Abstract
Human (h) DNA repair enzyme thymine DNA glycosylase (hTDG) is a key DNA glycosylase in the base excision repair (BER) pathway that repairs deaminated cytosines and 5-methyl-cytosines. The cell cycle checkpoint protein Rad9–Rad1–Hus1 (the 9-1-1 complex) is the surveillance machinery involved in the preservation of genome stability. In this study, we show that hTDG interacts with hRad9, hRad1 and hHus1 as individual proteins and as a complex. The hHus1 interacting domain is mapped to residues 67–110 of hTDG, and Val74 of hTDG plays an important role in the TDG–Hus1 interaction. In contrast to the core domain of hTDG (residues 110–308), hTDG(67–308) removes U and T from U/G and T/G mispairs, respectively, with similar rates as native hTDG. Human TDG activity is significantly stimulated by hHus1, hRad1, hRad9 separately, and by the 9-1-1 complex. Interestingly, the interaction between hRad9 and hTDG, as detected by co-immunoprecipitation (Co-IP), is enhanced following N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) treatment. A significant fraction of the hTDG nuclear foci co-localize with hRad9 foci in cells treated with methylating agents. Thus, the 9-1-1 complex at the lesion sites serves as both a damage sensor to activate checkpoint control and a component of the BER.
INTRODUCTION
The base excision repair (BER) pathway recognizes a large variety of spontaneous and induced DNA lesions (1–3). The first step of BER is carried out by a lesion-specific DNA glycosylase. These enzymes find lesions in the vast genomic DNA and excise the damaged bases to generate potentially mutagenic apurinic/apyrimidinic (AP) sites using a base-flipping mechanism (4). About one-third of germ-line mutations leading to genetic disease are at CpG sites (5), which are susceptible to spontaneous deamination yielding U/G and T/G mismatches. Several DNA glycosylases are involved in the repair of U/G mismatches. In addition to U/G mismatches, thymine DNA glycosylase (TDG) and methyl-binding domain IV (MBD4/Med1) recognize and initiate the repair of T/G mismatches and other toxic and mutagenic lesions. TDG belongs to a large glycosylase family that can excise uracil from DNA including MUG, UNG and SMUG1. The mechanism of substrate recognition of TDG (6,7) is distinguished from other DNA glycosylases in two respects. (i) TDG recognizes a wide array of DNA lesions including U/G, T/G, 3,N4-ethenocytosine (εC)/G, and T/O6-methylguanine (MeG) mispairs as well as several oxidized bases such as thymine glycol, 5-formyl-U, 5-hydroxy-U and 5-hydroxy-methyl-U (8–10). (ii) TDG removes U or T from U/G or T/G mismatches, respectively, only on double-stranded DNA, and with significant specificity for CpG sites (7,11).
The catalytic domain of human (h) TDG resides within residues 123–300 (9,12). The flexible N-terminal domain accounts for the nonspecific DNA binding and slows its dissociation from AP site (13). Human hTDG can be modified by the small ubiquitin-like modifier (SUMO) proteins at Lys-330, which affects its dissociation from DNA and its interaction with apurinic/apyrimidinic endonuclease (APE1) (13,14). The structure of hTDG (residues 112–339) conjugated to SUMO-1 is very similar to Escherichia coli mismatch DNA glycosylase (MUG) (15,16). TDG has been shown to interact with, and modulate the activity of, proteins involved in transcriptional regulation (17). These interacting proteins include the retinoic acid and retinoid X receptors (18), the estrogen receptor (19), the transcription factor c-Jun (19,20), the thyroid transcription factor-1 (21) and the CBP/p300 transcriptional co-activators (17). CBP/p300 also catalyze the acetylation of mouse TDG at lysine residues 70, 94, 95 and 98, which suppresses the stimulation of TDG by APE1 (17). Moreover, TDG interacts with nucleotide excision repair XPC–RAD23B protein complex, which enhances TDG turnover (22). Because Tdg−/− knockout mice are embryonically lethal (23), TDG must play an essential role during development. So far, TDG is the only glycosylase that is essential for mouse development. This indicates that other DNA glycosylases such as MBD4 and UNG2 cannot substitute TDGs function even if they have some overlapping substrate specificities.
DNA repair is coordinated with cell cycle progression and DNA-damage checkpoints (3,24). Cell cycle checkpoints are surveillance mechanisms that monitor the cell's state, preserve genome integrity and play roles in preventing carcinogenesis (25–28). The signal transduction pathways triggered by DNA damage involve many components, including sensors, transducers and effectors. Human ATM (ataxia telangiectasia mutated) and ATR (ATM- and Rad3-related protein) are phosphoinositol phosphate 3 (PI-3) kinase-related kinases (27). ATM becomes activated in response to double-stranded DNA breaks, while ATR responds to DNA lesions and DNA replication blockage. After stress, ATM or ATR transmits the DNA damage signal by phosphorylating Chk1, Chk2, p53 and other proteins, which in turn regulate the cell cycle, DNA repair and apoptosis. DNA damage signaling is dependent on Rad9–Rad1–Hus1 and Rad17 sensors. Rad9, Rad1 and Hus1 form a heterotrimeric complex (the 9-1-1 complex) that exhibits structural similarity with the homotrimeric clamp proliferating cell nuclear antigen (PCNA) (29–31). The 9-1-1 complex is loaded onto DNA by Rad17–RFC (32–34).
The 9-1-1 complex is directly associated with many DNA repair proteins, including factors involved in the BER pathway. We have shown that the 9-1-1 complex physically and functionally interacts with two DNA glycosylases: the MutY homologs (MYHs) (35,36) and Nei-like glycosylase 1 (NEIL1) (37). The 9-1-1 complex has been shown to interact with and stimulate other BER enzymes including APE1 (38), polymerase β (Polβ) (39), flap endonuclease 1 (FEN1) (40,41) and DNA ligase l (42,43). In the present report, we demonstrate that hTDG DNA glycosylase physically and functionally interacts with Rad9, Rad1 and Hus1 as individual proteins and as a complex. The interacting site of the 9-1-1 complex is localized to residues 67–110 of hTDG. Val74 of hTDG plays an important role in TDG–Hus1 interaction. In addition, the catalytic rates of hTDG(67–308) with U/G and T/G mismatches are similar to those of intact hTDG and are greater than those of the TDG-core containing residues 111–308. Interestingly, the interaction between hRad9 and hTDG is enhanced after cells are treated by methylation agents. The role of hTDG in DNA damage response may be related to the embryonic lethality of Tdg−/− knockout mice (23). Our findings support the model that checkpoint proteins require a series of ‘adaptors’ to recognize DNA damage and that the 9-1-1 complex is a component of the BER pathway.
MATERIALS AND METHODS
Human cell culture
Human HeLa S3 cells were purchased from American Type Cell Culture (ATCC), cultured in modified Ham's F-12 (Mediatech) and supplemented with 10% fetal bovine serum (FBS). At 80% confluence, cells were treated with 0.4 μM of N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) for different times. Cells were harvested and washed with PBS. Cell extracts were prepared as described (44,45). The protein concentration was determined by Bio-Rad protein assay based on the Bradford method (Bio-Rad).
Construction of expression plasmids for glutathione-S-transferase (GST)-fusions and His-hTDG protein in E. coli
The plasmid pET28-TDG-FL containing full-length cDNA encoded residues 1–410 of hTDG inserted into the NheI/SalI site of pET28c was obtained from Dr Primo Schär, University of Basel, Switzerland. The truncated hTDG constructs were amplified by polymerase chain reaction (PCR) method using template pET28-TDG-FL and primers listed in Table S1 in the supplementary Data. The PCR products were digested with NheI and SalI and ligated into the NheI–SalI-digested pET28c vector (EMD Biosciences) to yield pET28-TDG plasmids of TDG-core(111–308), TDG(56–308) and TDG-N(1–124). The PCR products were digested with BamHI and SalI and ligated into the BamHI–XhoI-digested pGEX-4T-2 vector (GE Health) or pET-21a vector (EMD Biosciences) to yield plasmids pGEX-TDG and pET21-TDG, respectively, of TDG-FL, TDGΔN(111–410), TDGΔC1(1–308), TDGΔN55(56–410) and TDG(67–308). The sequences of the cloned genes were confirmed by DNA sequencing.
