Abstract
Techniques for investigation of exogenous small interfering RNA (siRNA) after penetration of the cell are of substantial interest to the development of efficient transfection methods as well as to potential medical formulations of siRNA. A FRET-based visualization method including the commonplace dye labels fluorescein and tetramethylrhodamin (TMR) on opposing strands of siRNA was found compatible with RNA interference (RNAi). Investigation of spectral properties of three labelled siRNAs with differential FRET efficiencies in the cuvette, including pH dependence and FRET efficiency in lipophilic environments, identified the ratio of red and green fluorescence (R/G-ratio) as a sensitive parameter, which reliably identifies samples containing >90% un-degraded siRNA. Spectral imaging of siRNAs microinjected into cells showed emission spectra indistinguishable from those measured in the cuvette. These were used to establish a calibration curve for assessing the degradation state of siRNA in volume elements inside cells. An algorithm, applied to fluorescence images recorded in standard green and red fluorescence channels, produces R/G-ratio images of high spatial resolution, identifying volume elements in the cell with high populations of intact siRNA with high fidelity. To demonstrate the usefulness of this technique, the movement of intact siRNA molecules are observed after introduction into the cytosol by microinjection, standard transfection and lipofection with liposomes.
INTRODUCTION
RNA interference (RNAi) is a natural response of eukaryotic cells to double-stranded RNA (dsRNA) leading to silencing of homologous gene transcripts. While application of RNAi in reverse genetic approaches is nowadays common use, therapeutic use, in particular, of small interfering RNAs (siRNAs) is still fraught with challenges. In this exponentially growing field of RNAi, stability, degradation and delivery of siRNA receive increasing attention. A limited correlation between chemical siRNA stability on one hand and RNAi efficiency and duration on the other hand has been described, but the limits are not well understood (1). The argonaute-2 (Ago2) protein, a central component of the RNA-induced silencing complex (RISC), features a RNAse III type domain which cleaves the siRNA passenger strand (2,3). However, given that several thousand siRNA molecules penetrate into a cell during transfection, the comparatively low number and siRNA-cleavage activity of Ago-2-containing RISCs makes it unlikely that Ago-2 is a major factor in the degradation of exogenous siRNA. Thus there is considerable interest in discovering other degradation factors and their subcellular location, a process that would greatly benefit from knowledge on the subcellular location of their potential substrate, i.e. reserves of intact siRNAs after transfection.
The standard siRNA delivery protocol in mammalian cell culture includes incubation of siRNAs with cationic lipid transfection agents, resulting in siRNA-containing lipoplex particles of ill-defined size around ∼500 nm (4). While lipoplexes facilitate uptake (5), standard cationic transfection agents are toxic to the cells (6) and considered unsafe for in vivo applications. Potential adequate systemic delivery systems include ∼100 nm liposomes (7) and related formulations (8). The huge excess of siRNA molecules devoted to one cell in standard transfection protocols (107–109) over the targeted mRNA (101–104) clearly illustrates the potential benefits of new approaches to monitor delivery and transport processes into and inside the cell. It is unclear, in what manner the ensemble of siRNA molecules, once penetrated into the cell, contributes to RNAi, because they outnumber the ensemble of RISCs by several orders of magnitude. In this context, methods to determine the whereabouts of pools of intact siRNA in the cell should be extremely helpful.
To this end, we present here a FRET-based imaging approach where communicating dyes are located on opposing strand of the applied siRNA, allowing a non-destructive and non-invasive assessment of the degradation state of siRNAs in cultured cells. RNAi efficiency of siRNA duplexes is superior to single strands of siRNA by several orders of magnitude (9), clearly illustrating the necessity of tracing the whereabouts of the intact siRNA duplex in the cell. Standard methods, which trace siRNA conjugated to fluorophores inside cells, cannot distinguish between intact siRNA duplexes, single strands or even just the mere fluorophores left after siRNA degradation in the imaging process. Therefore, the use of FRET techniques, based on distance-dependent communication of two dyes, is an attractive way to approach this issue (10–12), because degradation of a double-labelled siRNA results in spatial separation of the dyes and a breakdown of FRET. The most important problems that needed to be addressed for judicious use of FRET-labelled siRNA inside cells include compatibility of dye-labelling with RNAi (13,14), possible changes of spectral properties of the dyes inside the cell due to lipophilic environment or pH variations, and conclusive and quantitative correlation of fluorescence emission from inside the cell with siRNA integrity or degradation status. We report here a judicious application of siRNA-labelling with a commonplace FRET dye pair, using different relative positions of the dyes to establish a calibration system that correlates FRET efficiency with siRNA integrity outside and inside a cell. This calibration can be used with a standard confocal microscope to visualize regions containing high amounts of intact siRNA inside the cell at high resolution.
MATERIALS AND METHODS
siRNAs
siRNAs against GFP (passenger: 5′-GCAAGCUGACCCUGAAGUUCAU-3′/guide 5′-P-GAACUUCAGGGUCAGCUUGCCG-3′) (15) and the rat ABCb1b transporter mRNA sequence (passenger: 5′-AAACUGUUGUCUGGUCAAGCC-3′/guide: 5′- GGCUUGACCAGACAACAGUUU-3′′/GenBank: NM_000927) containing fluorescein and tetramethylrhodamin (TMR) labels were purchased from IBA (Göttingen, Germany). The chemical structure of the TMR derivative conjugated to the 3′ or 5′, respectively, of the guide strand is shown in Supplementary Figure S1a. Figure S1b shows a similar derivative of fluorescein attached to the 5′ or 3′ of the passenger strand, with the exception of the ABCb1b passenger strand, where the respective structure formula, including a 3′-terminal dC as an attachment site, is shown in Figure S1c.
