Abstract
Proteins recognize specific DNA sequences not only through direct contact between amino acids and bases, but also indirectly based on the sequence-dependent conformation and deformability of the DNA (indirect readout). We used molecular dynamics simulations to analyze the sequence-dependent DNA conformations of all 136 possible tetrameric sequences sandwiched between CGCG sequences. The deformability of dimeric steps obtained by the simulations is consistent with that by the crystal structures. The simulation results further showed that the conformation and deformability of the tetramers can highly depend on the flanking base pairs. The conformations of xATx tetramers show the most rigidity and are not affected by the flanking base pairs and the xYRx show by contrast the greatest flexibility and change their conformations depending on the base pairs at both ends, suggesting tetramers with the same central dimer can show different deformabilities. These results suggest that analysis of dimeric steps alone may overlook some conformational features of DNA and provide insight into the mechanism of indirect readout during protein–DNA recognition. Moreover, the sequence dependence of DNA conformation and deformability may be used to estimate the contribution of indirect readout to the specificity of protein–DNA recognition as well as nucleosome positioning and large-scale behavior of nucleic acids.
INTRODUCTION
Recent analysis suggests that the entire human genome contains only 20 000–25 000 genes (1). And although the function of the noncoding region is still largely unknown, some regions are known to play important roles in the regulation of gene expression. The information contained within the DNA sequence is extracted by proteins that recognize specific DNA sequences in two ways (2–4). The first is a direct readout mechanism, in which recognition is mediated by direct contacts between amino acids and bases. The second is an indirect readout mechanism, in which proteins recognize DNA sequences based on their conformational properties–i.e. sequence-dependent conformational changes such as the bending and/or the deformability of the DNA (5–10)−with water molecules serving as bridges between the amino acids and bases. The sequence specificity of protein–DNA binding is commonly predicted using a sequence-based method that uses sequence information from observed binding sites. However, it is difficult to separate the direct and indirect contributions to the specificity using the sequence-based method.
To assess the respective contributions of the direct and indirect readout mechanisms, one needs to evaluate the specificity of each mechanism quantitatively. In thermodynamic terms, the free energy of a protein–DNA interaction measures the stability of each complex. In the case of the direct readout mechanism, we look for DNA sequences that optimize protein–DNA interactions (11). In the case of the indirect readout mechanism, we look for DNA sequences that optimize the conformation energy of the DNA [contribution of DNA conformation in refs (12) and (13)]. We have developed a method for quantifying the interaction energy and the specificity of the direct readout based on a statistical analysis of the structures of protein–DNA complexes. With this approach, we derived empirical potential functions for the specific interactions between amino acids and bases, and used these potentials to calculate the interaction energy, EPD, for the protein–DNA complex. By using a sequence-structure threading method, in which different DNA sequences are threaded on the protein–DNA framework and the energy for each sequence is calculated, we have been able to quantify the difference in the fitness of various DNA sequences against the protein–DNA template structure. This sequence-structure threading of random DNA sequences enabled us to calculate Z-scores defined as (EPD – <EPD>)/σ, where <EPD> is the average interaction energy and σ is the standard deviation. This normalized quantity serves as a measure of the specificity of the protein–DNA interaction within a complex, so that if the real genome sequence was threaded, we could predict DNA target sites for regulatory proteins (14).
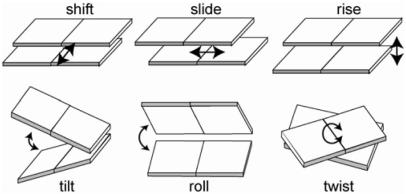
The specificity of the indirect readout mechanism has been quantified using structural data from protein–DNA complexes. To evaluate the indirect readout, we need to evaluate the internal energy of the DNA within the complex to determine how the sequences fit into the DNA structure within the complex. A simple way to describe the sequence-dependent conformation of DNA is to use six conformational parameters (shift, slide, twist, rise, roll and tilt, see Figure 1) to characterize the local geometry of each base-pair step (15). The internal DNA energy is then calculated as the sum of harmonic functions along the conformational coordinates, and the corresponding force parameters and equilibrium geometries are estimated from the observed distributions of these conformational variables in the structures of protein–DNA complexes (15,16). The Z-score that represents the specificity of the indirect readout mechanism can be calculated using sequence-structure threading, and we have shown that combining the direct and indirect readout energies derived from these statistical analyses leads to enhanced specificity (14).
Figure 1.
Six base-pair step parameters. The parameter values were calculated using the X3DNA software package (29).
The method used to quantify the specificities of the direct and indirect readout mechanisms based on the structures of known protein–DNA complexes is powerful, but there are inherent problems with this approach, as the amount of available structural data remains limited, and there may be a certain amount of bias in that data. One way to overcome these problems is to derive these potentials using unempirical computer simulations (17,18). The ensembles of base–amino acid interactions and DNA conformations can be produced in Monte Carlo and molecular dynamics (MD) simulations. If at equilibrium, the interactions or conformations reflect a Boltzmann distribution, we can derive the potentials of mean force, which are equivalent to the empirical statistical potentials, from the ensemble. We have already carried out such computer simulations to derive the potentials of mean force for the direct readout (17) and for indirect readouts (13), and showed that these potentials can reproduce very similar potentials obtained by statistical analyses of known crystal structures (17,18) and high specificities for the target DNA recognition by DNA-binding proteins (13). Thus computer simulations can provide insight into the mechanism of protein–DNA recognition.
Another advantage of computer simulations is that they can reveal details about the energetics and dynamics of the recognition process that are difficult to obtain through experimentation. The indirect readout mechanism, which includes the effects of sequence-dependent DNA conformation and deformability, is particularly difficult to analyze experimentally. In the present work, therefore, we attempted to use MD simulations to carry out detailed analyses of the sequence dependence of DNA conformation and deformability. To examine longer-range effects of the sequence dependence of base-pair step parameters, we considered all possible tetrameric steps (136 combinations) embedded at the center of DNA dodecamers and carried out 10 ns MD simulations for these DNAs in water. From the trajectories of the MD, we derived an ensemble of base-pair step parameters for every tetrameric step and examined the characteristics of the sequence-dependent conformational parameters. Here we report the results of those analyses and discuss the sequence-dependent conformational characteristics of base-pair step parameters and their relationship to structural features.
