation of the heparan sulfate biosynthetic machinery, together with the genetic tools available in Drosophila, makes this problem readily accessible in this model system.
The Simpson-Golabi-Behmel syndrome
In 1996 Pilia et al. reported that GPC3, which encodes one member of the glypican family, is mutated in patients with Simpson-Golabi-Behmel syndrome (SGBS) (25). SGBS is an X-linked disorder characterized by pre- and postnatal overgrowth, and a broad spectrum of clinical manifestations that vary from a very mild phenotype in carrier females, to infantile lethal forms in some males (reviewed in ref. 26). The list of clinical manifestations of SGBS can include a distinct facial appearance, macroglossia, cleft palate, syndactyly, polydactyly, supernumerary nipples, cystic and dysplastic kidneys, congenital heart defects, rib and vertebral abnormalities, and umbilical/inguinal hernias (27–30).
Most of the GPC3 mutations identified so far are point mutations or small deletions encompassing a varying number of exons (31–34). Given the lack of correlation between patient phenotype and the location of the mutations, it has been proposed that SGBS is caused by the lack of a functional GPC3 protein, with additional genetic factors being responsible for the intra- and interfamilial phenotypic variation (31). The generation of GPC3-deficient mice has provided a strong support to this hypothesis (35). These mice display several of the abnormalities found in SGBS patients, including developmental overgrowth, and cystic and dysplastic kidneys.
Starting from early stages of kidney development a persistent increase in the proliferation rate of epithelial cells in the ureteric bud/collecting system was observed (35). This finding supports the idea that GPC3 can act as a negative regulator of cell proliferation, which is obviously consistent with the overgrowth of the SGBS patients and the GPC3-deficient mice.
Some of the clinical features of the SGBS, such as syndactyly and multiple nipples, suggest that GPC3 is involved in the regulation of cell survival in certain tissues during development. Experimental evidence supporting this hypothesis has been provided by the demonstration that GPC3 can induce apoptosis in a cell type–specific manner (36).
SGBS shares several clinical features with Beckwith-Wiedemann syndrome (BWS), another overgrowth syndrome (37). Since overexpression of IGF-II is thought to be one of the contributing factors to BWS (38), it has been proposed that GPC3 negatively regulates IGF-II activity by competing for IGF-II binding with the IGF-I receptor (the signaling receptor for IGF-II), and that GPC3 loss-of-function mutations are equivalent to overexpression of IGF-II (31). Further support for this hypothesis was provided by the generation of mice overexpressing IGF-II (39). In addition to some of the phenotypic features of BWS, these mice display skeletal defects that are typical of SGBS. It is important to note, however, that the SGBS patients and the GPC3-null mice display severe kidney abnormalities that have not been reported in the IGF-II transgenic mice.
Additional support for the possibility that GPC3 is involved in IGF-II signaling comes from the finding that IGF-II receptor–deficient (IGF2R-deficient) mice display the same degree of developmental overgrowth as that of the GPC3-null mice (40, 41). The IGF2R is a well-characterized negative regulator of IGF-II. It binds this growth factor, and downregulates its activity by endocytosis and degradation (42). Not surprisingly, then, the IGF2R-deficient mice display increased levels of IGF-II in blood and tissues (40, 41). The GPC3-null mice, on the other hand, have normal levels of IGF-II (33). Furthermore, no direct interaction between IGF-II and GPC3 has been detected (23). It can be concluded, therefore, that if GPC3 inhibits IGF-II signaling, it does so by a mechanism that is fundamentally different from that of the IGF2R.