Construction of V44A and V74A hTDG mutants
Plasmids pET28-hTDG(V44A) and pET28-hTDG(V74A) were derived from pET28-TDG-FL by the QuickChange mutagenesis method as per the manufacturer's instructions (Stratagene). The mutagenesis of V44A employed two complementary oligonucleotides, Chang498 (sense strand) and Chang499 (antisense strand). The mutagenesis of V74A employed two complementary oligonucleotides, Chang510 (sense strand) and Chang511 (antisense strand). Plasmids pGEX-4T-hTDG(V44A) and pGEX-4T-hTDG(V74A) were generated from pGEX-4T-hTDG-FL by similar mutagenesis method. The mutants containing V44A and V74A were first screened by NarI and PstI cleavage, respectively, and then confirmed by DNA sequencing.
Purification of hTDG and deletion constructs expressed in E. coli
Rosetta cells (Invitrogen) harboring the expression plasmids were cultured in LB broth containing 100 μg/ml of ampicillin at 37°C. Protein expression was induced at A590 of 0.6 by the addition of isopropylthiogalactoside (IPTG) to a final concentration of 0.2 mM. The cells were grown at 20°C and then harvested 16 h later. Purification of hTDG was performed as previously described (9). After the Ni-Fast Flow resin (GE Health), His-tagged hTDG-Full, hTDG(V44A), hTDG(V74A), hTDG-ΔN and TDG-ΔN55 proteins were purified by 1 ml Hi-Trap-Q column (GE Health). hTDG-ΔC1, hTDG-core, hTDG(56–308), hTDG(67–308) and hTDG-N were purified by 1 ml Hi-Trap-SP column (GE Health). hTDG-ΔC1 and hTDG(V44A) were further purified by 1 ml Hi-Trap Heparin column (GE Health).
Purification of hRad9, hRad1, hHus1 and the yeast and human 9-1-1 complexes
His-tagged hHus1 expressed in E. coli BL21 Star cells (Stratagene) was purified by Ni-NTA resin (QIAGEN) and 1 ml Hi-Trap Heparin columns (GE Health) as described (36). The 9-1-1 complex of Schizosaccharomyces pombe expressed in E. coli was purified as described (36). Human Rad9, Rad1, Hus1 and the 9-1-1 complex were purified from Sf9 insect cells (Invitrogen) infected with baculoviruses as described previously (37).
GST pull-down assay
BL21 Star cells (Stratagene) harboring the GST expression plasmids were cultured in LB broth containing 100 μg/ml of ampicillin. Protein expression was induced as described above. The cell paste from a 500 ml culture was resuspended in 9 ml of buffer G (50 mM Tris–HCl, pH 7.4, 150 mM NaCl and 2 mM EDTA) containing 0.5 mM DTT and 0.1 mM PMSF and treated with lysozyme (1 mg/ml) for 30 min at 4°C. After sonication, the solution was centrifuged at 10 000g for 20 min and the supernatant was saved. The GST-tagged proteins were immobilized on glutathione-Sepharose 4B (GE Health) as described (45). GST fusion proteins (500 ng) were incubated with purified proteins (100 ng) in 0.2 ml volume of buffer G containing 0.1% (v/v) NP40 at 4°C with shaking overnight. After centrifugation at 1000g for 2 min, the pellets were washed five times with 1 ml of buffer G containing 0.1% (v/v) NP40. In the experiments involving the 9-1-1 complex, NP40 was reduced to 0.02% in the incubation and washing steps. Bound proteins were eluted by boiling in SDS loading buffer (30 mM Tris–HCl, pH 6.8, 5% (v/v) glycerol, 1% SDS, 0.5 mg/ml bromophenol blue and 1% β-mercapoethanol) and resolved on a 12% SDS–polyacrylamide gel. The proteins were subsequently analyzed by western blot using the corresponding antibodies as described below.
Co-immunoprecipitation
Cell extracts (1 mg) were precleared by adding 30 μl Protein G agarose (Invitrogen) for 2 h at 4°C. After centrifugation at 1000g, the supernatant was incubated with 4 μg of monoclonal anti-Rad9 antibody (Imgenex) overnight at 4°C. Protein G agarose (30 μl) was added and incubated for 4 h at 4°C. After centrifugation at 1000g, the supernatant was saved and the pellet was washed. The pellet fractions were resolved on a 12% SDS–PAGE and western blot analysis for hTDG was performed.
Western blotting and antibodies
Proteins were separated on SDS–polyacrylamide gels and transferred to nitrocellulose membranes. The membranes were blocked with PBS containing 0.1% Tween-20 and 10% nonfat dry milk, reacted with primary antibodies, and then incubated with horseradish peroxidase-linked second antibodies with wash between each step (46). Western blotting was detected by the enhanced chemiluminescence (ECL) analysis system (GE Health) according to the manufacturer's protocol. Human TDG monoclonal antibody was from Serotec. Human TDG polyclonal antibody was a gift of Dr Primo Schär, University of Basel, Switzerland. Monoclonal antibody of Rad9 was from Imgenex. His-tag, S-tag and FLAG-tag antibodies were from BD Bioscience, Santa Cruz Biotechnology and Sigma, respectively.
Immunofluorescence staining
Human HeLa cells cultured in Lab-Tek chamber slides (NUNC) overnight were treated with 0.4 μM of MNNG (VWR) or 40 μM of temozolomide (TMZ) (a gift from NCI) for 24 h. The cells were fixed with 4% formaldehyde for 15 min at room temperature, and permeabilized at room temperature in PBS 0.5% Triton X-100 for 10 min. After being blocked in PBS containing 15% FBS for 15 min at 37°C, the cells were reacted with hTDG polyclonal antibody and hRad9 monoclonal antibody (Imgenex) at 37°C for 30 min. Next, the cells were washed three times with PBS and incubated with Alexa Fluor 594 goat anti-mouse and Alexa Fluro 488 goat anti-rabbit antibodies (Invitrogen) at a 1:250 dilution in PBS for 30 min at 37°C. The cells were then washed three times in PBS. Nuclear DNA was counterstained with 4′,6′-diamidineno-a-phenylindole (DAPI) (Vector Laboratories). Images were captured by Nikon E400 fluorescent microscope with an attached CCD camera.
TDG glycosylase activity assay
The DNA substrate of hTDG for gel assay is a 44-mer duplex containing a T/G mismatch (Table S1 in the Supplementary Data). The strand containing the mispaired T was labeled at the 5′ end with [γ-32P]ATP by polynucleotide kinase, annealed with the other strand, and then filled-in with Klenow fragment as described by Lu et al. (47). The hTDG reaction (10 μl) contained 50 mM Tris–HCl (pH 8.0), 1 mM DTT, 50 μg/ml bovine serum albumin, 1 mM EDTA and 1.8 fmol (0.18 nM) of DNA substrate. hHus1, hRad1, hRad9 or the 9-1-1 complex was added immediately after hTDG and reactions proceeded at 37°C for 30 min. After adding 1.1 μl of 1 N NaOH and incubating at 90°C for 30 min, the reaction samples were supplemented with 5 μl of formamide dye (90% formamide, 10 mM EDTA, 0.1% xylene cyanol and 0.1% bromophenol blue) and 7 μl of the mixture was loaded onto a 14% polyacrylamide sequencing gel containing 7 M urea. The gel images were viewed on a PhosphorImager and quantified using the ImageQuant software (GE Health). The area at the product position in the control lane (no protein) was used to subtract background signal. The hTDG cleavage activity was calculated by the percentage of product over total DNA (product plus substrate bands).