METHODS
MD simulation
We generated a set of dodecamer B-DNA sequences, 5′-CGCG-n1n2n3n4-CGCG-3′, within which ni is one of bases. Each sequence has one of 136 unique tetranucleotides at its center, and the terminals are always the CGCG tetranucleotide, which increases the stability of the ensemble. This is in contrast to the analysis by Beveridge et al. (19) in which each oligomer was 15 bp long and built by repeating tetranucleotide sequences and capping the ends with a single G to avoid fraying (5′-G-n4-n1n2n3n4- n1n2n3n4-n1n2n3n4-G-3′). Initial DNA structures were built based on the Arnott B-DNA model (20) using the nucgen module in AMBER packages 6 and 7 (21,22). Using the Leap module in the package, the initial DNA structures were solvated with TIP3P water so that the DNA molecule was covered with at least a 9 Å layer of water in each direction in a truncated octahedral unit cell 60 × 60 × 60 Å3. To neutralize the system, 22 K+ ions were added at favorable positions, and then 17 K+ and 17 Cl− ions were added so that the salt concentration of the system would be 0.15 M.
All the minimizations and MD simulations were carried out using the AMBER packages 6 and 7 (21,22). We first used a 1000-step minimization for water molecules and ions, keeping the DNA structure fixed, followed by an additional 2500-step minimization for the entire system to remove large strains from the system. The cutoff used for the van der Waals interactions was 9.0 Å. Nucleic acids are highly charged molecules and interact strongly with their solvent and other solutes over long distances. Such long-range electrostatic forces greatly influence the delicate balance of structural forces in conformations of nucleic acids. In this study, we used the current state of the art technique, the particle mesh Ewald method (23) for the proper treatment of long-range electrostatic interactions. After the minimization, the entire system was linearly heated from 0 to 300 K with a weak harmonic restraint on the initial coordinates of the DNA (10 kcal/mol) during 20 ps of MD simulation under the NVT condition. We then carried out 100 ps of molecular simulation, keeping a weak DNA restraint on the equilibration of the system under the NPT condition at 300 K. MD simulation for each of the 136 unique sequences was then carried out to sample the DNA conformations for 10 ns under the NPT condition with a time constant of 0.2 ps for the pressure control. The temperature was controlled to be 300 K using Berendsen's algorithm (24) with a coupling time of 1 fs, which was set to be the same as the time step in the MD simulation. To obtain the ‘canonical’ ensemble, we used the smaller time constant for the temperature control than a typical value of 0.5–5 ps because simulations under such a condition produce an ensemble closer to the “canonical” ensemble in the configurational space (25,26), though fluctuation of the kinetic energy is known to be suppressed. We have also carried out a MD simulation with a coupling time of 1 ps for the heat bath, and obtained similar average conformational parameters, at least for base-pair step parameters (see Supplementary Data). The SHAKE algorithm (27) was used for bonds involving hydrogen. The force field parameters used for the MD were from Wang et al. (28) (parm99), which were improved from the parm94 (35). We used the final 9 ns trajectories, during which the conformation was sampled every picosecond, to finally obtain the ensemble of 9000 conformations.
Analysis of base-pair step parameters
To characterize the conformation and deformability of the DNA sequences, we focused on base-pair step parameters that can describe the DNA conformation when treating the base pair as a rigid body. The six rigid-body parameters describing base-pair steps (tilt, roll, twist, shift, slide and rise; see Figure 1) were calculated using the X3DNA package (29). The variances and covariances of the six parameters can be used to deduce stiffness matrices, F (15,16), which gives the extent of the deformability of the base-pair steps:
where M is the covariance matrix of the step parameters Θ = (θ1, …, θ6) and k and T are the Boltzmann's constant and absolute temperature, respectively. For convenience, we use reduced units where kT is unity because the value of kT does not change the relative deformability among distinct sequences and Z-scores as well. Together with the average conformational coordinates <θi>, conformational energy for a given DNA could be calculated as the sum of the energy of each base-pair step along the DNA chain. When computing the means and variances, we used an iterative procedure that discarded outliers of 3 times of the standard deviation (SD). Typically, the process converged in five or six iterations.
The energy for each base-pair step is given by
where ΔΘ = (Δθ1, …, Δθ6) and Δθi = θi − <θi>.
The distribution of the step parameters for each of the 136 tetrameric sequences shows sequence-specific deformability. The spread of such a distribution can be evaluated as the product of the eigenvalues of the covariance matrix (M) of the step parameters. This provides an estimate of the conformational entropy (30), which hereafter will be called S.
Evaluation of similarity among tetramers
Similarity of averaged conformations and covariance matrices among tetramers was evaluated by linear correlation C, which is given as
where x and y are vectors composed of the six step parameters or 21 elements of the covariance matrix (left, lower triangular portion) for each of the tetramers. Because the elements of the vectors have distinct dimensions, each element was normalized to the corresponding parameter for all the tetramers—i.e. they were normalized by (x–<x>)/σ where <> denotes the value of a parameter averaged over all the tetramers, and/σ is the SD. If two vectors are identical, the similarity value is 1.0.
Evaluation of fitness for nucleosome core structure
DNA sequences were threaded on known nucleosome core structures (PDB codes: 1kx3, 1kx4 and 1kx5) (31), and the fitness of each of the three co-crystallized DNA sequences to each of the three structures was evaluated in the form of a Z-score that was calculated as the deviation of the energy of the target sequence from the energy distribution of random DNA sequences. The energy for a threaded sequence is calculated as the sum of base-pair step energies, which are described by the stiffness matrix. We threaded 50 000 random DNA sequences on each of the nucleosome structures and calculated the Z-score of the co-crystallized DNA sequences.
RESULTS AND DISCUSSION
Basically, 1 to 2 ns trajectories, or 1000–2000 conformations, were sufficient to obtain reliable statistics. However, sequences such as xCGx and xTAx, which were found to be highly deformable, required ∼5–6 ns long trajectories for the convergence of the distributions of each of parameters which was measured by χ2 tests. So, we decided to use the entire trajectory of 9 ns for the analyses of all the 136 sequences.