Fluorescence emission scans
Solution fluorescence assays were performed in a FP-6500 fluorimeter (JASCO, Tokyo, Japan) equipped with ETC-273 temperature controller, a peltier element and an F-25 (Julabo, Seelbach, Germany) cooling unit. Emission spectra (3 nm bandwidth), recorded upon excitation at 488 nm (3 nm bandwidth) were corrected for differential wavelength PMT sensitivity. Unless indicated otherwise, spectra were recorded in 50 μl SUPRASIL cuvettes (HELLMA, Müllheim, Germany) in 1 × PBS (pH 7.4) at 20°C.
Digestion with double-strand specific nuclease V1
In situ degradation assays were conducted by recording emission spectra of 1.5 μM siRNA samples at 37°C in 40 mM Tris–HCl pH 7.4, 10 mM MgCl2 before and at certain time points after addition of 46 mU RNase V1 (Roche, Indianapolis, USA).
FRET efficiency
The theoretical FRET efficiency EFRET was calculated according to
where the distance between fluorophores was gauge from X-ray structures of standard A-type RNA helices as 17 Å for high-FRET and 74 Å for low-FRET siRNA, and the Förster radius for fluorescein and TMR R0 = 55 Å was taken from the literature (16). Experimental FRET efficiency Q of the siRNAs was determined as the extent of fluorescein fluorescence quenching using the equation:
where F1 is the fluorescence intensity at 520 nm upon excitation with 488 nm and F2 the fluorescence intensity at 520 nm of the donor-only sample, determined by complete digestion with RNase.
Spectral imaging of the microinjected cells
Spectral imaging was performed on a confocal laser scanning system (C1Si, Nikon, Badhoevedorp, The Netherlands) on an inverted microscope (TE2000-E, Nikon), which was equipped with an oil-immersion objective lens (CFI Plan Apo 40×). The cells were excited with an Argon laser at the 12% intensity in standby modus at 488 nm and the fluorescence was detected with 32-channel PMTs which were corrected for differential wavelength sensitivity. A spectral gain of 180 and pixel dwell of 6 µs was used.
R/G-ratio imaging in confocal standard mode
Cells were imaged on a DM IRBE TCS MP1 confocal microscope (Leica, Wetzlar, Germany) with an oil-immersion objective (63× magnification; NA 1.32) and an Argon laser for excitation at 488 nm, operated with software version 2.61_Build_1537. A triple dichroic filter (TD 488/544/633) was used as a main excitation beam splitter. The image size width and height were 158.7 μm. Ten images of 512 × 512 8-bit-pixels were recorded with a confocal plane distance of 20 μm. z-Stacks were composed from these images and the pixels thereon considered volume elements (voxels). The following standard settings were applied to all samples: Gain: 830/Offset: 53.7/Pinhole 228 µm/Excitation: 488 nm/Laser intensity: 100%/PMT1: 510–540 nm (green channel)/PMT2: 570–600 nm (red channel). In order to avoid pixel saturation, certain additional images were recorded at lower gain values (750, 600 and 500).
The green and red channel images (standard settings) were added on a voxel basis and the resulting image (termed GPR) was compared to a manually chosen threshold of 200 to segment the cells from the background. Background values RBG and GBG were calculated as the mean intensities of the red and green channel, respectively.
Using the voxels from the red and green channels inside cells, R and G, the R/G ratio was calculated according to the equation:
Starting from the data set obtained at highest PMT gain, R/G values from pixel pairs reaching saturation in either channel were substituted with the corresponding values from unsaturated pixel pairs from data sets obtained at lower PMT gains. Distributions of the calculated R/G ratios were visualized in histograms of 256 bins. For visualization, only a subset of pixels of the ratio image were red/green coloured. These pixels were determined by application of neighbourhood-thresholding segmentation to the GPR image and a subsequent dilatation algorithm. The threshold was calculated as the sum of the mean and the SD. R/G pixels were composed as follows; pixels with a value below 1.1 were coloured (‘solid’) green (degraded), pixels with a value above 2.1 were coloured (‘solid’) red and pixels of intermediate values were coloured in a transition from green over yellow to red.
Remaining pixels were determined to be either background, and accordingly coloured black, or to belong to the cell, in which case they were coloured in grey. Discrimination of background and cell was done as follows: Each channel was first thresholded separately, using the sum of mean and standard deviation as threshold value. The results were added and smoothed by subsequent application of first a median, then a mean filter and finally thresholded using a fixed threshold value of 10. The threshold value was determined by visual inspection yielding the most accurate segmentation of the cells shape.
Gene knockdown
To measure the RNAi effect against a GFP mRNA, siRNAs were co-transfected with a GFP-expression vector (pEGFP-N1, BD Clontech, Heidelberg, Germany), and the resulting GFP fluorescence compared to cells transfected with the vector only. One day before transfection 2 × 105 human embryonic kidney cells (HEK293) were plated in 0.5 ml Dulbecco's Modified Eagle Medium (Biochrom, Berlin, Germany) containing 10% foetal calf serum (FCS) (Biowest, Nuaillé, France) yet without any antibiotics per well of a 24-well cell culture plate. On the following day cells had a confluence of ∼80–90%. A total of 30 pmol of the annealed duplex siRNA against GFP and 400 ng pEGFP-N1 were mixed in 50 μl OptiMEM I medium (Invitrogen, Karlsruhe, Germany). 1 μl Oligofectamine (Invitrogen) was diluted in 50 μl OptiMEM I. Diluted Oligofectamine reagent was added to the diluted siRNA/plasmid mixture and incubated for 15 min at room temperature. Transfection mixture was added dropwise onto the cells. After 24 h cells were harvested and washed once in PBS. GFP expression was analysed by flowcytometry (FACS Canto, BD, Heidelberg, Germany) in the FITC channel (530/30 nm). Therefore, live cells were identified and gated by typical forward and side scatter characteristics. 10 000 cells were analysed and geometric mean fluorescence intensity (MFI) of all gated cells was determined as a measure for the strength of GFP expression. The presented results are the mean value of three different wells.