Because hTDG is strongly inhibited by its AP DNA product (7), we used single turnover kinetics and saturating enzyme conditions to compare the activities of different hTDG constructs under the exact conditions as described (8). The reaction (550 μl) contained 5 μM hTDG, 20 mM HEPES (pH 7.5), 0.2 mM EDTA, 2.5 mM MgCl2, 0.1 M NaCl, 0.1 mg/ml bovine serum albumin and 500 nM of 19-mer DNA substrate containing a T/G or U/G mismatch (Table S1 in the Supplementary Data) and proceeded at 22°C. Samples (100 μl) taken at specific time points were quenched with 50 μl of quench solution (0.3 M NaOH and 0.03 M EDTA), incubated at 85°C for 15 min, and then analyzed by high-pressure liquid chromatography (HPLC) to determine the reaction progress (8). Rate constants were determined by fitting the data to a single-exponential equation using nonlinear regression with Grafit 5 (48).
RESULTS
The human 9-1-1 complex interacts with hTDG
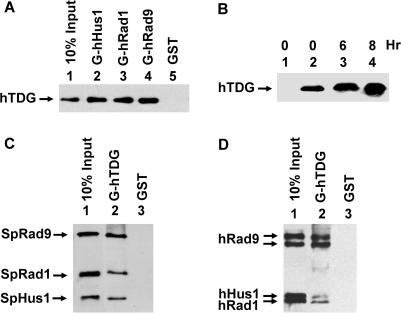
We have shown that the 9-1-1 complex physically and functionally interacts with the human and S. pombe MYHs (35,36) and NEIL1 (37). Both MYH and NEIL1 glycosylases are involved in repair of oxidized bases. To determine whether the 9-1-1 complex interacts with other DNA glycosylases responsible for other types of DNA damage, we tested hTDG which is involved in repairing DNA lesions derived from deamination. First, we used the GST pull-down assay to show the physical interactions of hTDG with hRad9, hHus1 and hRad1. GST-hHus1, GST-hRad1 or GST-hRad9 fusion protein bound to glutathione-Sepharose was incubated with purified hTDG protein (residues 1–410). As shown in Figure 1A, hTDG could be pulled down to a similar extent by GST-hHus1, GST-hRad1 and GST-hRad9. The individual proteins used in Figure 1A were expressed separately in E. coli, thus hTDG can interact with hHus1, hRad1 and hRad9 when they are not in a complex.
Figure 1.
Physical interactions of hTDG with hHus1, hRad1, hRad9 and the 9-1-1 complex. (A) hTDG binds to all subunits of the 9-1-1 complex. GST-hHus1 (lane 2), GST-hRad1 (lane 3), GST-hRad9 (lane 4) and GST alone (lane 5) were immobilized on glutathione-sepharose and incubated with purified hTDG-full (100 ng). The pellets were fractionated by a 12% SDS–PAGE followed by western blot analysis with the hTDG antibody. Lane 1 contains 10 ng (10% of the total input) of hTDG. (B) Co-IP of hTDG by hRad9 antibody in extracts prepared from HeLa cells following MNNG treatment. HeLa cells were treated with 0.4 μM of MNNG for indicated time. Immunoprecipitation were performed with antibody against hRad9 in extracts and the western blot was detected by hTDG polyclonal antibody (lanes 2–4). Lane 1 is a negative control in which the immunoprecipitation was performed with antibody against FLAG. (C) Binding of the Schizosaccharomyces pombe 9-1-1 complex to GST-hTDG. GST-tagged hTDG (lane 2) or GST beads (lane 3) were incubated with the purified S. pombe 9-1-1 complex (100 ng) expressed in E. coli. The SpHus1, SpRad1 and SpRad9 proteins were tagged with a C-terminal His, N-terminal His and C-terminal S-tag, respectively. Lane 1 contains 10 ng (10% of the total input) of the 9-1-1 complex. The western blot was detected by a mixture of the antibodies against His-tag and S-tag. (D) Binding of the human 9-1-1 complex to GST-hTDG. GST-tagged hTDG (lane 2) or GST beads (lane 3) were incubated with the purified human 9-1-1 complex (100 ng) expressed in baculovirus system. The hHus1, hRad1 and hRad9 proteins were all tagged with FLAG. Lane 1 contains 10 ng (10% of the total input) of the 9-1-1 complex. The western blot was detected by FLAG antibody.
To test whether hTDG interacts with the 9-1-1 complex, GST-hTDG fusion protein was bound to glutathione-Sepharose and incubated with S. pombe 9-1-1 complex expressed in E. coli or the human 9-1-1 complex expressed in the baculovirus system. Figure 1C and D show that all three subunits of both complexes could be pulled down by GST-hTDG. The interaction between hTDG and the 9-1-1 complex was also demonstrated by co-immunoprecipitation (Co-IP). We used hRad9 antibody to Co-IP hTDG from HeLa cell extracts. As shown in Figure 1B (lane 2), hTDG could be immunoprecipitated by hRad9 antibody.
Mapping the 9-1-1 interacting domain within hTDG
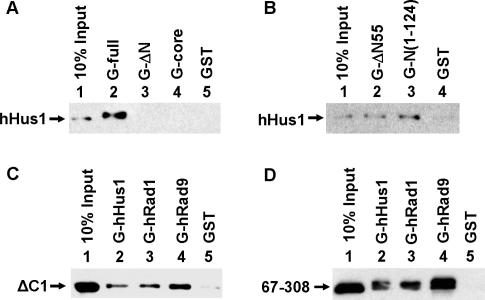
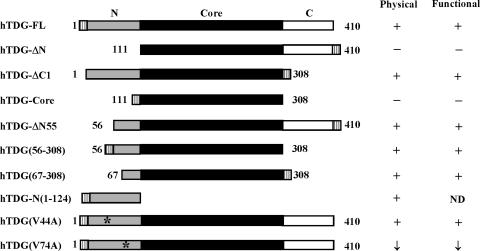
By using truncated hTDG proteins, we determined the region of hTDG engaged in the physical interaction with the 9-1-1 complex. The results are shown in Figure 2 and summarized in Figure 3. In Figure 2A and B, GST-hTDG fusion proteins bound to glutathione-Sepharose were used to pull down His-tagged hHus1 expressed in E. coli. Both hTDG-ΔN(111–410) and hTDG-core(111–308) exhibited no interaction with hHus1 (Figure 2A), however, hTDG-N(1–124) and hTDG-ΔC1(1–308) could interact with hHus1 (Figure 2B, lane 3 and data not shown). As shown in Figure 2C, hTDG-ΔC1 could be pulled down by GST-hHus1, GST-hRad1 and GST-hRad9 immobilized on glutathione-Sepharose beads. Thus, the 9-1-1 complex interacts with hTDG through the N-terminal domain (residues 1–110). To narrow down the hHus1 interacting domain on hTDG, we made three additional N-terminal deletion constructs. hTDG-Δ55 (Figure 2B, lane 2), hTDG(56–308) (data not shown) and hTDG(67–308) (Figure 2D, lane 2) could interact with hHus1. hTDG(67–308) could also interact with hRad1 and hRad9 (Figure 2D, lanes 3 and 4). Therefore, residues 67–110 of hTDG are essential for the interaction of hTDG with the 9-1-1 components.
Figure 2.
Determination of regions within hTDG involved in binding to the 9-1-1 components. (A and B) Interactions of His-tagged hHus1 with GST-hTDG constructs. His-tagged hHus1 (100 ng) expressed in E. coli was incubated with GST-TDG constructs or GST alone immobilized on beads. The pellets were fractionated on a 12% SDS–PAGE followed by western blot analysis with the His antibody. Lane 1 contains 10 ng His-hHus1 (10% of the total input). (C and D). Binding of the hTDGΔC1 and hTDG(67–308) deletion mutants, respectively, to GST-hRad9, GST-hRad1 and GST-Hus1. Immobilized GST-hHus1 (lane 2), GST-hRad1 (lane 3), GST-hRad9 (lane 4) and GST alone (lane 5) were incubated with 100 ng each of the constructs. Lane 1 of (C) or (D) contains 10 ng hTDGΔC1 or hTDG(67–308), respectively.
Figure 3.