Does the potential involvement of GPC3 in IGF-II signaling contradict the results of the genetic studies in Drosophila that implicated glypicans in the regulation of signaling by Wnt and TGF-β family members? Not necessarily. Given the complex clinical features of SGBS, and the differences in the kidney phenotype between the IGF-II transgenic and the GPC3-deficient mice, it is possible that some of the abnormalities observed in SGBS and the GPC3-null mice are the consequence of the involvement of GPC3 in the regulation of other growth factors in addition to IGF-II. Experimental evidence supporting this hypothesis has been provided by two recent reports (43, 44). In one of them, GPC3-deficient mice were mated with mice that were heterozygotes for BMP4. Interestingly, the offspring displayed polydactyly and rib malformations with high penetrance. These abnormalities were not observed in either parental strain (43). The second report used a branching morphogenesis assay to compare the response of embryonic kidney explants from GPC3-null and wild-type mice to BMP2, BMP7, and FGF-7, three polypeptides that are known to be involved in kidney morphogenesis (45–47). The results showed that GPC3 deficiency abrogated the inhibitory activity of BMP2 on branching morphogenesis, while converting BMP7-induced inhibition to stimulation, and enhancing the stimulatory effects of FGF-7. It was proposed, therefore, that GPC3 plays a role in BMP2, BMP7, and FGF-7 signaling during kidney development (44).
In summary, based on these results it is reasonable to speculate that, depending on the tissue and the developmental stage, GPC3 can regulate different kinds of growth factors. This versatility may be due to the fact that some of interactions of GPC3 do not depend on the HS chains but on the core protein, as suggested by recently published experimental evidence showing that the elimination of the HS chains does not abolish the capacity of this glypican to suppress growth in a colony-forming assay (36). In addition, since GPC3 can be secreted (48), it is possible that the secreted form of GPC3 acts via a mechanism distinct from that of the form attached to the cell surface.
The role of glypicans in cancer
Given the ability of glypicans to regulate the activity of growth and survival factors, recent reports associating changes in glypican expression with tumor progression were not surprising.
One of the first studies that established a connection between glypicans and cancer showed that GPC1 expression is significantly increased in a large proportion of pancreatic cancers (49). Furthermore, it was also reported that transfection of antisense GPC1 inhibits the mitogenic response of cultured pancreatic cancer cells to FGF2 and heparin-binding EGF-like growth factor (49) and decreases the tumorigenicity of the transfected cells (50). Although the precise mechanism by which the reduction of GPC1 expression inhibits tumor growth remains to be determined, the results are clearly consistent with in vitro experiments that have shown that GPC1 can facilitate the interaction of FGF2 with the FGF receptor and stimulate FGF2 signaling (24).
Another series of studies has implicated GPC3 in tumor progression. Lin et al. (51) showed that GPC3 expression is downregulated in a significant proportion of ovarian cancer cell lines. In all cases where GPC3 expression was lost, the promoter region of the GPC3 gene was hypermethylated, and no mutations were found in the coding region. GPC3 expression was restored by treatment with a demethylating agent. In addition, the authors demonstrated that ectopic expression of GPC3 inhibits colony-forming activity in several ovarian cancer cell lines.
Another recent study associating GPC3 with cancer built upon differential mRNA display data in which normal rat mesothelial cells were compared with mesothelioma cell lines. Murthy et al. (52) found that GPC3 was consistently downregulated in the tumor cell lines. Moreover, a similar downregulation was found in primary rat mesotheliomas and in cell lines derived from human mesotheliomas. As with ovarian cancer, no mutations in the GPC3 coding sequence were found, but most of the cell lines displayed aberrant methylation in the GPC3 promoter region. As reported previously (36), this study showed that ectopic expression of GPC3 in mesothelioma cell lines inhibits their colony-forming activity.
Another differential mRNA study generated results that are opposite to the ones described above. This study reported that, whereas GPC3 is not expressed in human normal adult liver, its expression is upregulated in most hepatocarcinomas (53). Similarly, we have found that, while normal colon does not express GPC3, a significant proportion of colorectal tumors do (J. Filmus et al., unpublished observations). Since it is highly expressed in embryonic liver and intestine and is silenced in the corresponding normal adult tissues, GPC3 behaves — in these organs, at least — as an oncofetal protein. In general, oncofetal proteins do not seem to play a critical role in tumor progression but have been used as tumor markers or as targets for immunotherapy (54, 55). In this regard, we note that Hsu et al. (53) reported that GPC3 is more frequently upregulated in hepatocarcinomas than α-fetoprotein, another oncofetal protein that has been extensively used in the clinic as a tumor marker (56), and which is thought to be a potential target for immunotherapy (57). It remains to be seen whether the oncofetal behavior of GPC3 can be exploited for clinical use and whether the re-expression of this glypican plays a role in the progression of liver and colorectal tumors.