Formulation of siRNA in liposomes
siRNA-containing liposomes were prepared by using a dual asymmetric centrifuge (DAC) (17). For this, 18 mg of a mixture of hydrogenated egg PC (EPC-3, Lipoid AG, Ludwigshafen, Germany), cholesterol (Sigma) and N-(carbonyl-methoxypolyethylene glycol2000)-1,2-distearoyl-sn-glycero-3-phosphoethanolamine (Lipoid AG) (65/32/3 mol%) were dissolved in 500 µl of ethanol and transferred into a 10 ml glass vial (MedChrom, Eppelheim, Germany). The organic solvent was removed under a constant stream of nitrogen. The resulting thin lipid film was dried under vacuum overnight at room temperature. Fifty microlitre of siRNA-containing PBS solution was added, the vials were clamped and the phospholipids were incubated for 5 min at room temperature to allow hydration and swelling of the lipids. The hydrated and swollen lipid-siRNA mixture was then homogenized by using a DAC (DAC 150, Hauschild GmbH, Hamm, Germany) for 6 × 5 min at 3500 r.p.m., 272 μl of buffer were added to disperse the cream-like liposomal formulation (vesicular phospholipids gel) to conventional liposome dispersion. For dispersion, the vial was shaken for 5 min in horizontal shaker. Non-entrapped siRNA was removed from the dispersion by filtrating the dispersion through a column containing sephadex G-50 Fine (Amersham, Sweden). The volume was measured and the mixture was adjusted to a lipid concentration of 5.6 mmol containing ∼560 pmol siRNA per ml.
Growth and fixation
REB4 cells (18) were grown in 45% v/v α-MEM (supplemented with 2.2 g/l NaHCO3); 45% v/v HAMs F10; 10% v/v FCS; 100 µg/ml PenStrep Mix; 1 ng/ml bFGF at 37°C and 5% CO2 atmosphere. For amplification, cells were passaged into and grown in 75 cm2 flasks. Seeding density for RBE4 was 40 000 cells per cm2. Freezing medium was 35% v/v α-MEM (supplemented with 2.2 g/l NaHCO3); 35% v/v HAMs F10; 20% v/v FCS; 10% v/v dimethyl sulphoxide. For the confocal experiments the cells were grown on sterilized cover slips (Lab-Tek TM II Nalge Nunc, New York, USA) placed on the bottom of 12-well plates. Cover slips were treated with 3 µg/cm2 rat tail collagen prior to passaging to improve adherence on the glass cover slips. The bottom of the well plates was treated with a thin layer of 2% agarose and the cover slips were placed onto the hot agarose layer. For confocal imaging, the cover slips with the cells were briefly dipped into PBS prior to fixation in freshly prepared fixation buffer (4% formaldehyde in growth medium without PenStrep and FCS) for 10–20 min, followed by washing in PBS buffer for three times.
Transfection with oligofectamin and liposomal formulations
A standard protocol for HeLa cells (19) was adapted to Rat Brain Endothelial RBE4 cells (18), which were transfected at 70% confluency in a 12-well culture plate. Two days prior to transfection, antibiotics were omitted in the fresh growth medium (0.7 ml per well), which was replaced with 0.364 ml of transfection medium (growth medium without PenStrep and FCS) just prior to transfection. Six microlitre (30 pmol; 1.8 × 1013 molecules, ∼108 molecules per cell) of annealed siRNA were mixed with 100 μl of transfection medium, and 6 μl Oligofectamine (Invitrogen) mixed with 24 μl transfection medium. After pre-incubation at room temperature for 5 min, these solutions were combined and the resulting mixture was incubated for an additional 15 min at room temperature, then added to one well, and incubated for at least 3 h before 3× transfection medium was added. For lipofection with liposomally formulated siRNAs, cells were grown to 70% confluency on the cover slips coated with collagen as in the transfection assay. Per well of a 12-well culture plate, ∼30 pmol of siRNA in liposomes were mixed with transfection medium to a final volume of 500 μl. Liposomes were distributed evenly by gentle agitation. Transfection medium was replaced with 3× transfection medium after 3 h of lipofection.
Microinjection
One day prior to microinjection, ∼5 × 105 cells were plated on small glass cover slips (Ø 12 mm) placed into a small petri dish (Nunclon) containing 2 ml growth medium. Cells were transferred into a petri dish filled with 2 ml of a carbonate-free α-MEM medium containing 25 mM HEPES (without PenStrep and FCS) immediately prior to injection. For injection, capillaries were pulled with an automatic, high precision puller (P-87 Brown Flaming Micropipette Puller, Sutter Instrument Company, Novato CA, USA) and loaded with 50 µM labelled siRNAs in PBS. In each experiment, 50 cells were injected into the cytoplasm, each cell with 200 fl of the siRNA solution (100 μM, corresponding to ∼1.2 × 107 siRNA molecules per cell). Time needed for precise injection of a sample of 50 cells was ∼15 min. The microinjection was performed using an automated computer-assisted Zeiss AIS injection system (Carl Zeiss Jena, Germany) equipped with an Eppendorf 5242 pneumatic injector (Eppendorf, Hamburg, Germany) for precise control of the injection pressure. Working pressure for injection in adherent cells was ∼200 hPa and the injection time per individual cell 0.2 s. After the microinjection procedure cells were immediately fixed as described above.