Comparison of averaged conformations of crystal and MD-derived structures among dimeric steps
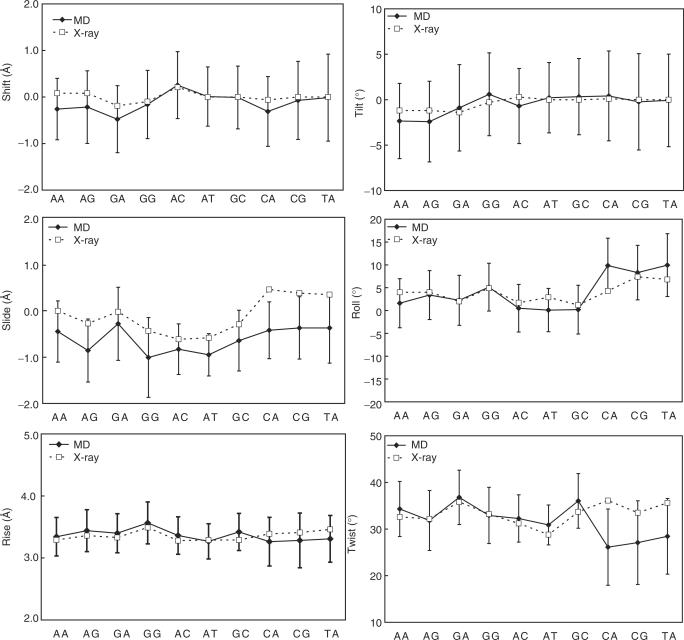
In our analysis of base-pair step parameters, we excluded highly deviated structures from the statistics and considered only structures that formed hydrogen bonds between base pairs at the central 10 internal sequence positions within dodecamers. We first compared the averaged conformations of the crystal and MD-derived structures after grouping the 136 tetrameric sequences into 10 unique dimeric steps according to the central dimer. As shown in Figure 2 and Table 1, the MD-derived conformations basically agreed with the crystal conformations collected from 239 protein–DNA complex structures (32). With respect to the tilt angle, RR sequences (except for GG) took negative values in both the MD and crystal conformations. The roll angles in the MD-derived conformations showed the largest value for RY, followed by RR and YR. This order was also observed in the crystal.
Figure 2.
Averaged step parameters for the indicated dimeric steps. The solid and dotted lines denote the averaged values for the MD and crystal structures (32), respectively. Error bars indicate the SD for the MD structures.
Table 1.
Averaged base-pair step parameters over the last 9 ns of 10-ns-long molecular dynamics simulationsa
| Step | N | Twist | (s.d.) | Tilt | (s.d.) | Roll | (s.d.) | Shift | (s.d.) | Slide | (s.d.) | Rise | (s.d.) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | 143987 | 34.3 | (5.9) | −2.3 | (4.1) | 1.6 | (5.4) | −0.26 | (0.66) | −0.45 | (0.66) | 3.34 | (0.31) |
| AG | 143994 | 31.8 | (6.4) | −2.4 | (4.4) | 3.4 | (5.4) | −0.22 | (0.78) | −0.86 | (0.68) | 3.44 | (0.34) |
| GA | 143994 | 36.8 | (5.8) | −0.9 | (4.8) | 2.2 | (5.5) | −0.48 | (0.72) | −0.29 | (0.79) | 3.40 | (0.32) |
| GG | 143997 | 32.9 | (6.0) | 0.6 | (4.6) | 5.1 | (5.2) | −0.16 | (0.73) | −1.01 | (0.86) | 3.56 | (0.34) |
| AC | 143997 | 32.3 | (5.1) | −0.7 | (4.1) | 0.5 | (5.2) | 0.26 | (0.72) | −0.83 | (0.54) | 3.36 | (0.30) |
| AT | 89966 | 30.9 | (4.3) | 0.2 | (3.9) | 0.1 | (4.7) | 0.01 | (0.64) | −0.96 | (0.45) | 3.27 | (0.28) |
| GC | 89998 | 36.0 | (5.9) | 0.3 | (4.2) | 0.2 | (5.3) | −0.01 | (0.67) | −0.65 | (0.65) | 3.42 | (0.30) |
| CA | 143996 | 26.1 | (8.2) | 0.4 | (4.9) | 9.8 | (6.0) | −0.31 | (0.75) | −0.43 | (0.61) | 3.26 | (0.39) |
| CG | 89994 | 27.1 | (9.0) | −0.2 | (5.3) | 8.3 | (6.0) | −0.07 | (0.84) | −0.38 | (0.67) | 3.28 | (0.44) |
| bTA1 | 83897 | 28.4 | (8.1) | −0.1 | (5.1) | 9.9 | (6.9) | −0.01 | (0.94) | −0.38 | (0.76) | 3.31 | (0.38) |
| TA | 89951 | 27.1 | (9.4) | −0.2 | (5.1) | 10.0 | (6.8) | −0.01 | (0.91) | −0.16 | (1.10) | 3.29 | (0.38) |
aN is the number of sampled dimeric step conformations. For example, an AA step appears in 16 tetramers, and each tetramer has 9000 conformations sampled at every 1 ps. Structures which do not form at least 10 out of 12 bp are excluded from the analysis. s.d. stands for standard deviation.
bTA1 is a subset of TA in which conformations with slide >2 Å are excluded.
The greatest difference between the crystal and MD-derived structures was observed in the twist for YR steps and slide parameters for all the dimeric steps. The twist angles in the YR steps from MD were smaller than those in the crystal. This underwind was very apparent in several earlier simulations of DNA in solution (33,34), in which the parm94 force field (35) was used, but the improved force field, parm99 (22) used here showed good agreements with the crystal data except for YR steps. At the moment, it is unclear whether the underwind in the YR steps is still due to a defect in the force field or is a natural property of these steps. With respect to the slide parameters, values from MD-derived structures were always smaller than those obtained from the crystal structures, though they were closer to those from NMR structures (36). It is noteworthy that at the current state of the art, the MD-derived structure also gives NOE volumes that are closer to those obtained experimentally than does the crystal structure or the canonical B-DNA (36).