Conclusions
It is now well established that glypicans can regulate the activity of a wide variety of growth and survival factors. In the future it will be important to define more precisely which growth and survival factors are regulated by each glypican in vivo. In addition, the biochemical basis of such regulation will have to be determined. A combination of genetics, biochemistry, and cell biology will be required to attain a better understanding of glypican function and to develop improved therapies for the pathologies in which these molecules are implicated.
Acknowledgments
We thank Sophie Ku for her assistance in the preparation of this manuscript, and Howard Song for a critical reading of it. Work in J. Filmus’s laboratory has been supported by the Canadian Institute of Health Research, and the March of Dimes. Work in S.B. Selleck’s laboratory has been supported by the NIH, the March of Dimes, and the Human Frontiers Science Program.
References
- 1.Filmus, J., and Song, H.H. 2000. Glypicans. In Proteoglycans. R.V. Iozzo, editor. Marcel Dekker Inc. New York, New York, USA. 161–176.
- 2.David G, et al. Molecular cloning of a phosphatidylinositol-anchored membrane heparan sulfate proteoglycan from human lung fibroblasts. J Cell Biol. 1990;111:3165–3176. doi: 10.1083/jcb.111.6.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stipp CS, Litwac ED, Lander AD. Cerebroglycan: an integral membrane heparan sulfate proteoglycan that is unique to the developing nervous system and expressed specifically during neuronal differentiation. J Cell Biol. 1994;124:149–160. doi: 10.1083/jcb.124.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filmus J, Church J, Buick RN. Isolation of a cDNA corresponding to a developmentally regulated transcript in rat intestine. Mol Cell Biol. 1988;8:4243–4249. doi: 10.1128/mcb.8.10.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe K, Yamada H, Yamaguchi Y. K-glypican: a novel GPI-linked heparan sulfate proteoglycan that is highly expressed in developing brain and kidney. J Cell Biol. 1995;130:1207–1218. doi: 10.1083/jcb.130.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veugelers M, et al. Characterization of glypican-5 and chromosomal localization of human GPC5, a new member of the glypican gene family. Genomics. 1997;40:24–30. doi: 10.1006/geno.1996.4518. [DOI] [PubMed] [Google Scholar]
- 7.Saunders S, Paine-Saunders S, Lander AD. Expression of the cell surface proteoglycan glypican-5 is developmentally regulated in kidney, limb, and brain. Dev Biol. 1997;190:78–93. doi: 10.1006/dbio.1997.8690. [DOI] [PubMed] [Google Scholar]
- 8.Veugelers M, et al. Glypican-6, a new member of the glypican family of cell surface proteoglycans. J Biol Chem. 1999;274:26968–26977. doi: 10.1074/jbc.274.38.26968. [DOI] [PubMed] [Google Scholar]
- 9.Paine-Saunders S, Viviano BL, Saunders S. GPC6, a novel member of the glypican gene family, encodes a product structurally related to GPC4 and is colocalized with GPC5 on human chromosome 13. Genomics. 1999;57:455–458. doi: 10.1006/geno.1999.5793. [DOI] [PubMed] [Google Scholar]
- 10.Nakato H, Futch TA, Selleck SB. The division abnormally delayed (dally) gene: a putative integral membrane proteoglycan required for cell division patterning during postembryonic development of the nervous system in Drosophila. Development. 1995;121:3687–3702. doi: 10.1242/dev.121.11.3687. [DOI] [PubMed] [Google Scholar]