RESULTS
Spectral characterization of FRET-labelled siRNA in the cuvette
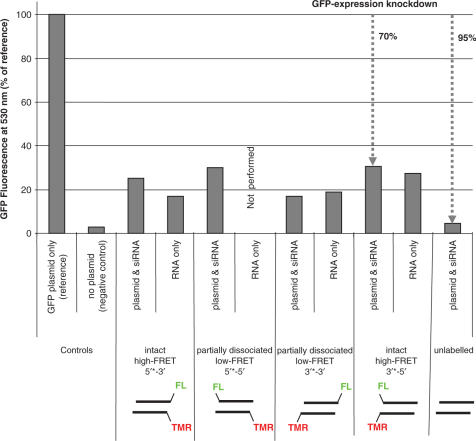
The concept of using a FRET dye pair to monitor RNA integrity has been proposed and partially implemented by several groups (10–12). An important aspect to be emphasized in such studies is the site of dye conjugation and the chemistry employed, because certain dye labels were reported to impede knockdown efficiency (13,14,20,21). We labelled siRNA at different sites with the well-characterized FRET pair fluorescein and TMR, which has a Förster radius of ∼55 Å (16). In previous investigations on FRET in siRNA, the dyes were either tethered to the 5′′ and the 3′ end of the same strand (5′–3′) (12) or to the respective 5′ ends of opposing strands (5′*–5′, where * denotes the guide strand) (11). Some controversial data on functionality of dye-labelled siRNA in the literature (13,14,20–22) instigated us to test the RNAi function of permutations of attachment sites with labels on opposite strands of siRNAs. Four permutations, in which the guide strand was labelled with TMR and the passenger strand was labelled with fluorescein, were submitted to a GFP reporter gene knockdown assay. In agreement with previous reports (20,21), 5′-dye labels were found to impede knockdown efficiency somewhat in comparison to an unlabelled siRNA (Figure 1). However, and in some contrast to earlier reports, all of the permutations resulted in a knockdown efficiency still better than 70%. The latter value is a conservative estimate, since the siRNA by itself produces an increased background fluorescence in the GFP channel, thus biasing the apparent knockdown values towards lower efficiency (see lanes labelled ‘RNA only’ in Figure 1).
Figure 1.
Knockdown efficiencies of FRET-labelled siRNAs. Knockdown efficiencies of GFP in HEK293 cells 24 h after transfection with plasmid DNA encoding GFP alone, or co-transfected with GFP-specific siRNAs. GFP expression was determined by flowcytometry. Geometric MFI of GFP plasmid alone was set as 100% reference. Note the knockdown efficiency of dye-labelled siRNAs is underestimated, because the fluorescein dye labels on the siRNAs contribute to the overall fluorescence activity, as seen in several ‘RNA only’ samples.
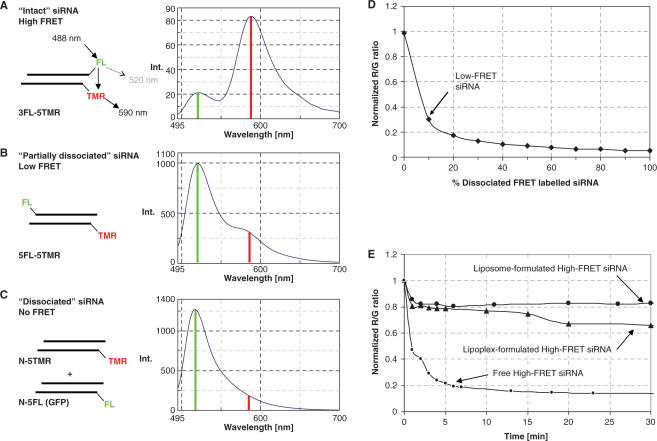
Spectral properties of the siRNA constructs were investigated in the cuvette upon fluorescein excitation at 488 nm. The respective emission spectra (Figure 2A–C) clearly reflected differential FRET efficiencies as a consequence of the spatial separation of the dyes: constructs with communicating dyes in proximity (5′*–3′), or on the opposite ends of the duplex (5′*–5′) showed high and low FRET efficiency, respectively, in zcomparison to an siRNA mixture containing the respective dyes in different molecules which showed no FRET at all. According to their labelling type and the resulting FRET efficiencies, siRNAs were dubbed ‘high-FRET’, ‘low-FRET’ and ‘no-FRET’, respectively. Digestion with double-strand specific nuclease V1 was monitored in real time in the cuvette, and the end-point of this degradation provided the ‘donor-only’ fluorescence values needed to calculate FRET efficiencies, which correlated well with values calculated from the estimated distance between the dyes: 0.98 versus 0.97, 0.17 versus 0.18 and 0, respectively. FRET properties of ‘no-FRET’ siRNA were very similar to those of heat-denatured ‘high-FRET’ siRNA as observed in fluorescence melting curves (Supplementary Table S1 and Supplementary Figure 2). Since the ‘no-FRET’ sample showed the same spectral properties as the fully degraded samples, it was further on used as a model compound for ‘dissociated’ siRNA, where the term dissociated indicates that the same spectral properties may also result from dissociation of both RNA strands without involving actual RNA cleavage. Similarly, the ‘high FRET’ compound was the ‘intact’ siRNA, whose degradation status constitutes the actual object under investigation. Of note, the TMR emission cannot serve as a stand-alone indicator of siRNA integrity, because the emission shoulder of the dequenched fluorescein produces a fluorescence increase at the typical TMR emission wavelength in degraded samples. Rather than calculating FRET efficiencies, the relative fluorescence emissions of the TMR and fluorescein (R/G ratio) were further on used as a measure of siRNA integrity. This ratio, which consistently exceeded a value of three in three different ‘high-FRET’ siRNAs, is strongly dependent on the content of degraded siRNA in a sample, as shown in the calibration curve obtained from mixtures of different ratios of intact and ‘dissociated’ siRNA. The curve shown in Figure 2D is normalized to 1 to account for these subtle variations. Its steep initial slope provides a dynamic range from 0% to 10% ‘dissociated’ RNA, which is useful for tracing populations containing high proportions of intact siRNA, while the small changes in between 10% and 100% ‘dissociated’ siRNA do not allow to measure precisely the degradation state of such mixtures. A small intermediate region of moderate changes of around 10% corresponds to the R/G value of the ‘low-FRET siRNA’, which was further on used as a model compound emulating the spectral properties of ‘partially dissociated’ siRNA, providing an additional calibration point to mark the lower end of the dynamic R/G range.
Figure 2.