Comparison of the deformability of crystal and MD-derived dimeric steps
The distribution of step parameters approximates the conformational entropy of the dimeric steps (30). We therefore calculated the entropy S (see Methods section) for each of the 10 dimeric steps in the MD-derived conformations and compared them with those obtained from the crystal data (30). Although DNA deformability often has been discussed on the basis of crystal structures, it is not clear how well crystal structures reflect the deformability of DNA in solution. Table 2 shows that S values from the MD and the crystal data were remarkably well correlated (correlation coefficient of 0.90) though the values of S derived from the MD were about 10 times larger than those derived from crystal data. The magnitude of S cannot be directly compared because an ‘effective temperature’ T for the ensemble of crystal data is not known and not trivial to determine (37,38). The effective temperature was reported to be 295 K so that the normal modes of the homo-polymer of DNA corresponded to those of an elastic rod with the elastic constants comparable with those of generic DNA (38,39) while lower temperatures of 166 or 232 K were used so that fluctuation strength of a MD ensemble and that of a crystal ensemble was equal (37). Following the latter assumption, the effective temperature for the crystal was deduced to be 204 K in our results. Nonetheless, it is evident that AT is the most rigid and TA is the most deformable among both the MD-derived and crystal structures (4,30), and the MD and crystal structures show the same tendency in that the most rigid deformability is RY, followed by RR, and YR is the most flexible. It is noteworthy that this tendency is derived from completely different physicochemical features in the MD-derived and crystal structures. Whereas the MD ensembles reflect thermal fluctuation, the crystal ensembles are a collection of conformations of the same base-pair step from different crystal structures. Nonetheless, they show a high correlation, indicating that the deformability of DNA upon protein binding is proportional to the extent of the thermal fluctuation of the DNA conformations. The analysis by Olson et al. (15,30) was actually carried out based on the assumption that the large collection of DNA conformations within a protein–DNA complex could well reflect the deformability of the DNA. Our MD results appear to support their intuitive, but reasonable, assumption about the nature of DNA deformability.
Table 2.
Sequence-dependent dimeric step deformability (Sa) based on structures from MD simulations and crystal structures
| Steps | S (MD) | S/SAT (MD) | S (crystal) | S/SAT (crystal) | S (crystal2) | S/SAT (crystal2) |
|---|---|---|---|---|---|---|
| AA | 11.2 | 2.0 | 1.0 | 1.4 | 0.7 | 0.4 |
| AG | 15.9 | 2.9 | 2.4 | 3.4 | 1.8 | 1.1 |
| GA | 15.7 | 2.8 | 1.9 | 2.7 | 1.4 | 0.9 |
| GG | 16.8 | 3.0 | 3.3 | 4.7 | 3.1 | 1.9 |
| AC | 9.4 | 1.7 | 0.9 | 1.3 | 0.8 | 0.5 |
| AT | 5.5 | 1.0 | 0.7 | 1.0 | 1.6 | 1.0 |
| GC | 12.3 | 2.2 | 3.3 | 4.7 | 2 | 1.3 |
| CA | 31.9 | 5.8 | 6.3 | 9.0 | 4.5 | 2.8 |
| CG | 45.2 | 8.2 | 4.9 | 7.0 | 3.1 | 1.9 |
| bTA1 | 57.3 | 10.4 | 7.2 | 10.3 | 6.6 | 4.1 |
| TA | 90.5 | 16.4 |
The largest and smallest values are highlighted in bold.
aS is the deformability of the dimeric step, which is taken as conformational entropy with the unit deg3Å3. The deformability is calculated as the product of the eigenvalues of the covariance matrix of six step parameters. S(crystal) and S(crystal2) were obtained from Olson et al. (30). S(crystal2) is a subset of S(crystal) in which overrepresented structures were excluded. S/SAT in the third, fifth and seventh columns are S values normalized by S of AT.
bTA1 is a subset of TA in which conformations with shift >2 Å were excluded.
Comparison of the averaged conformations of crystal and MD-derived structures among tetrameric steps
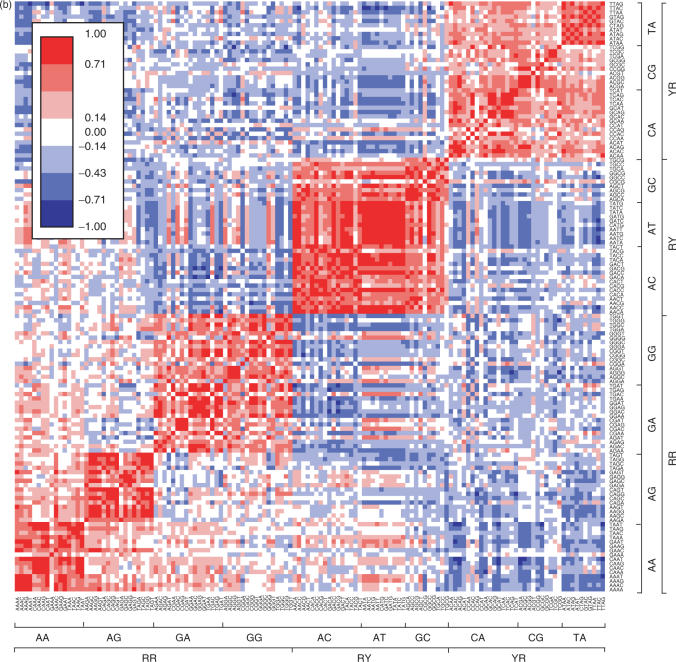
To more rigorously examine sequence-dependent conformation and deformability, we analyzed the 136 different tetrameric steps shown in Supplementary Table 1. Similarities among the 136 sequences based on the six step parameters (six dimensions) are plotted in Figure 3a. Tetramers assuming similar conformations are shown in red in the figure. With respect to the dimers, the most clearly separated are the xYRx group. Thirteen of the 16 tetrameric sequences having a central YR step assumed a similar averaged conformation; ACAT, ACGT and ATAT were exceptions. Interestingly, those three had conformations similar to those having a central RR step, especially the xGGx and xAGx sequences. xRYx sequences also showed similarity among themselves. By contrast, the xRRx sequences each showed several small red blocks in Figure 3a. For example, the conformations of the xGAx sequences were similar to one another, except that those of the AGAx sequences were strongly affected by the fourth base. On the other hand, AGGx and GGGx assumed similar conformations, regardless of the fourth base.
Figure 3.
Similarity among tetrameric steps. (a) Similarity among the averaged conformations. (b) Similarity among the correlation matrices. High and low similarities are shown by red and blue colors, respectively.