- 11.Baeg GH, Lin X, Khare N, Baumgartner S, Perrimon N. Heparan sulfate proteoglycans are critical for the organization of the extracellular distribution of Wingless. Development. 2001;126:87–94. doi: 10.1242/dev.128.1.87. [DOI] [PubMed] [Google Scholar]
- 12.Litwack ED, Stipp CS, Kumbasar A, Lander AD. Neuronal expression of glypican, a cell surface glycosylphosphatidylinositol-anchored heparan sulfate proteoglycan, in the adult rat nervous system. J Neurosci. 1994;14:3713–3724. doi: 10.1523/JNEUROSCI.14-06-03713.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, Choo B, Wong Z-M, Filmus J, Buick RN. Expression of OCI-5/Glypican 3 during intestinal morphogenesis: regulation by cell shape in intestinal epithelial cells. Exp Cell Res. 1997;235:3–12. doi: 10.1006/excr.1997.3629. [DOI] [PubMed] [Google Scholar]
- 14.Litwack ED, et al. Expression of the heparan sulfate proteoglycan glypican-1 in the developing rodent. Dev Dyn. 1998;211:72–87. doi: 10.1002/(SICI)1097-0177(199801)211:1<72::AID-AJA7>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 15.Pellegrini M, et al. Gpc3 expression correlates with the phenotype of the Simpson-Golabi-Behmel syndrome. Dev Dyn. 1998;213:431–439. doi: 10.1002/(SICI)1097-0177(199812)213:4<431::AID-AJA8>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Padgett RW, St. Johnston RD, Gelbeart WM. A transcript from a Drosophila factor-beta family. Nature. 1987;325:81–84. doi: 10.1038/325081a0. [DOI] [PubMed] [Google Scholar]
- 17.Padgett RW, Wozney JM, Gelbart WM. Human BMP sequences can confer normal dorsal-ventral patterning in the Drosophila embryo. Proc Natl Acad Sci USA. 1993;90:2905–2909. doi: 10.1073/pnas.90.7.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson SM, et al. dally, a Drosophila glypican, controls cellular responses to the TGF-beta-related morphogen Dpp. Development. 1997;124:4113–4120. doi: 10.1242/dev.124.20.4113. [DOI] [PubMed] [Google Scholar]
- 19.Tsuda M, et al. The cell-surface proteoglycan Dally regulates Wingless signaling in Drosophila. Nature. 1999;400:276–280. doi: 10.1038/22336. [DOI] [PubMed] [Google Scholar]
- 20.Toyoda H, Kinoshita-Toyoda A, Selleck SB. Structural analysis of glycosaminoglycans in Drosophila and Caenorhabditis elegans and demonstration that tout velu, a Drosophila gene related to EXT tumor suppressors, affects heparan sulfate in vivo. J Biol Chem. 2000;275:2269–2275. doi: 10.1074/jbc.275.4.2269. [DOI] [PubMed] [Google Scholar]
- 21.Baeg GH, Perrimon N. Functional binding of secreted molecules to heparan sulfate proteoglycans in Drosophila. Curr Opin Cell Biol. 2000;12:575–580. doi: 10.1016/s0955-0674(00)00134-4. [DOI] [PubMed] [Google Scholar]
- 22.Lin X, Perrimon N. Dally cooperates with Drosophila Frizzled 2 to transduce Wingless signaling. Nature. 1999;400:281–284. doi: 10.1038/22343. [DOI] [PubMed] [Google Scholar]
- 23.Song HH, Shi W, Filmus J. OCI-5/rat glypican-3 binds to fibroblast growth factor-2 but not to insulin-like growth factor-2. J Biol Chem. 1997;272:7574–7577. doi: 10.1074/jbc.272.12.7574. [DOI] [PubMed] [Google Scholar]
- 24.Steinfeld R, Van Den Berghe H, David G. Stimulation of fibroblast growth factor receptor-1 occupancy and signaling by cell surface-associated syndecans and glypican. J Cell Biol. 1996;133:405–416. doi: 10.1083/jcb.133.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pilia G, et al. Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat Genet. 1996;12:241–247. doi: 10.1038/ng0396-241. [DOI] [PubMed] [Google Scholar]