Graphic depiction of hTDG constructs and the summary of physical and functional interactions of these constructs with the 9-1-1 complex. The intact hTDG contains 410 amino acid residues. Deletion constructs are marked with the residue's numbers at their N- and C-termini. Stripped boxes are for His-tag, gray boxes are for the N-terminal domain, black boxes are for the core domain, and white boxes are for the C-terminal domain. Point mutations are marked with stars. The physical interactions of hTDG and the 9-1-1 complex are derived from results of Figures 1, 2 and 7 as well as data not shown. The functional interactions of hTDG and hHus1 are derived from results of Figures 4, 5 and 7 as well as data not shown. ‘+’ for positive, ‘–’ for negative and ‘↓’ for reduced interactions with either hHus1 or the 9-1-1 complex. ND represents no enzyme activity.
The hTDG activity can be enhanced by hHus1, hRad1 and hRad9, and the 9-1-1 complex TDG
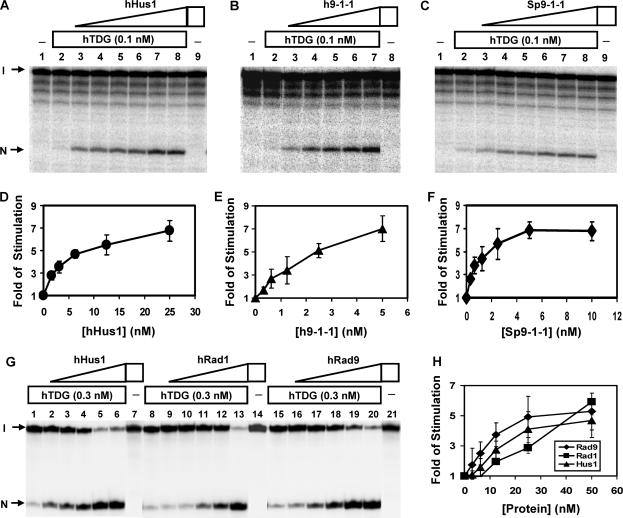
Because hTDG physically interacts with hHus1, hRad1 and hRad9 as individual proteins and as a complex, we tested whether the glycosylase activity of hTDG can be enhanced by hHus1, hRad1, hRad9 or the 9-1-1 complex. We used hHus1 and the 9-1-1 complex of S. pombe expressed in E. coli (36) as well as purified hHus1, hRad1, hRad9 and the 9-1-1 complex expressed in the baculovirus-transfected insect cells (37). We added increasing amounts of purified hHus1, hRad1, hRad9 and the 9-1-1 complex to the hTDG glycosylase reactions with DNA substrate containing a T/G mismatch. As shown in Figure 4A (lanes 3–8), the hTDG activity was enhanced significantly by hHus1 protein expressed in bacteria. The difference between hTDG (0.1 nM) alone and hTDG with 25 nM of hHus1 was approximately 7-fold (Figure 4D). Human Hus1 alone at 25 nM did not have glycosylase activity on the substrate containing a T/G mismatch (Figure 4A, lane 9). A similar stimulation effect on the hTDG glycosylase activity was observed separately with hHus1, hRad1 and hRad9 expressed in the baculovirus system (Figure 4G and H). The human 9-1-1 complex expressed in the baculovirus system and the 9-1-1 complex of S. pombe expressed in E. coli also stimulated the hTDG activity (Figure 4B, C, E and F). Interestingly, the 9-1-1 complex has a stronger stimulation effect on hTDG activity than hHus1, hRad1 and hRad9, separately.
Figure 4.
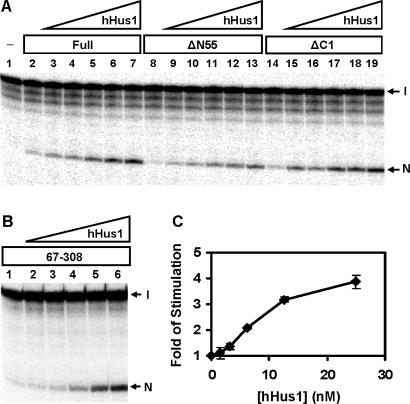
Human TDG glycosylase activity can be stimulated by hHus1, hRad1, hRad9 and the 9-1-1 complex. (A) Human Hus1 expressed in bacteria enhances the activities of TDG. Lane 1, T/G-containing DNA substrate. Lane 2, 1.8 fmol (0.18 nM) of DNA substrate was incubated with hTDG (0.1 nM). Lanes 3–8 are similar to lane 2 but with added 1.563, 3.13, 6.25, 12.5, 25 and 50 nM hHus1, respectively. Lane 9, DNA was incubated with 50 nM hHus1. The products were separated on a 14% DNA sequencing gel. Arrows mark the intact DNA substrate (I) and the cleavage product (N) after NaOH treatment. (B) hTDG glycosylase activity is enhanced by human (h9-1-1) complex. Reaction conditions are similar to (A) except that lanes 3–7 contained 0.313, 0.625, 1.25, 2.5 and 5 nM human 9-1-1 complex expressed in the baculovirus-transfected insect cells, respectively. Lane 8, DNA was incubated with 5 nM human 9-1-1 complex. (C) hTDG glycosylase activity is enhanced by S. pombe (Sp9-1-1) complex. Reaction conditions are similar to (A) except that lanes 3–8 contained 0.313, 0.625, 1.25, 2.5, 5 and 10 nM S. pombe 9-1-1 complex, respectively. Lane 9, DNA was incubated with 10 nM S. pombe 9-1-1 complex. (D–F). Quantitative analyses of the fold of stimulation of hHus1, human 9-1-1, S. pombe 9-1-1, respectively, on hTDG glycosylase activity from three experiments. In the presence of 0.1 nM hTDG-full, ∼1% of DNA was cleaved. The error bars reported are the SD of the averages. (G) hTDG activity was stimulated by hHus1, hRad1 and hRad9 expressed in the baculovirus-transfected insect cells. Lanes 1, 8 and 15, 1.8 fmol (0.18 nM) of T/G-containing DNA substrate was incubated with hTDG (0.3 nM). Lanes 2–6 are similar to lane 1 but with added 3.13, 6.25, 12.5, 25 and 50 nM hHus1, respectively. Lanes 9–13 are similar to lane 8 but with added 3.13, 6.25, 12.5, 25 and 50 nM hRad1. Lanes 16–20 are similar to lane 15 but with added 3.13, 6.25, 12.5, 25 and 50 nM hRad9, respectively. Lanes 7, 14 and 21, DNA substrate was incubated with 50 nM of hHus1, hRad1 and hRad9, respectively. (H) Quantitative analyses of the fold of stimulation of hRad9 (diamonds), hRad1 (squares) and hHus1 (triangles) on hTDG glycosylase activity from three experiments. In the presence of 0.3 nM hTDG-full, ∼12% of DNA was cleaved.
Functional interaction is parallel with the physical interaction between hTDG and hHus1
The above physical interaction results indicate that the 9-1-1 complex interacts with hTDG through the N-terminal domain (residues 67–110). hTDG(67–308) is the smallest construct of hTDG which contains glycosylase activity and retains interaction with hHus1. We then test the stimulation effects of hHus1 to His-tagged hTDG deletion constructs. The activities of hTDGΔN55(56–410), hTDGΔC1(1–308) and hTDG(67–308) could be enhanced by hHus1 (Figure 5). However, hHus1 did not enhance the hTDGΔN(111–410) and hTDG-core(111–308) activity (data not shown). Thus, the functional interaction between the 9-1-1 complex and hTDG is parallel to their physical interaction.
Figure 5.
Human Hus1 can stimulate hTDG-ΔN55, hTDGΔC and hTDG(67–308) activities. (A) The stimulation effect of hHus1 expressed in bacteria on the glycosylase activities of TDG, hTDG-ΔN55 and hTDGΔC1. Lane 1, T/G-containing DNA substrate. Lane 2, 1.8 fmol (0.18 nM) of DNA substrate was incubated with hTDG (0.1 nM). Lanes 3–7 are similar to lane 2 but with added 1.563, 3.125, 6.25, 12.5 and 25 nM hHus1, respectively. Lanes 8–13 are similar to lanes 2–7 except using hTDG-ΔN55. Lanes 14–19 are similar to lanes 2–7 except using hTDG-ΔC1. The products were separated on a 14% DNA sequencing gel. Arrows mark the intact DNA substrate (I) and the cleavage product (N) after NaOH treatment. (B) Human Hus1 can stimulate hTDG(67–308) activity. Lane 1, 1.8 fmol (0.18 nM) of DNA substrate was incubated with hTDG(67–308) (0.1 nM). Lanes 2–6 are similar to lane 1 but with added 1.563, 3.125, 6.25, 12.5 and 25 nM hHus1, respectively. (C) Quantitative analyses of the fold of stimulation of hHus1 on the glycosylase activity of hTDG(67–308) from three experiments. In the presence of 0.1 nM hTDG(67–308), ∼4% of DNA was cleaved.