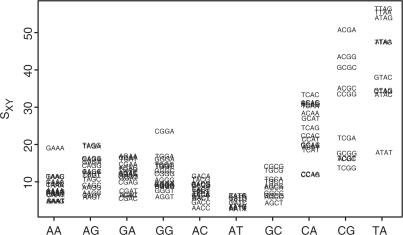
Comparison of the deformability of crystal and MD-derived tetrameric steps
Figure 4 shows that the S values for the xRYx steps are the smallest, followed by the xRRx and xYRx steps. But it is noteworthy that the S values of the xYRx steps are widely distributed and that some xYRx sequences (e.g. CCAG, CCAA and TCGG) have small S values that are comparable to those of the xRYx and xRRx steps. This means that classification of bases into just 10 dimeric steps cannot entirely account for the deformability of DNA.
Figure 4.
Deformability of tetrameric steps measured by conformational entropy Sxy. The conformational entropy was calculated as the square root of the eigenvector product of the covariance matrix. Larger values indicate that the tetramers have larger conformational spaces. See Supplementary Data for the enlarged figure and raw values of the deformability.
We next calculated the similarity of the covariance matrices for the six step parameters among the 136 tetrameric sequences. As expected, when grouped according to the central dinucleotide, xRRx, xRYx and xYRx steps show a clear blocked color (red) in Figure 3b. xRRx steps were further divided into three groups, xAAx, xAGx and others (xGAx and xGGx). xYRx steps were somewhat less similar to one another, though the conformations of xYRx showed a high degree of similarity, indicating that the deformability of xYRx steps was strongly affected by the flanking base pairs. Among the tetrameric sequences, it was the xRYx steps that were least dependent on the flanking base pairs and had similar covariance matrices; this was especially true for xATx sequences, which were the stiffest.
As mentioned above, the conventionally used approach of describing DNA conformation and deformability based on dimer sequences is not sufficient for sequence characterization, though consideration in terms of dimer sequences does enable one to roughly infer the physicochemical properties.
Propagation of step parameter correlation in sequence
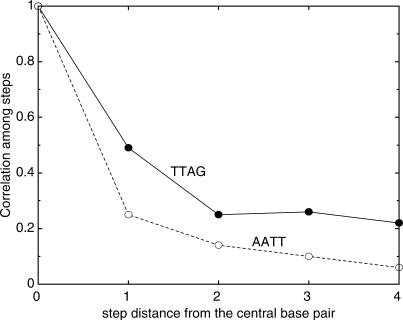
To determine the extent to which DNA conformation is affected by a particular base-pair step, we calculated the correlation between the central base pair and i ± n steps, where n is 1 to 4 (Figure 5) as done by Lankas et al. (38). In the figure, the highest values of the correlation coefficients among step parameters, whether positive or negative, is plotted against step distance from the central step. The most rigid tetramer, AATT, shows a sharp drop in correlation at the next step as previously observed (38), whereas it was found that with the most flexible, TTAG, the correlation propagates farther than with AATT. This longer correlation was commonly found for the other flexible tetrameric sequences such as TTAA, ACGA and so on, indicative of longer range effects of the step parameters on conformation.
Figure 5.
Highest correlation coefficients between step i and i + n, where n = 1, 2, 3 or 4, for the most rigid and most flexible tetrameric sequences.
Nucleosome positioning
In eukaryotic cells, the DNA is packaged into a chromatin structure so that it can fit into the cell nucleus. For regulatory proteins to bind to their target DNA sequences, the sequences need to be accessible either as an outward-facing segment on the nucleosome surface or within linkers between nucleosomes. So how are such situations set up? Recently, Segal et al. (40) showed that the positions of nucleosomes can be predicted by considering the patterns of appearance of dinucleotides in the 147 bp that are wrapped up in each nucleosome core. Using the positioning preference of the AA/TT, TA and GC dinucleotides, they successfully predicted nucleosome positions with ∼50% accuracy. We suggest this accuracy might be improved by considering longer DNA sequences—i.e. tetranucleotides. For instance, we have shown here that when sandwiched by different sequences, dinucleotides, especially TA, show distinct deformabilities, which should be considered when predicting nucleosome positioning.
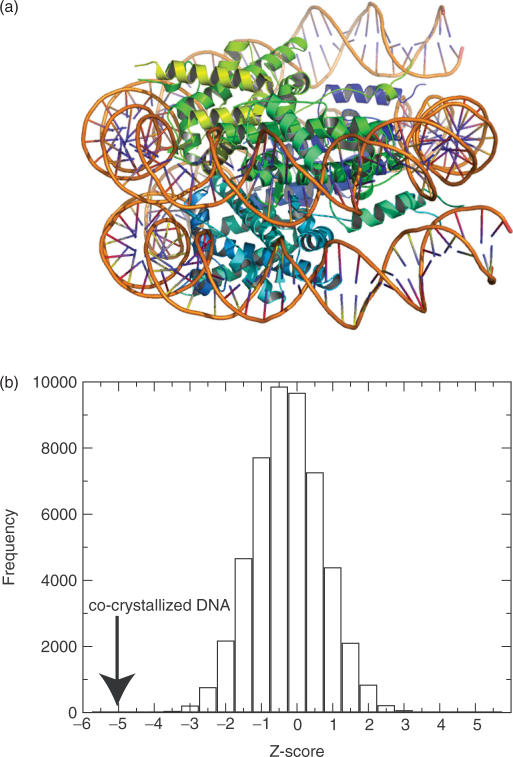
As a preliminary test, we evaluated the fitness of co-crystallized DNA sequences against histones (31) (PDB code: 1kx3, 1kx4 and 1kx5) by threading the DNA sequences on each of the three nucleosome core structures. Compared with the energies of randomly generated DNA sequences of the same length, the energies of the co-crystallized sequences were clearly lower than those of the random sequences, suggesting that the physicochemical properties of the co-crystallized DNA sequences are suitable for wrapping the histone proteins (Table 3 and Figure 6). With that in mind, we anticipate being able to determine which parts of genome sequences adopt a nucleosome structure. Although we considered just a smooth deformation of DNA, it should be noted that a recent MD study of DNA minicircles suggested that kinks in DNA or not a smooth deformation may enhance DNA flexibility (41). In the study, the kinking is likely to occur at YR steps, especially at CG steps. The CG steps are in tetrameric context ACGA, ACGC, ACGG, CCGG and GCGC. In our result, such tetrameric steps containing CG at the center are highly flexible among xCGx tetrameric sequences. The occurrence of kinking may be associated with the high deformability and depend on the flanking sequences.
Table 3.