- 26.Neri G, Gurrieri F, Zanni G, Lin A. Clinical and molecular aspects of the Simpson-Golabi-Behmel syndrome. Am J Med Genet. 1998;79:279–283. doi: 10.1002/(sici)1096-8628(19981002)79:4<279::aid-ajmg9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 27.Behmel A, Plochl E, Rosenkranz W. A new X-linked dysplasia gigantism syndrome: identical with the Simpson dysplasia syndrome? Hum Genet. 1984;67:409–413. doi: 10.1007/BF00291401. [DOI] [PubMed] [Google Scholar]
- 28.Garganta CL, Bodurtha JN. Report of another family with Simpson-Golabi-Behmel syndrome and a review of the literature. Am J Med Genet. 1992;44:129–135. doi: 10.1002/ajmg.1320440202. [DOI] [PubMed] [Google Scholar]
- 29.Golabi M, Rosen L. A new X-linked mental retardation overgrowth syndrome. Am J Med Genet. 1984;17:345–358. doi: 10.1002/ajmg.1320170128. [DOI] [PubMed] [Google Scholar]
- 30.Gurrieri F, Cappa M, Neri G. Further delineation of the Simpson-Golabi-Behmel (SGB) syndrome. Am J Med Genet. 1992;44:136–137. doi: 10.1002/ajmg.1320440203. [DOI] [PubMed] [Google Scholar]
- 31.Hughes-Benzie RM, et al. Simpson-Golabi-Behmel Syndrome: genotype/phenotype analysis of 18 affected males from 7 unrelated families. Am J Med Genet. 1996;66:227–234. doi: 10.1002/(SICI)1096-8628(19961211)66:2<227::AID-AJMG20>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 32.Lindsay S, et al. Large scale deletions in the GPC3 gene may account for a minority of cases of Simpson-Golabi-Behmel syndrome. J Med Genet. 1997;34:480–483. doi: 10.1136/jmg.34.6.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veugelers M, et al. Mutational analysis of the GPC3/GPC4 glypican gene cluster on Xq26 in patients with Simpson-Golabi-Behmel syndrome: identification of loss-of-function mutations in the GPC3 gene. Hum Mol Genet. 2000;9:1321–1328. doi: 10.1093/hmg/9.9.1321. [DOI] [PubMed] [Google Scholar]
- 34.Xuan JY, Hughes-Benzie RM, Mackenzie AE. A small interstitial deletion in the GPC3 gene causes Simpson-Golabi-Behmel syndrome in a Dutch-Canadian family. J Med Genet. 1999;36:57–58. [PMC free article] [PubMed] [Google Scholar]
- 35.Cano-Gauci DF, et al. Glypican-3-deficient mice exhibit the overgrowth and renal abnormalities typical of the Simpson-Golabi-Behmel syndrome. J Cell Biol. 1999;146:255–264. doi: 10.1083/jcb.146.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duenas Gonzales A, et al. OCI-5/GPC3, a glypican encoded by a gene that is mutated in the Simpson-Golabi-Behmel overgrowth syndrome, induces apoptosis in a cell line-specific manner. J Cell Biol. 1998;141:1407–1414. doi: 10.1083/jcb.141.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weng EY, Mortier GR, Graham JM. Beckwith-Wiedemann Syndrome. Clin Pediat. 1995;34:317–326. doi: 10.1177/000992289503400605. [DOI] [PubMed] [Google Scholar]
- 38.Weksberg R, Squire JA. Molecular biology of Beckwith-Wiedemann syndrome. Med Pediatric Oncol. 1997;27:462–469. doi: 10.1002/(SICI)1096-911X(199611)27:5<462::AID-MPO13>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 39.Eggenschwiler J, et al. Mouse mutant embryos overexpressing IGF-II exhibit phenotypic features of the Beckwith-Wiedemann and Simpson-Golabi-Behmel syndromes. Genes Dev. 1997;11:3128–3142. doi: 10.1101/gad.11.23.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang ZQ, Fung MR, Barlow DP, Wagner EF. Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene. Nature. 1994;372:464–467. doi: 10.1038/372464a0. [DOI] [PubMed] [Google Scholar]
- 41.Lau MM, et al. Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes Dev. 1994;8:2953–2963. doi: 10.1101/gad.8.24.2953. [DOI] [PubMed] [Google Scholar]