Fluorescent labelling of siRNAs and concept of R/G ratio in the cuvette. (A) Orientation of dye labels in ‘high-FRET’ or ‘intact’ siRNA with fluorescein at the 3′ end of the sense strand and with TMR at the 5′ end of the antisense strand. Upon excitation at 488 nm, some fluorescein dyes emit fluorescence at 520 nm (indicated by a green bar) while the majority transfers the excitation by FRET to TMR, which shows a fluorescence maximum at 590 nm (red bar). Inversion of sense and antisense strands with respect to dye labelling produces identical emission spectra. (B) Orientation of dye labels in a model ‘low-FRET’ siRNA, which is being used as a model for ‘partially dissociated’ siRNA populations. Both dyes are attached to the 5′-ends of the respective strands. Due to enlarge distance between the dyes, the reduced FRET efficiency leads to strongly increased fluorescence at 520 nm in the emission spectrum. (C) A spectrum with no evidence of FRET is produced by a mixture of two ‘no-FRET’ siRNAs with different sequences, one 5′ -labelled with TMR and directed against the rat ABCb1b-transporter protein, the other 5′-labelled with fluorescein and directed against GFP (see Materials and Methods section for sequences). (D) Calibration curve which allows deducing the amount of dissociated siRNA in a sample of high-FRET siRNA, obtained from the emission spectra of defined mixtures of high-FRET and no-FRET siRNAs. The R/G ratio (emission at 590 nm/emission at 520 nm) of ‘intact’ siRNA was normalized to 1. (E) Degradation kinetics of formulations of intact siRNA in lipoplexes (triangles), liposomes (large dots) and neat (small dots).
The R/G ratio concept was further developed in the cuvette to trace degradation of intact siRNA in different formulations typical for applications in either transfection of cultured cells or in intravenous applications to animals. Lipoplexes formed from ‘intact’ siRNA and oligofectamin revealed particular degradation kinetics upon incubation with RNase V1 (Figure 2E): the R/G ratio rapidly dropped to ∼80% of its initial value and then abruptly stabilized, while in degradation of free siRNA it continued to drop quickly out of the dynamic range. Complete digestion of lipoplexed siRNA was effectively delayed to several hours (data not shown). The initial drop-off in the R/G ratio likely corresponds to degradation of siRNA, which is accessible on the lipoplex surface or not complexed at all.
For comparison, we have formulated ‘intact’ siRNA into so-called stealth liposomes which are covered with a polyethylene layer to prevent rapid clearance from the blood stream (23). A novel DAC technique allows formulation of liposomes with a narrow size distribution of around 100 nm on a very small scale and with high-inclusion yield (17). Presumably due to strong light scattering, addition of empty liposomes to a solution of ‘intact’ siRNA reduces the overall fluorescence by a factor of ∼2 (data not shown). The background fluorescence of empty liposomes increases the baseline and consequently decreases the R/G ratio. Hence, R/G values in the following reactions were normalized to the initial value. After a brief initial drop to 80% of the initial R/G value, freshly formulated siRNA-liposomes showed complete resistance to nuclease V1, as indicated by an R/G ratio that remained stable even after 2 h (Figure 2E). However, formulations of several weeks of age slowly release the siRNA, as shown by gel filtration. Accordingly, the R/G ratio in a V1 degradation of 10-week-old siRNA liposomes rapidly dropped out of the dynamic range (data not shown).
Investigations of pH dependence showed a strong decrease of overall fluorescence observed in the emission spectra taken at low pH in the cuvette for free siRNA as well as in liposomes. Investigations of siRNA labelled with either dye showed that this was due to the known sensitivity of fluorescein towards low pH, while TMR fluorescence was only moderately diminished. In summary, the R/G ratio in ‘intact’ and ‘partially dissociated’ siRNA was effectively increased in acidic conditions, showing a minimum around neutral pH (Supplementary Figure S3). Consequently, siRNA populations in certain organelles e.g. maturating endosomes and lysosomes are expected to show an increased R/G ratio, but contribute little to the overall fluorescence emission.
Spectral analysis of cells after microinjection of high-FRET siRNAs, low-FRET siRNAs and no-FRET siRNAs
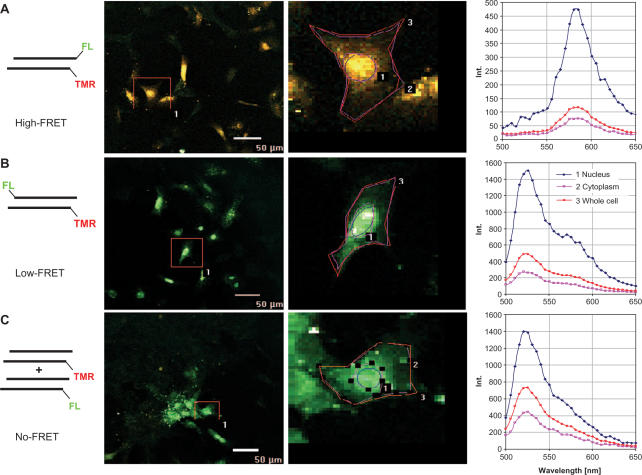
To validate the R/G ratio concept in tissue culture experiments, RBE4 cells were microinjected with ∼107 molecules of either ‘intact’, ‘partially dissociated’ or ‘dissociated’ siRNAs in a volume of ∼200 fl. After injection, cells were immediately fixed and then imaged by confocal spectroscopy in spectral mode, i.e. emission spectra of the different siRNA inside the cells ranging from 500 to 650 nm were recorded in 32 bins of 5 nm upon excitation at 488 nm (24). Areas of interest were defined corresponding to different zones and/or organelles of the cell. As can be seen in Figure 3, R/G ratios of the ‘intact’ siRNAs vary somewhat, depending on whether the zones of interest comprise the entire cell (4.5), the cytoplasm (3.9) or the nucleus only (<5). In other microinjected cells the R/G ratios of intact siRNA varied between ∼3 and ∼5 in accordance with the variations observed in the cuvette. Overall, the excellent agreement with the respective spectral properties in the cuvette show that the R/G ratio can be meaningfully interpreted inside cells, despite the potential changes of dye properties due to intracellular variations of lipophilicity and pH. Moreover, the validity of the calibration curve in Figure 2D inside the cell is clearly demonstrated. However, the definition of zones of interest (Figure 3), necessary to obtain enough fluorescence signal intensity for spectral analysis, constitutes an important limitation in that it provides poor spatial resolution.