Sequence fitness (Z-score) for given template tertiary structuresa
| Structure | |||
|---|---|---|---|
| Sequence | 1kx3 | 1kx4 | 1kx5 |
| 1kx3 | −5.2 | −1.5 | −1.9 |
| 1kx4 | −2.9 | −5.2 | −3.5 |
| 1kx5 | −2.6 | −2.0 | −5.0 |
aFigures in the table are Z-scores defined as (E − <E>)/σ, where <E> is the averaged energy of 50 000 random DNA sequences and σ is the SD of the energies.
Figure 6.
(a) Nucleosome core structure (31) (PDB: 1kx3) drawn using Pymol software (50). (b) Energy distribution of random DNA sequences threaded on the structure (1k×3). The co-crystallized DNA energy is indicated by an arrow, showing the high fitness of the template structure.
Role of indirect readout
It should be worth discussing the contribution of indirect and direct readouts to the specific DNA binding of proteins. Do they compensate each other in protein–DNA recognition? To address this question, we quantified the specificity of protein–DNA recognition by direct readout and indirect readout (3,12). The result showed that some DNA-binding proteins mainly use direct readout, some proteins do mainly indirect and others do both. When we added both the contributions with a weight, we observed the specificity for the target DNA increased for almost all the DNA-binding proteins tested, suggesting that direct and indirect readout mechanism are complementary with each other (14).
Protein–DNA recognition is also discussed in terms of enthalpy and entropy (42). The direct and indirect readouts here are another point of view for understanding protein–DNA recognition. In the direct readout, we have not considered the contributions of enthalpy and entropy of water/solute and solute/solute interactions separately, but the statistical potential includes the effects of enthalpy and entropy of water/solute and solute/solute interactions implicitly through many different configurations of water/solute and solute/solute interactions in protein–DNA complexes. As in the direct potential, the potential of mean force for the DNA conformation derived from all-atom MD simulations includes the effects of enthalpy and entropy of water/solute and solute/solute interactions implicitly. The aim of present study is not to dissect the enthalpy and entropy contributions in details, but such analysis will be presented elsewhere.
Related works
Lankas et al. (38) inferred the deformability of 10 unique dimeric steps by carrying out MD simulations with two 18 bp DNA oligomers. They showed stiffness parameters that were similar in magnitude to the crystal data reported by Olson et al. (15). Moreover, the calculated stiffness parameters we obtained in the present study were generally well correlated with theirs, giving correlation coefficients of 0.47 for shift, 0.57 for slide, 0.97 for rise, 0.94 for tilt, 0.94 for roll and 0.87 for twist. The relatively poor correlation of slide probably reflects changes in the force field used because the slide value showed a correlation with the χ angle of the backbone (correlation coefficient was 0.52) based on which parameterization in the parm99 force field was modified from parm94. On the other hand, the poor correlation for shift is probably due to an effect of the tetrameric sequences on all sequence patterns that could not be considered in the earlier work, rather than a difference in the force field used. As shown in Figure 4, even tetramers with the same central dimer can have different deformabilities.
In another related work, the Ascona B-DNA Consortium also reported the results of MD simulations of the 136 unique tetranucleotide sequences. But instead of considering those sequences sandwiched between CGCG tetramers, as in the present study, they considered 39 DNA oligomers containing the 136 unique sequences (19,43) and analyzed the sampled DNA conformations in terms of the torsion angles of DNA backbone (α, β, γ, ε, ζ). They concluded from measurements of the Kullback–Leibler Distance that the AT and CG dinucleotides are least affected by the flanking base pairs, while GG, GA and AG show the largest effects (43). That result is consistent with our present finding, and the earlier one by the Olson group (30), that AT is least affected by flanking base pairs. The characteristics of other dinucleotides are distinct, however. In the present study, RR dimers such as GG, GA and AG were moderately affected by the flanking base pairs, while YR dimers such as TA, CG and CA were greatly affected (see Figure 3 and Table 1 in Supplementary Data).
There are several possible explanations for the difference between those earlier works and the present one. For instance, (i) the force field used by the Consortium was parm94, whereas we used parm99; (ii) there was a difference in oligomer DNA calculated and (iii) there was a difference in the focusing parameters (backbone and step parameters), among others. In the Ascona B-DNA Consortium results, all 10 dimeric steps showed a large undertwist (a twist angle of less than 30°, as compared to the canonical angle of 36°) as a result of a defect in the force field (22). By using the parm99 force field, this shortcoming was largely overcome in the present study, though we did generally observe a slight undertwist, and xYRx sequences still showed a large undertwist. On the other hand, our results are consistent with those of the Consortium with respect to roll–i.e. xYRx sequences have relatively high roll angles. And with respect to slide, both the Consortium and we show negative values, whereas analysis of the crystal structures shows slide to be nearly zero. Even though the values we obtained in the present study using parm99 are closer to zero than those obtained by the Consortium using parm94, it appears there is still room for improvement in the force field. It should be noted that parm99 force field as well as parm94 seem to still produce over-populations of the α/γ = (g+, t) backbone which were seen in relatively long (more than 10 ns) simulations (19,44) and a new force field was provided (45). Taking this into account, it cannot be excluded that the results for some tetrameric sequences might be an artifact. However, since 96.4% of backbone conformations we analyzed took the canonical backbone conformation, that figure is comparable with a figure of 95% which was obtained by the analysis of many crystal structures (46) and the over-populations were seen in quite long simulations, we believe that tetrameric sequence deformability observed in this study reflects the sequence context.
The sequence dependence of the DNA structure has also been studied using other computational approaches. Matsumoto and Olson (39) carried out a normal mode analysis at the base-pair level and successfully obtained large-scale force fields by starting from small-scale force fields. They used knowledge-based harmonic energy functions deduced from the crystal structures of protein–DNA complexes (15), which can be easily replaced with those derived in the present study; moreover, they can be substituted with tetramer versions of the functions to more precisely describe the sequence-dependent motions of DNA.