- 42.Ludwig T, et al. Mouse mutants lacking the type 2 IGF receptor (IGF2R) are rescued from perinatal lethality in Igf2 and Igf1r null backgrounds. Dev Biol. 1996;177:517–535. doi: 10.1006/dbio.1996.0182. [DOI] [PubMed] [Google Scholar]
- 43.Paine-Saunders S, Viviano BL, Zupicich J, Skarnes WC, Saunders S. Glypican-3 controls cellular responses to Bmp4 in limb patterning and skeletal development. Dev Biol. 2000;225:179–187. doi: 10.1006/dbio.2000.9831. [DOI] [PubMed] [Google Scholar]
- 44.Grisaru S, Cano-Gauci D, Tee J, Filmus J, Rosenblum ND. Glypican-3 modulates BMP- and FGF-mediated effects during renal branching morphogenesis. Dev Biol. 2001;231:31–46. doi: 10.1006/dbio.2000.0127. [DOI] [PubMed] [Google Scholar]
- 45.Qiao J, et al. FGF-7 modulates ureteric bud growth and nephron number in the developing kidney. Development. 1999;126:547–554. doi: 10.1242/dev.126.3.547. [DOI] [PubMed] [Google Scholar]
- 46.Kuure S, Vuolteenaho R, Vainio S. Kidney morphogenesis: cellular and molecular regulation. Mech Dev. 2000;92:31–45. doi: 10.1016/s0925-4773(99)00323-8. [DOI] [PubMed] [Google Scholar]
- 47.Piscione TD, et al. BMP-2 and OP-1 exert direct and opposite effects on renal branching morphogenesis. Am J Physiol. 1997;273:F961–F975. doi: 10.1152/ajprenal.1997.273.6.F961. [DOI] [PubMed] [Google Scholar]
- 48.Filmus J, Shi W, Wong ZM, Wong MJ. Identification of a new membrane-bound heparan sulfate proteoglycan. Biochem J. 1995;311:561–565. doi: 10.1042/bj3110561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kleef J, et al. The cell surface heparan sulfate proteoglycan glypican-1 regulates growth factor in pancreatic carcinoma cells and is overexpressed in human pancreatic cancer. J Clin Invest. 1998;102:1662–1673. doi: 10.1172/JCI4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kleeff J, et al. Stable transfection of a glypican-1 antisense construct decreases tumorigenicity in PANC-1 pancreatic carcinoma cells. Pancreas. 1999;19:281–288. doi: 10.1097/00006676-199910000-00009. [DOI] [PubMed] [Google Scholar]
- 51.Lin H, Huber R, Schlessinger D, Morin PJ. Frequent silencing of the GPC3 gene in ovarian cancer cell lines. Cancer Res. 1999;59:807–810. [PubMed] [Google Scholar]
- 52.Murthy SS, et al. Expression of GPC3, an X-linked recessive overgrowth gene, is silenced in malignant mesothelioma. Oncogene. 2000;19:410–416. doi: 10.1038/sj.onc.1203322. [DOI] [PubMed] [Google Scholar]
- 53.Hsu HC, Cheng W, Lai PL. Cloning and expression of a developmentally regulated transcript MXR7 in hepatocellular carcinoma: biological significance and temporospatial distribution. Cancer Res. 1997;57:5179–5184. [PubMed] [Google Scholar]
- 54.Coggin JH., Jr The implications of embryonic gene expression in neoplasia. CRC Critical Reviews in Oncology/Hematology. 1992;5:37–55. doi: 10.1016/s1040-8428(86)80052-x. [DOI] [PubMed] [Google Scholar]
- 55.Matsuura H, Hakomori S-I. The oncofetal domain of fibronectin defined by monoclonal antibody FDC-6: its presence in fibronectins from fetal and tumor tissues and its absence in those from normal adult tissues and plasma. Proc Natl Acad Sci USA. 1985;82:6517–6521. doi: 10.1073/pnas.82.19.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus: a prospective study of 22,707 men in Taiwan. Lancet. 1981;2:1129–1132. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 57.Vollmer CM, et al. Alpha-fetoprotein-specific genetic immunotherapy for hepatocellular carcinoma. Cancer Res. 1999;59:3064–3067. [PubMed] [Google Scholar]