Figure 3.
Spectral properties of siRNAs in cells are identical to those in the cuvette. True colour fluorescence imaging of cells microinjected with intact siRNA (A), ‘partially dissociated’ siRNA (B) and ‘dissociated’ siRNA (C) upon excitation at 488 nm. Left column, survey images; middle column, zoom of a single cell with defined ROIs; right column, spectra obtained from the respective ROIs: whole cell (open red circles), nucleus (solid blue diamonds), cytoplasm (open pink squares).
Confocal microscopy in standard mode and R/G imaging
For higher spatial resolution, microinjected cells were imaged with a confocal microscope in high spatial resolution mode. Because of the shape of the fluorescence emission bands, R/G ratios of the same sample vary, depending on the bandwidth of the collected emission light. Thus, R/G values calculated from spectra recorded from cells in the confocal spectral mode were binned with a bandwidth of 5 nm, meaning that value at 580 nm is generated by photons of wavelengths from 578 to 582 nm. Broadening the bins lowers the spectral resolution, but improves sensitivity, because more photons are collected in the same bin. In the following, a standard procedure called R/G imaging is developed, where fluorescence light is collected with a bandwidth of 30 nm through the use of a band-pass filter setting in standard confocal mode. Since the spectral equivalence of siRNAs in the cuvette and in the cells has been demonstrated above, it is no longer necessary to record a full fluorescence emission spectrum as in Figure 3, because the fluorescence wavelength at two selected wavelengths is sufficient to characterize the intactness of the siRNA. Hence, we now resorted to recording two superimposable images, a green one of the fluorescein emission peak (510–540 nm) and a red one of the TMR emission peak (570–600 nm). Figure 4 shows both channels of the cells microinjected with three siRNAs types, which will now be used as calibration standards, marking the R/G values at which siRNAs can still be considered ‘intact’.
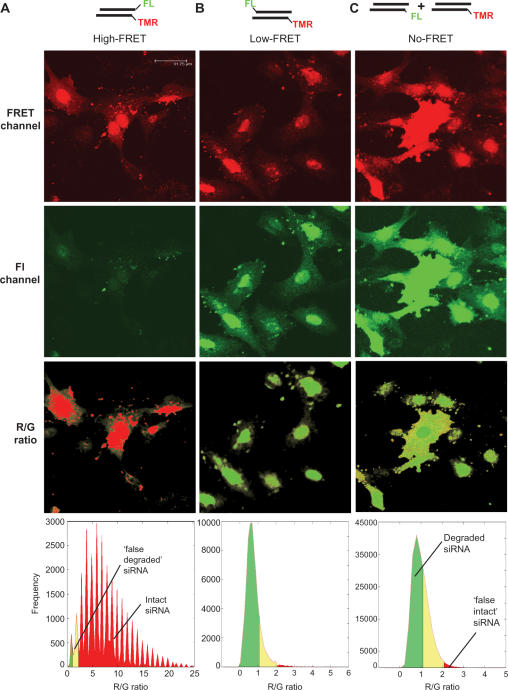
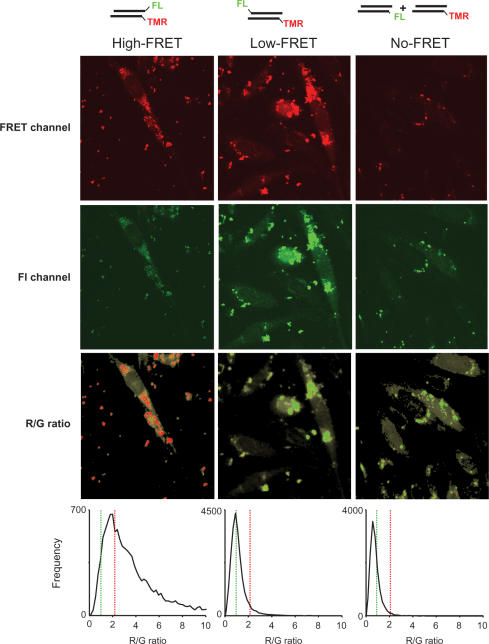
Figure 4.
Basis for R/G imaging algorithm. Cells microinjected with intact siRNA (A), ‘partially dissociated’ siRNA (B) and ‘dissociated’ siRNA (C) upon excitation at 488 nm were imaged at high spatial resolution in a red channel (570–600 nm; upper row) and a green channel (510–540 nm; second row from the top). R/G ratio images (second row from the bottom), obtained after image processing show low fluorescent parts of cells in light grey and assign a colour-coded degradation status to each voxel. Red voxels contain >90% intact siRNA. Pixels identified as belonging to the cell by fluorescence thresholding are in grey. Statistical occurrence of R/G-ratio values is analysed in histograms (lower row) for each microinjected siRNA. The spikes in the left histogram for intact siRNA result from background correction during the data treatment algorithm; the percentile distributions are not significantly affected by this.