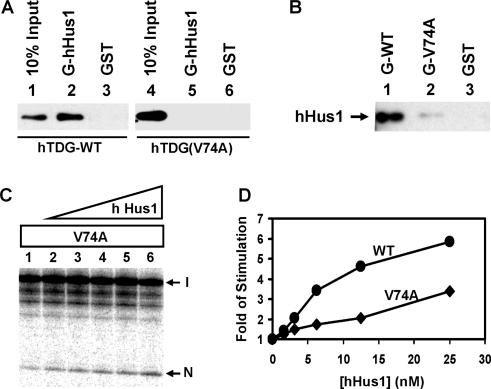
Computational analysis reveals that residues 67–110 of hTDG are highly conserved among other vertebrate TDG proteins (Figure 6). We have shown that V315 of hMYH (marked with a star in Figure 6) and I261 of SpMYH are important for their interactions with Hus1 (36). V315 of hMYH is conserved with mammalian TDG proteins (Figure 6). We thus constructed the hTDG(V74A) mutant. hTDG(V74A) protein did not associate with the GST-hHus1 immobilized on beads (Figure 7A). In the reciprocal pull-down assay, the interaction between hHus1 and GST-hTDG(V74A) mutant was much reduced as compared with wild-type hTDG immobilized on beads (Figure 7B). The purified hTDG(V74A) mutant protein was shown to exhibit similar glycosylase activity as the wild-type enzyme (compare Figure 7C, lane 1 with Figure 5A, lane 2). Consistent with its reduced physical interaction with hHus1 (Figure 7A and B), hTDG(V74A) required greater amounts of hHus1 for stimulation (Figure 7C, lanes 2–6 and Figure 7D, diamonds). As a control, the hTDG(V44A) interacted with hHus1 similarly as the wild-type hTDG (data not shown). Therefore, V74A of hTDG plays an important role in hHus1 interaction.
Figure 6.
Alignment of the Hus1 interacting domains on vertebrate TDGs and hMYH. Sequences are: Homo sapiens TDG (hTDG, accession No. AAI04478), mouse Mus musculus TDG (mTDG, accession No. NP_766140), rat Rattus norvegicus TDG (rTDG, accession No. AAI29089), chicken Gallus gallus TDG (chTDG, accession No. NP_990081), frog Xenopus laevis TDG (xTDG, accession No. AAH77465) and Homo sapiens MYH (hMYH, accession No. U63329). A star marks V74 of hTDG and V315 of hMYH (36), which are important for hHus1 binding. Identical amino acid residues are shaded in black and conserved residues are boxed in gray.
Figure 7.
Val74 of hTDG is important for interaction with hHus1. (A) Binding of the hTDG wild-type (WT) and hTDG(V74A) mutant to GST-hHus1. Immobilized GST-hHus1 (lanes 2 and 5) and GST alone (lanes 3 and 6) were incubated with 100 ng of hTDG wild-type (lanes 2 and 3) and hTDG(V74A) (lanes 5 and 6) proteins. The pellets were fractionated by a 12% SDS–PAGE followed by western blot analysis with the hTDG antibody. Lanes 1 and 4 contain 10 ng (10% of the total input) of hTDG-WT and hTDG(V74A), respectively. (B) Interactions of His-tagged hHus1 with GST-hTDG (WT) and GST-hTDG(V74A). His-tagged hHus1 (100 ng) was incubated with GST-TDG constructs or GST alone immobilized on beads. The pellets were fractionated on a 12% SDS–PAGE followed by western blot analysis with the His antibody. (C) The stimulation effect of hHus1 expressed in bacteria on the glycosylase activities of hTDG(V76A). The experiment was performed similarly as Figure 4A except using hTDG(V74A). Lane 1, 1.8 fmol (0.18 nM) of DNA substrate was incubated with hTDG(V74A) (0.1 nM). Lanes 2–6 are similar to lane 1 but with added 1.563, 3.125, 6.25, 12.5 and 25 nM hHus1, respectively. (D) Quantitative analyses of the fold of stimulation of hHus1 on the glycosylase activity of hTDG-WT (circles) and hTDG(V74A) (diamonds) from two experiments.
hTDG(67–308) has full catalytic activity
It has been reported that the efficient hydrolysis of thymines from T/G mismatches requires the N-terminal sequence (9,12). Therefore, we determine whether hTDG(67–308) can efficiently excise T from a T/G mismatch by comparison with hTDG-full and hTDG-core. Like many DNA glycosylases, TDG exhibits essentially no enzymatic turnover due to strong binding to its product (49). Thus, we measure the rate constant of hTDG with both U/G- and T/G-containing DNA under single turnover kinetics and saturating enzyme conditions in which the concentration of hTDG is 10-fold excess over DNA concentration under the exact condition of Bennett et al. (8) by the HPLC method. hTDG(67–308) had 21-fold higher rate for U/G-containing DNA than T/G-containing substrate (Table 1). hTDG(67–308) removes U from U/G mispairs with a rate of kmax = 2.15 min−1, similar to that of intact hTDG (Table 1). The activity of hTDG(67–308) against T/G mispairs is 2-fold slower than that of intact TDG (0.104 min−1 versus 0.19 min−1, Table 1). In contrast, the smaller hTDG-core (111–308) has lower activity than hTDG-full, 3.4-fold lower for U/G and 15-fold lower for T/G mispairs (Table 1). Thus, residues 67–110 are necessary for full catalytic activity. In contrast to previous reports (9,12), the core domain TDG(111–308) has significant activity on T/G mispairs with kmax = 0.013 min−1. These observations indicate that hTDG(67–308) acts like intact enzyme.
Table 1.
The rate constantsof hTDG contructs with DNA substrates containing an U/G mismatch or T/G mismatch
| Construct | U/G DNA | T/G DNA |
|---|---|---|
| Kmax(min−1) | Kmax (min−1) | |
| hTDG-full | 2.70 ± 0.15a | 0.19 ± 0.04a |
| hTDG(67-308) | 2.15 ± 0.01 | 0.104 ± 0.008 |
| hTDG-core | 0.80 ± 0.20 | 0.013 ± 0.001 |
aRate constants in this study are comparable to those derived from Bennett et al. (8).
DNA damage stimulates the interaction of hTDG–hRad9
To investigate the effect of DNA damage on the interaction between hTDG and the 9-1-1 complex, we performed Co-IP experiments with extracts from MNNG-treated HeLa cells. In these experiments, hTDG could be immunoprecipitated by hRad9 antibody from HeLa extracts (Figure 1B, lane 2). Interestingly, the interaction between hRad9 and hTDG was enhanced after MNNG treatment (Figure 1B, lanes 3 and 4). This result indicates that hTDG–hRad9 interaction is enhanced following treatment with a DNA methylation agent.
Co-localization between hTDG and the 9-1-1 complex
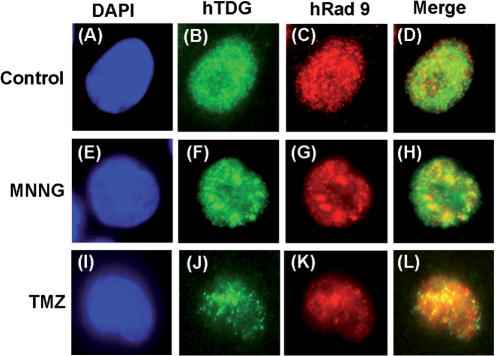
Next, we tested whether hTDG and hRad9 translocated to the same nuclear foci following DNA damage. Using an indirect immunofluorescent method, we showed that hRad9 foci were co-localized to some of hTDG foci after MNNG and TMZ treatments (Figure 8). Several large yellow foci were observed in MNNG-treated cells (Figure 8H). In TMZ-treated cells, hTDG and hRad9 formed discrete nuclear foci (Figure 8J and K). A significant fraction of the hTDG nuclear foci were found to co-localize with hRad9 foci in TMZ-treated cells (Figure 8L). This data indicates that hTDG and the 9-1-1 complex accumulate in repair foci following DNA damage.