Packer and coworkers (47–49) computationally studied sequence-dependent DNA structure using sequences ranging from dimers to octamers as a function of two principal degrees of freedom, slide and shift–i.e. given set of slide and shift values, the remaining four parameters were optimized with respect to the stacking and backbone energies. They concluded that AA, AT and TA are moderately context dependent and that CG, GC and GG are all strongly context dependent. In this case, we have to be cautious when comparing their results with ours because the meanings of context dependency differ. They examined the effect of the flanking dinucleotide (NN) at the 5′ or 3′ position against a particular dinucleotide (e.g. AANN or NNAA), not the effect of the flanking base pairs on both sides of the dimer, as we did (e.g. NAAN). When we re-examined their data [Table 3 in ref. (48)] on the basis of the curvature distribution with respect to the central dimer, we reached the unexpected conclusion that AT is most strongly flanking sequence dependent, while CA, AA and CG are the least, which is explicitly different from the conventional knowledge. We observed a low context dependency for AT and a high context dependency for CG and CA steps, but not for AA in terms of the volume of the conformational space. These discrepancies could be due to the degrees of freedom considered. In interpreting their results, we considered flexibility with respect to one degree of freedom (only the slide of the central step); in interpreting our own findings, we considered the flexibility as the volume of all six degrees of freedom.
Some limitations and future works
We have presented a framework for describing the flanking sequence-dependent, elastic constants of DNA, and it should be noted that there are some limitations to this framework. The spread of the distribution of base-pair step parameters was calculated under the assumption that the distributions are Gaussian functions. This is based on the framework of linear elasticity in which the energy function of deformation can be harmonic, and the force constant can be derived from the Gaussian distribution of the parameters. This assumption was applicable as a first approximation; however, two highly flexible tetramers, TTAA and TTAG, clearly had two Gaussian distributions for shift, and we selected a major distribution with a shift of <2.0 Å for the analysis. In such cases, the harmonic treatment may be oversimplified.
In addition, although we have shown the sequence-dependent deformability and conformation of DNA, we have not discussed what causes them to differ among DNA sequences. That said, we recently observed that the extent of the deformability likely correlates with hydration in the minor groove. That will be reported elsewhere.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online for the averaged base-pair step parameters over the last 9 ns of 10-ns-long MD simulations for each of 136 tetrameric sequences.
ACKNOWLEDGEMENTS
This work was supported in part by a Grant-in-Aid for Scientific Research 18031042 (H.K.) from Ministry of Education, Culture, Sports, Science and Technology in Japan. F.S. thanks Advanced Technology Institute Inc. (ATI) for financial support. We appreciate Dr W.K. Olson for providing data of protein–DNA complex structures. A part of MD calculations was carried out using ITBL computer facilities at JAEA. Funding to pay the Open Access publication charges for this article was provided by Japan Science and Technology Agency.
Conflict of interest statement. None declared.
REFERENCES
- 1.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Dickerson RE. The DNA helix and how it is read. Sci. Am. 1983;249:94–111. [Google Scholar]
- 3.Sarai A, Kono H. Protein-DNA recognition patterns and predictions. Annu. Rev. Biophys. Biomol. Struct. 2005;34:379–398. doi: 10.1146/annurev.biophys.34.040204.144537. [DOI] [PubMed] [Google Scholar]
- 4.El Hassan MA, Calladine CR. Conformational characteristics of DNA: empirical classifications and a hypothesis for the conformational behaviour of dinucleotide steps. Philos. Trans. Roy. Soc. (Ser. A) 1997;355:43–100. [Google Scholar]
- 5.Lamoureux JS, Stuart D, Tsang R, Wu C, Glover JN. Structure of the sporulation-specific transcription factor Ndt80 bound to DNA. EMBO J. 2002;21:5721–5732. doi: 10.1093/emboj/cdf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamoureux JS, Maynes JT, Glover JN. Recognition of 5′-YpG-3′ sequences by coupled stacking/hydrogen bonding interactions with amino acid residues. J. Mol. Biol. 2004;335:399–408. doi: 10.1016/j.jmb.2003.10.071. [DOI] [PubMed] [Google Scholar]
- 7.Napoli AA, Lawson CL, Ebright RH, Berman HM. Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: recognition of pyrimidine-purine and purine-purine steps. J. Mol. Biol. 2006;357:173–183. doi: 10.1016/j.jmb.2005.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen S, Gunasekera A, Zhang X, Kunkel TA, Ebright RH, Berman HM. Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: alteration of DNA binding specificity through alteration of DNA kinking. J. Mol. Biol. 2001;314:75–82. doi: 10.1006/jmbi.2001.5090. [DOI] [PubMed] [Google Scholar]
- 9.Hegde RS. The papillomavirus E2 proteins: structure, function, and biology. Annu. Rev. Biophys. Biomol. Struct. 2002;31:343–360. doi: 10.1146/annurev.biophys.31.100901.142129. [DOI] [PubMed] [Google Scholar]
- 10.Huang DB, Phelps CB, Fusco AJ, Ghosh G. Crystal structure of a free kappaB DNA: insights into DNA recognition by transcription factor NF-kappaB. J. Mol. Biol. 2005;346:147–160. doi: 10.1016/j.jmb.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 11.Kono H, Sarai A. Strucure-based prediction of DNA target sites by regulatory proteins. Proteins: Struct. Funct. Genet. 1999;35:114–131. [PubMed] [Google Scholar]
- 12.Gromiha MM, Siebers JG, Selvaraj S, Kono H, Sarai A. Intermolecular and intramolecular readout mechanisms in protein-DNA recognition. J. Mol. Biol. 2004;337:285–294. doi: 10.1016/j.jmb.2004.01.033. [DOI] [PubMed] [Google Scholar]
- 13.Arauzo-Bravo M, Fujii S, Kono H, Ahmad S, Sarai A. Sequence-dependent conformational energy of dna derived from molecular dynamics simulations:toward understanding the indirect readout mechanism in protein-DNA recognition. J. Am. Chem. Soc. 2005;127:16074–16089. doi: 10.1021/ja053241l. [DOI] [PubMed] [Google Scholar]
- 14.Sarai A, Siebers J, Selvaraj S, Gromiha MM, Kono H. Integration of bioinformatics and computational biology to understand protein-DNA recognition mechanism. J. Bioinform. Comput. Biol. 2005;3:1–15. doi: 10.1142/s0219720005000965. [DOI] [PubMed] [Google Scholar]