Differences between the cells microinjected with ‘intact’ and ‘dissociated’ siRNAs are obvious by mere visual inspection of the green channel in Figure 4, due to quenched fluorescein emission in intact siRNA. Images of ‘partially dissociated’ or ‘dissociated’ siRNAs showed dequenched emission in the green channel, but distinction between the two siRNA types was not possible with the naked eye. The spectroscopic information on the status of siRNA integrity in every single volume element (voxel), as obtained from successive imaging of confocal layers (z-stack imaging), was processed into R/G images by an algorithm outlined below and detailed in the method section. Voxels are first segregated as belonging to cells as regions of interest (ROI) by using a threshold level of fluorescence in both channels to exclude extracellular space from further analysis. Pixels thus identified as belonging to the cell are coloured grey. After renewed thresholding inside the ROI, to identify voxels containing significant amounts of dyes, these are further processed by dividing the background-corrected red channel image by the corresponding background-corrected green channel image on a voxel basis. The histograms in Figure 4 show the distributions of thus obtained R/G values from the cells microinjected with the three siRNAs.
The R/G ratio in confocal imaged ‘intact’ siRNA shows a distribution with a maximum of around 7.5, which, as a consequence of the differences in the optical detection parameters, is at variance with values determined in the cuvette or in spectral mode cell imaging. Mean values of ∼1, obtained from either ‘partially dissociated’ low-FRET siRNA or ‘dissociated’ no-FRET siRNA did not differ significantly. Using threshold R/G values of 1.1 and 2.1 the histograms were segmented into three zones, coloured red, yellow and green in Figure 3. The red segment in the histogram of intact siRNA (lower left in Figure 4) contains >93% of voxels, meaning that imaging these voxels in red accounts for the vast majority of all ‘intact siRNA’. The fraction of voxels with an R/G value higher than 2.1 in the ‘dissociated’, and ‘partially dissociated’ siRNA samples (red areas in the respective histograms), show that the probability of assigning ‘false intact’ siRNA to a red voxel is below 3%. Moreover, errors in the localization of clusters of such voxels are exponentially reduced with the number of voxels in a potential cluster: the likelihood that N adjacent voxels are all wrongly identified as intact is ∼(0.03)N and therefore negligible for N > 1.
Inversely, imaging voxels below 1.1 in green accounts for >63% of voxels in the ‘dissociated’ siRNA sample, and creates <3% ‘false dissociated’ (green areas in the respective histograms). For the intermediate segment of yellow voxels, assignment to either category is error-prone. This segment comprises ∼35% in the ‘dissociated’ samples, but below 5% in the ‘intact’ samples. Its delineation by threshold values of 1.1 and 2.1 is based on >10 histograms as in Figure 3, neither of which produced ‘false intact’ or ‘false dissociated’ voxels in excess of 3% upon application of these thresholds. The fraction of yellow voxels varied significantly (5–35%) among the various ‘dissociated’ and ‘partially dissociated’ samples, underscoring the above mentioned difficulty to precisely quantify samples containing 10% to 100% degraded siRNA (Figure 2D).
Applications of R/G ratio imaging to transfection and lipofection
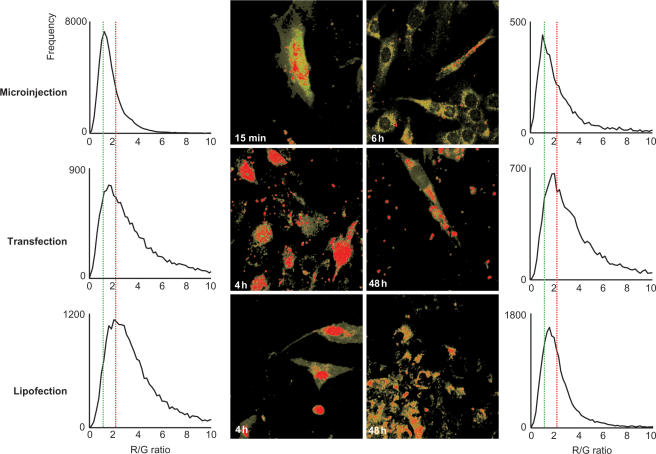
To test the validity of the above calibration in standard transfection experiments, cells were transfected with oligofectamin-lipoplexes of the three siRNAs types used in the microinjection experiments. Application of R/G imaging to cells 48 h after standard transfection was expected to show negligible or no red voxels for the cells transfected with no-FRET and low-FRET siRNAs, and a mixture of red and green voxels for the cells transfected with high-FRET siRNAs. In the later case, the green voxels would correspond to the cell regions, where the dyes are spatially separated, either by cleavage of the dye from the RNA, or from actual degradation of the siRNA, or by dissociation of sense- and antisense strands. Figure 5, depicting R/G images of cells 48 h post-transfection, shows that this is indeed the case: R/G imaging of cells containing no-FRET and low-FRET siRNAs produces exclusively green images (images of siRNAs labelled with either fluorescein or TMR can be found in Supplementary data, Figure S4). Since the ‘partially dissociated’ siRNA bears the spectral properties of a sample containing 10% ‘dissociated’ siRNA, the red voxels in Figure 4 resulting from ‘intact’ siRNA, identify populations >90% integrity. Comparison with images obtained from microinjection of 107 molecules per cell (Figure 3) shows roughly 106 intact siRNA molecules remaining, corresponding to ∼1% of the initial 108 molecules/cell in the transfection mixture (see Materials and Methods section). At 48 h, which typically coincides with efficient mRNA down regulation in RNAi experiments, all fluorescence, including the identified intact siRNA, is located in the cytoplasm, and none of it co-localized with DAPI stain (Supplementary Figure S5). Because a rapid influx of intact siRNA into the nucleus from the cytoplasm was evident after microinjection (Figure 3), this transport event was visualized by R/G-imaging of cells after microinjection, standard oligofectamin transfection and transfection with liposomally formulated siRNAs at early and late time points (Figure 6). Because microinjection is a direct delivery method without any lag time for the uptake of siRNA, the early time point here was chosen right after completion of the microinjections into the cytoplasm, i.e. a maximum of 15 min after the first cell was injected. Already at this early time point, intact siRNA is found at high concentrations in the nucleus, while 4 h later, a mixture of intact and dissociated siRNA is found completely relocated to the cytoplasm (Figure 4B). Because of the slower and more continuous uptake in oligofectamin-mediated transfection, as well as in lipofection with liposomally formulated siRNA, the cells are kept in contact with siRNA-containing medium for up to 4 h. At 4 h, a large population of intact siRNA is evident in the nucleus in both cases. However, about 4-fold higher concentrations of liposomally formulated siRNA were necessary to detect uptake, and these formulations were inactive in the knockdown assay (data not shown). After 48 h, the nucleus neither contains any intact siRNA nor any discernable fluorescence signal at all, regardless of the transfection method.