Figure 8.
Co-localization of hTDG and hRad9 in HeLa cells following MNNG and TMZ treatments. (A–D). Control cells without treatment. (E–H). Cells were treated with 0.4 μM of N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) for 24 h. (I–L). Cells were treated with 40 μM of TMZ for 24 h. (A), (E) and (I), DAPI stain; (B), (F) and (J), immunofluorescent staining with antibody against hTDG; (C), (G) and (K), immunofluorescent staining with antibody against hRad9; (D), merge of (B) and (C); (H), merge of (F) and (G) and (L), merge of (J) and (K). Co-localization of hTDG (green) and hRad9 (red) was visualized as yellow. Human TDG and Rad9 antibodies used are from Serotec and Imgenex, respectively.
DISCUSSION
It has been suggested that the BER pathway involves highly coordinated processes governed by protein–protein and protein–DNA interactions (50–53). In this study, we show that hTDG interacts with hRad9, hRad1 and hHus1 as individual proteins and as a complex. The glycosylase activity of hTDG is stimulated by hHus1, hRad1, hRad9, separately and the 9-1-1 complex. Because the functional interaction is parallel with the physical interaction between hTDG and the 9-1-1 complex, the 9-1-1 complex may stimulate hTDG by a direct contact with hTDG. Recently, the 9-1-1 complex has been shown to interact with and stimulate the enzymes involved in BER including MYH (35,36), NEIL1 (37), APE1 (38), Polβ (39), FEN1 (40,41) and DNA ligase l (42,43). Thus, the 9-1-1 complex is not only a DNA damage sensor (27) but is also involved in the BER pathway. The 9-1-1 complex stimulates the activities of APE1 and ligase 1 without encircling the DNA (38,43). This is consistent with our finding that individual subunits of the 9-1-1 complex can stimulate the glycosylase activities of TDG (this study), MYH (36) and NEIL1 (37). However, the 9-1-1 complex may function more efficiently than individual subunits (Figure 4).
We have shown that the 9-1-1 complex physically and functionally interacts with hNEIL1 (37) and MYH in S. pombe and human cells (35,36). Human TDG is the third glycosylase to interact with the 9-1-1 complex. Unlike MYH that interacts with the 9-1-1 complex mainly via the Hus1 subunit (35,36), hTDG and hNEIL1 interact with hHus1, hRad1 and hRad9 equally. Because these three glycosylase have distinct substrate specificities, the 9-1-1 complex needs to be channeled to different BER pathways in response to different DNA damage signals. Both hMYH and hNEIL1 co-localize with hRad9 following H2O2 treatment and hTDG co-localizes with hRad9 following treatments with methylating agents.
A consensus PCNA-binding motif [QXX(L/V)XXF(F/Y)] is found in many proteins involved in DNA replication, DNA repair, DNA methylation and chromatin assembly (54,55). Recently the list of proteins that interact with 9-1-1 has been expanding; however, the consensus-binding motif to the 9-1-1 complex is not clear. Our mapping analyses indicate that the 9-1-1 complex interacting domain is localized to residues 67–110 of hTDG and Val74 of hTDG plays an important role in TDG–Hus1 interaction. A comparison of the hHus1 interacting domains of hTDG and hMYH reveals several conserved amino acids including a conserved Val (marked with a star in Figure 6). It is significant that both V74 of hTDG and V315 of hMYH (36) play important roles in term of their respective interactions with hHus1. Thus, the interactions between DNA glycosylases and the 9-1-1 complex appear to involve hydrophobic interactions. However, this conserved Val is not present in chicken and Xenopus TDG sequences (Figure 6). Thus, the protein networks in TDG-directed BER may vary among different organisms. The crystal structure of a catalytic core of TDG (residues 112–339) conjugated to SUMO lacks residues 67–111 (15,16). Our preliminary NMR data (Fitzgerald,M., Lu,A.-L. and Drohat,A.C., unpublished data) indicate that a significant portion of hTDG(56–308) is disordered while hTDG-core (110–308) is well structured. This indicates that the N-terminal domain (residues 56–110) is unstructured. The Hus1 interacting domain of hNEIL1 (residues 290–350) may also be flexible because no identifiable density beyond residues 290 can be detected in the crystal structure of hNEIL1 containing residues 2–342 (56). The Hus1 interacting domain of hMYH (residues 295–350) is not present in the bacterial MutY structure (57,58). Based on both secondary structure (59) and disorder prediction (60), the region containing residues 295–350 of hMYH is also likely unstructured. Thus, a common feature of the Hus1-binding motif appears to have a disorder structure in solution. It is possible that this region becomes structured in the presence of the 9-1-1 complex and then this conformational change promotes the catalytic activities of DNA glycosylases. We have shown here that this disordered region (residues 67–110) of hTDG contributes to greater catalytic activity as compared to the core domain (residues 110–308) (Table 1). It is interesting to note that Hus1 interacting regions of hTDG and hMYH overlap with their APE1 interacting domains (17,45). It has been pointed out that many disordered segments fold on binding to their biological targets and permit protein promiscuity (61).
Mammalian cell cycle checkpoints have been recognized as key tumor-suppressor mediators that prevent the accumulation of mutations, which promote carcinogenesis (62,63). Checkpoints also fulfill a broad range of additional functions in development. It has been shown that targeted deletion of many murine checkpoint genes, including Atr (64), Chk1 (65,66), Hus1 (67), Rad9 (68) and Rad17 (69), resulted in embryonic lethality. It is interesting to note that Tdg−/− knockout mice are also embryonically lethal (23). Although TDG plays other roles beside DNA repair, its role in DNA damage response may be related to the embryonic lethality of Tdg−/− knockout mice (23).
Methylating agents are widely used as anticancer drugs. For example, TMZ has been used as a chemotherapy agent for brain tumor (70). Although both MNNG and TMZ are methylating agents, the distribution patterns of the foci of hTDG and hRad9 are different when cells are treated with these agents (Figure 8). The reason is not clear. The biological effects of methylation agents, such as cell cycle arrest and apoptosis, are mostly linked to O6-methylguanine (MeG), which can also be formed by tobacco smoke and methylnitrosourea. The MeG lesions can be repaired in a saturable manner by the suicide enzyme MeG methyltransferase (MGMT) (71). However, MGMT is inactivated in most solid tumor cells, and the persistence of MeG causes cytotoxicity. In vivo mutagenesis assays have shown that MeG mispairs with T during replication to give mutagenic T/MeG mispairs (72,73). Mismatch repair (MMR) enzymes MSH2/MSH6, TDG and MBD4 can recognize or excise T from T/MeG mispairs (74–78), and have an effect on the toxicity of methylating agents. MMR-deficient and MBD4-deficient cells have been shown to be resistant (or tolerant) to methylating agents (74,75,77). Given the ability of TDG to act on T/MeG (76,78), TDG may affect the cytotoxicity of methylating agents. Two models have been proposed to address the cytotoxicity of MeG: (i) a direct recognition of T/MeG mispairs by repair proteins initiates the signaling pathway leading to apoptosis and (ii) the repair enzymes remove the mispaired T on the daughter strand, but the persistence of MeG on the parental strand promotes futile cycles of excision and resynthesis, which ultimately result in apoptosis. The interaction between TDG and the 9-1-1 complex may play a role in this signaling pathway.