- 15.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Go M, Go N. Fluctuations of an a-helix. Biopolymers. 1976:1119–1127. doi: 10.1002/bip.1976.360150608. [DOI] [PubMed] [Google Scholar]
- 17.Pichierri F, Aida M, Gromiha MM, Sarai A. Free-energy maps of base-amino acid interactions for DNA-protein recognition. J. Am. Chem. Soc. 1999;121:6152. [Google Scholar]
- 18.Sayano K, Kono H, Gromiha MM, Sarai A. Multicanonical monte carlo calculation of free-energy map for base-amino acid interaction. J. Comp. Chem. 2000;21:954–962. [Google Scholar]
- 19.Beveridge DL, Barreiro G, Byun KS, Case DA, Cheatham TE, III, Dixit SB, Giudice E, Lankas F, Lavery R, et al. Molecular dynamics simulations of the 136 unique tetranucleotide sequences of DNA oligonucleotides. I. Research design and results on d(CpG) steps. Biophys. J. 2004;87:3799–3813. doi: 10.1529/biophysj.104.045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnott S, Hukins DW. Refinement of the structure of B-DNA and implications for the analysis of x-ray diffraction data from fibers of biopolymers. J. Mol. Biol. 1973;81:93–105. doi: 10.1016/0022-2836(73)90182-4. [DOI] [PubMed] [Google Scholar]
- 21.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham Iii TE, DeBolt S, Ferguson D, Seibel G, Kollman P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;91:1. [Google Scholar]
- 22.Cheatham III TE, Young MA. Molecular dynamics simulation of nucleic acids: successes, limitations, and promise. Biopolymers. 2000;56:232. doi: 10.1002/1097-0282(2000)56:4<232::AID-BIP10037>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 23.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 24.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 25.Nose S. Constant temperature molecular dynamics methods. Prog. Theor. Phys. Suppl. 1991;103:1–46. [Google Scholar]
- 26.Morishita T. Fluctuation formulas in molecular-dynamics simulations with the weak coupling heat bath. J. Chem. Phys. 2000;113:2976. [Google Scholar]
- 27.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comp. Phys. 1977;23:327–341. [Google Scholar]
- 28.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049. [Google Scholar]
- 29.Lu XJ, Olson K. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson WK, Colasanti AV, Li Y, Ge W, Zhang G, Zhurkin VB. Sponer J, Lankas F, editors. DNA simulation benchmarks as revealed by X-ray structures. Computational studies of RNA and DNA. 2006;2:235–257. [Google Scholar]
- 31.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 32.Colasanti AV. Ph.D. Thesis. NJ, USA: the State University of New Jersey, New Brunswick; 2006. Conformational States of Double Helical DNA, Rutgers. [Google Scholar]
- 33.Cheatham TE, III, Kollman PA. Observation of the A-DNA to B-DNA transition during unrestrained molecular dynamics in aqueous solution. J. Mol. Biol. 1996;259:434–444. doi: 10.1006/jmbi.1996.0330. [DOI] [PubMed] [Google Scholar]
- 34.Young MA, Ravishanker G, Beveridge DL. A 5-nanosecond molecular dynamics trajectory for B-DNA: analysis of structure, motions, and solvation. Biophys. J. 1997;73:2313–2336. doi: 10.1016/S0006-3495(97)78263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179. [Google Scholar]
- 36.Arthanari H, McConnell KJ, Beger R, Young MA, Beveridge DL, Bolton PH. Assessment of the molecular dynamics structure of DNA in solution based on calculated and observed NMR NOESY volumes and dihedral angles from scalar coupling constants. Biopolymers. 2003;68:3–15. doi: 10.1002/bip.10263. [DOI] [PubMed] [Google Scholar]
- 37.Becker NB, Wolff L, Everaers R. Indirect readout: detection of optimized subsequences and calculation of relative binding affinities using different DNA elastic potentials. Nucleic Acids Res. 2006;34:5638–5649. doi: 10.1093/nar/gkl683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lankas F, Sponer J, Langowski J, Cheatham TE., III DNA basepair step deformability inferred from molecular dynamics simulations. Biophys. J. 2003;85:2872–2883. doi: 10.1016/S0006-3495(03)74710-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsumoto A, Olson WK. Sequence-dependent motions of DNA: a normal mode analysis at the base-pair level. Biophys. J. 2002;83:22–41. doi: 10.1016/S0006-3495(02)75147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lankas F, Lavery R, Maddocks JH. Kinking occurs during molecular dynamics simulations of small DNA minicircles. Structure. 2006;14:1527–1534. doi: 10.1016/j.str.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 42.Privalov PL, Dragan AI, Crane-Robinson C, Breslauer KJ, Remeta DP, Minetti CA. What drives proteins into the major or minor grooves of DNA? J. Mol. Biol. 2007;365:1–9. doi: 10.1016/j.jmb.2006.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dixit SB, Beveridge DL, Case DA, Cheatham TE, III, Giudice E, Lankas F, Lavery R, Maddocks JH, Osman R, et al. Molecular dynamics simulations of the 136 unique tetranucleotide sequences of DNA oligonucleotides. II: sequence context effects on the dynamical structures of the 10 unique dinucleotide steps. Biophys. J. 2005;89:3721–3740. doi: 10.1529/biophysj.105.067397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varnai P, Zakrzewska K. DNA and its counterions: a molecular dynamics study. Nucleic Acids Res. 2004;32:4269–4280. doi: 10.1093/nar/gkh765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez A, Marchan I, Svozil D, Sponer J, Cheatham Iii TE, Laughton CA, Orozco M. Refinement of the amber force field for nucleic acids. Improving the description of {alpha}/{gamma} conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Djuranovic D, Hartmann B. DNA fine structure and dynamics in crystals and in solution: the impact of BI/BII backbone conformations. Biopolymers. 2004;73:356–368. doi: 10.1002/bip.10528. [DOI] [PubMed] [Google Scholar]
- 47.Packer MJ, Dauncey MP, Hunter CA. Sequence-dependent DNA structure: dinucleotide conformational maps. J. Mol. Biol. 2000;295:71–83. doi: 10.1006/jmbi.1999.3236. [DOI] [PubMed] [Google Scholar]
- 48.Packer MJ, Dauncey MP, Hunter CA. Sequence-dependent DNA structure: tetranucleotide conformational maps. J. Mol. Biol. 2000;295:85–103. doi: 10.1006/jmbi.1999.3237. [DOI] [PubMed] [Google Scholar]
- 49.Gardiner EJ, Hunter CA, Packer MJ, Palmer DS, Willett P. Sequence-dependent DNA structure: a database of octamer structural parameters. J. Mol. Biol. 2003;332:1025–1035. doi: 10.1016/j.jmb.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 50.DeLano WL. Pymol package. http://www.pymol.org/ (10 April 2007, date last accessed).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.