The 9-1-1 complex, Rad17 and ATM/ATR are proposed to act at an early step to sense DNA damage (27,79). There are two models to address how these sensors are recruited to the damaged sites. In the first model, these checkpoint proteins may detect a common intermediate, such as single-stranded DNA coated by replication protein A (RPA), which is processed by various DNA repair pathways (28). RPA has been shown to directly interact with the 9-1-1 complex (80). In the second model, these checkpoint proteins may require a series of ‘adaptors’ to recognize DNA damage. Such adaptor proteins may be DNA damage recognition proteins involved in BER, mismatch repair, nucleotide excision repair and double-strand break repair (81–85). Our findings that three DNA glycosylases interacts with the 9-1-1 complex support the model that checkpoint proteins require a series of ‘adaptors’ to recognize DNA damage. In this model, a DNA glycosylase first recognizes specific DNA lesions, and then recruits Rad9–Rad1–Hus1 to stimulate BER and to initiate the signal response pathway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr Evan Y. Lee (University of California, Irvine, CA, USA) and Dr Alan E. Tomkinson (University of Maryland, Baltimore, MD, USA) for kindly providing the plasmids. We thank Dr Alan E. Tomkinson and Dr Aziz Sancar (University of North Carolina at Chapel Hill) for providing the baculovirus vectors. We appreciate Dr Howard Lieberman (Columbia University) for kindly supplying the 9-1-1 proteins for the initial work. We thank Dr Primo Schär (University of Basel, Switzerland) providing hTDG polyclonal antibody and the hTDG clone and the Development Therapeutics Program of National Cancer Institute for providing temozolomide. This work was supported by National Institute of Health grants CA78391 to A.L., GM072711 to A.C.D. and a NIH training grant (GM066706) to M.E.F. Funding to pay the Open Access publication charges for this article was provided by CA78391.
Conflict of interest statement. None declared.
REFERENCES
- 1.Friedberg EC. DNA damage and repair. Nature. 2003;421:436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 3.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 4.Mol CD, Parikh SS, Putnam CD, Lo TP, Tainer JA. DNA repair mechanisms for the recognition and removal of damaged DNA bases. Annu. Rev. Biophys. Biomol. Struct. 1999;28:101–128. doi: 10.1146/annurev.biophys.28.1.101. [DOI] [PubMed] [Google Scholar]
- 5.Krawczak M, Ball EV, Cooper DN. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am. J. Hum. Genet. 1998;63:474–488. doi: 10.1086/301965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardeland U, Bentele M, Lettieri T, Steinacher R, Jiricny J, Schar P. Thymine DNA glycosylase. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:235–253. doi: 10.1016/s0079-6603(01)68103-0. [DOI] [PubMed] [Google Scholar]
- 7.Waters TR, Swann PF. Kinetics of the action of thymine DNA glycosylase. J. Biol. Chem. 1998;273:20007–20014. doi: 10.1074/jbc.273.32.20007. [DOI] [PubMed] [Google Scholar]
- 8.Bennett MT, Rodgers MT, Hebert AS, Ruslander LE, Eisele L, Drohat AC. Specificity of human thymine DNA glycosylase depends on N-glycosidic bond stability. J. Am. Chem. Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardeland U, Bentele M, Jiricny J, Schar P. Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. J. Biol. Chem. 2000;275:33449–33456. doi: 10.1074/jbc.M005095200. [DOI] [PubMed] [Google Scholar]
- 10.Hardeland U, Bentele M, Jiricny J, Schar P. The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin S, Karran P. Incision at DNA G.T mispairs by extracts of mammalian cells occurs preferentially at cytosine methylation sites and is not targeted by a separate G.T binding reaction. Biochemistry. 1993;32:13032–13039. doi: 10.1021/bi00211a012. [DOI] [PubMed] [Google Scholar]
- 12.Gallinari P, Jiricny J. A new class of uracil-DNA glycosylase related to human thymine-DNA glycosylase. Nature. 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- 13.Steinacher R, Schar P. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr. Biol. 2005;15:616–623. doi: 10.1016/j.cub.2005.02.054. [DOI] [PubMed] [Google Scholar]
- 14.Hardeland U, Steinacher R, Jiricny J, Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, et al. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature. 2005;435:979–982. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- 16.Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, et al. Crystal structure of SUMO-3-modified thymine-DNA glycosylase. J. Mol. Biol. 2006;359:137–147. doi: 10.1016/j.jmb.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 17.Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 18.Um S, Harbers M, Benecke A, Pierrat B, Losson R, Chambon P. Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J. Biol. Chem. 1998;273:20728–20736. doi: 10.1074/jbc.273.33.20728. [DOI] [PubMed] [Google Scholar]
- 19.Chen D, Lucey MJ, Phoenix F, Lopez-Garcia J, Hart SM, Losson R, Buluwela L, Coombes RC, Chambon P, et al. T:G mismatch-specific thymine-DNA glycosylase potentiates transcription of estrogen-regulated genes through direct interaction with estrogen receptor alpha. J. Biol. Chem. 2003;278:38586–38592. doi: 10.1074/jbc.M304286200. [DOI] [PubMed] [Google Scholar]
- 20.Chevray PM, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc. Natl Acad. Sci. USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Missero C, Pirro MT, Simeone S, Pischetola M, Di Lauro R. The DNA glycosylase T:G mismatch-specific thymine DNA glycosylase represses thyroid transcription factor-1-activated transcription. J. Biol. Chem. 2001;276:33569–33575. doi: 10.1074/jbc.M104963200. [DOI] [PubMed] [Google Scholar]
- 22.Shimizu Y, Iwai S, Hanaoka F, Sugasawa K. Xeroderma pigmentosum group C protein interacts physically and functionally with thymine DNA glycosylase. EMBO J. 2003;22:164–173. doi: 10.1093/emboj/cdg016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst) 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 24.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 25.Canman CE. Replication checkpoint: preventing mitotic catastrophe. Curr. Biol. 2001;11:121–124. doi: 10.1016/s0960-9822(01)00057-4. [DOI] [PubMed] [Google Scholar]
- 26.Longhese MP, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P. DNA damage checkpoint in budding yeast. EMBO J. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 28.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 29.Burtelow MA, Roos-Mattjus PM, Rauen M, Babendure JR, Karnitz LM. Reconstitution and molecular analysis of the hRad9-hHus1-hRad1 (9-1-1) DNA damage responsive checkpoint complex. J. Biol. Chem. 2001;276:25903–25909. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- 30.Shiomi Y, Shinozaki A, Nakada D, Sugimoto K, Usukura J, Obuse C, Tsurimoto T. Clamp and clamp loader structures of the human checkpoint protein complexes, Rad9-Rad1-Hus1 and Rad17-RFC. Genes Cells. 2002;7:861–868. doi: 10.1046/j.1365-2443.2002.00566.x. [DOI] [PubMed] [Google Scholar]
- 31.Venclovas C, Thelen MP. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl Acad. Sci. USA. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5' recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc. Natl Acad. Sci. USA. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang DY, Lu AL. Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast. Schizosaccharomyces pombe. J. Biol. Chem. 2005;280:408–417. doi: 10.1074/jbc.M406800200. [DOI] [PubMed] [Google Scholar]
- 36.Shi G, Chang D-Y, Cheng CC, Guan X, Venclovas C, Lu A-L. Physical and functional interactions between MutY homolog (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem. J. 2006;400:53–62. doi: 10.1042/BJ20060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan X, Bai H, Shi G, Theriot CA, Hazra TK, Mitra S, Lu A-L. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates NEIL1 glycosylase. Nucleic Acids Res. 2007;35:2463–2472. doi: 10.1093/nar/gkm075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gembka A, Toueille M, Smirnova E, Poltz R, Ferrari E, Villani G, Hubscher U. The checkpoint clamp, Rad9-Rad1-Hus1 complex, preferentially stimulates the activity of apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta in long patch base excision repair. Nucleic Acids Res. 2007;35:2596–2608. doi: 10.1093/nar/gkl1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toueille M, El Andaloussi N, Frouin I, Freire R, Funk D, Shevelev I, Friedrich-Heineken E, Villani G, Hottiger MO, et al. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 2004;32:3316–3324. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedrich-Heineken E, Toueille M, Tannler B, Burki C, Ferrari E, Hottiger MO, Hubscher U. The two DNA clamps Rad9/Rad1/Hus1 complex and proliferating cell nuclear antigen differentially regulate flap endonuclease 1 activity. J. Mol. Biol. 2005;353:980–989. doi: 10.1016/j.jmb.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, Brandt P, Rossi ML, Lindsey-Boltz L, Podust V, Fanning E, Sancar A, Bambara RA. The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc. Natl Acad. Sci. USA. 2004;101:16762–16767. doi: 10.1073/pnas.0407686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smirnova E, Toueille M, Markkanen E, Hubscher U. The human checkpoint sensor and alternative DNA clamp Rad9-Rad1-Hus1 modulates the activity of DNA ligase I, a component of the long-patch base excision repair machinery. Biochem. J. 2005;389:13–17. doi: 10.1042/BJ20050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W, Lindsey-Boltz LA, Sancar A, Bambara RA. Mechanism of stimulation of human DNA ligase I by the Rad9-Rad1-Hus1 checkpoint complex. J. Biol. Chem. 2006;281:20865–20872. doi: 10.1074/jbc.M602289200. [DOI] [PubMed] [Google Scholar]
- 44.Gu Y, Parker A, Wilson TM, Bai H, Chang DY, Lu AL. Human MutY homolog (hMYH), a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins hMSH2/hMSH6. J. Biol. Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- 45.Parker A, Gu Y, Mahoney W, Lee S-H, Singh KK, Lu A-L. Human homolog of the MutY protein (hMYH) physically interacts with protein involved in long-patch DNA base excision repair. J. Biol. Chem. 2001;276:5547–5555. doi: 10.1074/jbc.M008463200. [DOI] [PubMed] [Google Scholar]
- 46.Towbin HT, Staehlin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gel to nitrocellulose sheets procedure. Proc. Natl Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu A-L, Tsai-Wu J-J, Cillo J. DNA determinants and substrate specificities of Escherichia coli MutY. J. Biol. Chem. 1995;270:23582–23588. doi: 10.1074/jbc.270.40.23582. [DOI] [PubMed] [Google Scholar]
- 48.Leatherbarrow RJ. Staines, U.K: Erithacus Software Ltd; 1998. GraFit 5.0. [Google Scholar]
- 49.Waters TR, Gallinari P, Jiricny J, Swann PF. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
- 50.Lu A-L, Li X, Gu Y, Wright PM, Chang D.-Y. Repair of oxidative DNA damage. Cell Biochem. Biophy. 2001;35:141–170. doi: 10.1385/CBB:35:2:141. [DOI] [PubMed] [Google Scholar]
- 51.Mitra S, Izumi T, Boldogh I, Bhakat KK, Hill JW, Hazra TK. Choreography of oxidative damage repair in mammalian genomes. Free Radic. Biol. Med. 2002;33:15–28. doi: 10.1016/s0891-5849(02)00819-5. [DOI] [PubMed] [Google Scholar]
- 52.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal a basic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 53.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat. Struct. Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 54.Warbrick E. PCNA binding through a conserved motif. BioEssays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 55.Zhang P, Mo JY, Perez A, Leon A, Liu L, Mazloum N, Xu H, Lee MY. Direct interaction of proliferating cell nuclear antigen with the p125 catalytic subunit of mammalian DNA polymerase. δ. J. Biol. Chem. 1999;274:26647–26653. doi: 10.1074/jbc.274.38.26647. [DOI] [PubMed] [Google Scholar]
- 56.Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zinc less finger motif required for glycosylase activity. Proc. Natl Acad. Sci. USA. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guan Y, Manuel RC, Arvai AS, Parikh SS, Mol CD, Miller JH, Lloyd S, Tainer JA. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat. Struct. Biol. 1998;5:1058–1064. doi: 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- 59.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 60.Obradovic Z, Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK. Predicting intrinsic disorder from amino acid sequence. Proteins. 2003;53(Suppl. 6):566–572. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- 61.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 62.Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 63.Hartwell L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell. 1992;71:543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- 64.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 66.Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, Ikeda K, Nakayama K, Nakanishi M, et al. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(-/-) mice. Genes Dev. 2000;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- 67.Weiss RS, Enoch T, Leder P. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 2000;14:1886–1898. [PMC free article] [PubMed] [Google Scholar]
- 68.Hopkins KM, Auerbach W, Wang XY, Hande MP, Hang H, Wolgemuth DJ, Joyner AL, Lieberman HB. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol. Cell Biol. 2004;24:7235–7248. doi: 10.1128/MCB.24.16.7235-7248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Budzowska M, Jaspers I, Essers J, de Waard H, van Drunen E, Hanada K, Beverloo B, Hendriks RW, de Klein A, et al. Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. EMBO J. 2004;23:3548–3558. doi: 10.1038/sj.emboj.7600353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levin N, Lavon I, Zelikovitsh B, Fuchs D, Bokstein F, Fellig Y, Siegal T. Progressive low-grade oligodendrogliomas: response to temozolomide and correlation between genetic profile and O6-methylguanine DNA methyltransferase protein expression. Cancer. 2006;106:1759–1765. doi: 10.1002/cncr.21809. [DOI] [PubMed] [Google Scholar]
- 71.Pegg AE. Mammalian O6-alkylguanine-DNA alkyltransferase: regulation and importance in response to alkylating carcinogenic and therapeutic agents. Cancer Res. 1990;50:6119–6129. [PubMed] [Google Scholar]
- 72.Bhanot OS, Ray A. The in vivo mutagenic frequency and specificity of O6-methyguanine in ΦX174 replicative form DNA. Proc. Natl Acad. Sci. USA. 1986;83:7348–7352. doi: 10.1073/pnas.83.19.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Loechler EL, Green CL, Essigmann JM. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc. Natl Acad. Sci. USA. 1984;81:6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cortellino S, Turner D, Masciullo V, Schepis F, Albino D, Daniel R, Skalka AM, Meropol NJ, Alberti C, et al. The base excision repair enzyme MED1 mediates DNA damage response to antitumor drugs and is associated with mismatch repair system integrity. Proc. Natl Acad. Sci. USA. 2003;100:15071–15076. doi: 10.1073/pnas.2334585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Griffin S, Branch P, Xu YZ, Karran P. DNA mismatch binding and incision at modified guanine bases by extracts of mammalian cells: implications for tolerance to DNA methylation damage. Biochemistry. 1994;33:4787–4793. doi: 10.1021/bi00182a006. [DOI] [PubMed] [Google Scholar]
- 76.Lari SU, Al Khodairy F, Paterson MC. Substrate specificity and sequence preference of G:T mismatch repair: incision at G:T, O6-methylguanine:T, and G:U mispairs in DNA by human cell extracts. Biochemistry. 2002;41:9248–9255. doi: 10.1021/bi020239n. [DOI] [PubMed] [Google Scholar]
- 77.Modrich P, Lahue RS. Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 78.Sibghat U, Gallinari P, Xu YZ, Goodman MF, Bloom LB, Jiricny J, Day RS., III Base analog and neighboring base effects on substrate specificity of recombinant human G:T mismatch-specific thymine DNA-glycosylase. Biochemistry. 1996;35:12926–12932. doi: 10.1021/bi961022u. [DOI] [PubMed] [Google Scholar]
- 79.Melo JA, Cohen J, Toczyski DP. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu X, Shell SM, Zou Y. Interaction and colocalization of Rad9/Rad1/Hus1 checkpoint complex with replication protein A in human cells. Oncogene. 2005;24:4728–4735. doi: 10.1038/sj.onc.1208674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brown KD, Rathi A, Kamath R, Beardsley DI, Zhan Q, Mannino JL, Baskaran R. The mismatch repair system is required for S-phase checkpoint activation. Nat. Genet. 2003;33:80–84. doi: 10.1038/ng1052. [DOI] [PubMed] [Google Scholar]
- 82.Giannattasio M, Lazzaro F, Longhese MP, Plevani P, Muzi-Falconi M. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J. 2004;23:429–438. doi: 10.1038/sj.emboj.7600051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lavin MF. The Mre11 complex and ATM: a two-way functional interaction in recognizing and signaling DNA double strand breaks. DNA Repair (Amst) 2004;3:1515–1520. doi: 10.1016/j.dnarep.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 84.Wang Y, Qin J. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc. Natl Acad. Sci. USA. 2003;100:15387–15392. doi: 10.1073/pnas.2536810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Mol. Cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.