Figure 5.
Validation of R/G imaging after transfection in cell culture. FRET channel, green channel and R/G imaging of cells 48 h post-transfection shows red voxels indicating residual intact siRNA only in samples transfected with intact siRNA, but not in those transfected with ‘partially dissociated’ or ‘dissociated’ siRNA, respectively. Pixels identified as belonging to the cell by fluorescence thresholding are in grey. Histograms of R/G statistics, onto which the threshold values of 1.1 and 2.1 are indicated by green and red bars, respectively, are shown below the R/G images.
Figure 6.
R/G imaging in cell culture. R/G images of cells after delivery of intact siRNA by microinjection (upper row), transfection with oligofectamin (middle row) and liposomes (lower row), respectively. Images in the left column are taken at early time points after siRNA application, and images in the right column are taken at late time points. Pixels identified as belonging to the cell by fluorescence thresholding are in grey. Histograms of R/G statistics are shown next to the respective R/G images.
DISCUSSION
The central objective of this work was the establishment of an imaging technique for intact siRNA in a cell. For the illustration of its potential, our imaging technique has been applied to processes of common interest, such as transfection in cell culture and formulation of siRNA in liposomes. These processes do not occur under physiological conditions and may as such cause atypical behaviour of the cells. For the following discussion, one should bear in mind that the observed processes are clearly relevant to fields such as ‘reverse genetics’ by RNAi or potential therapeutic applications of siRNA. However, since they may not reflect a native state of the cell, interpretation of these data in terms of the RNAi-related pathways must be approached with caution.
Dye conjugation chemistry and RNAi efficiency
In this work, a labelling type of siRNA with two standard dyes was investigated, both of which feature some drawbacks, including in particular the susceptibility of fluorescein to bleaching and to pH variations. In addition, the emission of TMR coincides with background fluorescence upon excitation at 488 nm in tissue (data not shown) rendering transfer to animals difficult. However, the GFP reporter gene assay employed here shows only little impediment of RNAi by the dyes, despite some dispute in the literature about the compatibility of siRNA end labelling with knockdown (13,14,20–22). Although 5′-labels appear to have a stronger negative effect than 3′-labels, as reported by Chiu and Rana (21) and Ohrt et al. (20), all of our constructs show high enough activity to easily justify their use as models of functionally active siRNAs in cells. Of note, two publications describing 5′-modifications as detrimental to knockdown (21) and (20) used 5′-amino groups to which dyes were conjugated by N-hydroxysuccinimide (NHS)- or tetrafluorophenol-chemistry, while the siRNAs presented here feature peptide bonds instead. Considering in addition, that these authors did not use fluorescein or TMR dyes, the reasons for discrepancies may well be found in the exact structure of the chemical entity conjugated to the 5′-phosphate, meaning that, while other dyes, such as Cy3 and Cy5, may have more favourable spectral properties, their effect on RNAi is not yet entirely clarified. Indeed, hydrolysis of this linker may occur before or during incorporation of the siRNA into RISC. In this case, our method would observe the intact siRNA as a prodrug, which is converted to its active form by cellular metabolism.
Features of the imaging techniques
Fluorescein and TMR are inexpensive; their spectral properties are contained in the standard settings of any fluorescence microscope, and these spectral properties, in particular the communication by FRET, favour the detection of intact, high-FRET siRNA, while being rather insensitive to any amount >10% ‘dissociated’ siRNA in a sample. In the cuvette, monitoring the R/G ratio allowed facile observation of siRNA degradation by nuclease, and partial or complete protection therefrom by formulation in lipoplexes and liposomes, respectively. The degradation kinetics of both formulations are of interest in the design of efficient transfection agents and in the field of siRNA delivery in vivo, which is of growing interest (8).
R/G imaging enables tracing of populations of un-degraded siRNAs in cells over extended periods of time. This task was hitherto afflicted with potential artefacts resulting from tracing of mere fluorophores, dissociated from the oligo during its degradation. Further commonplace criticisms of tracing dye-labelled oligos in cells have been addressed here: neither variations in pH nor a lipophilic environment (such as lipoplexes or liposomes) disturb the of R/G ratio beyond our interpretation limits. The validity of transferring the R/G concept from the cuvette into cells was thoroughly documented by in-cell spectral imaging, showing virtually identical spectral properties in both environments. However, in the actual R/G-imaging process, the spectral resolution of fluorescence light emitted inside cells was foregone in favour of high spatial resolution, obtainable with any confocal fluorescence microscope.
Potential applications of the imaging technique
The usefulness of our approach was demonstrated by observations of three different modes of siRNA internalization into cells: microinjection, lipoplex-mediated transfection and liposome-mediated transfection, each of which results in nuclear accumulation of intact siRNA soon after its application. After a certain lag period, a distribution in the cytoplasm is observed, but no more nuclear fluorescence is in evidence. Perinuclear localization is commonly reported for numerous nucleic acids (25), but this localization now appears to be a hallmark of a rather late stage in various processes of siRNA uptake. Transport in and out of the nucleus after microinjection has already been reported for antisense agents several years ago (26), and has only very recently been observed for siRNA (12,20). We show here that this process occurs after standard transfection with lipoplexes and after lipofection with liposomes as well. Previously reported application of (3′–5′) double labelling of an siRNA allowed assessment of integrity of only the labelled strand, but did not address potential strand unwinding during transport (12). Our combination of (3′–5′*) labelling and R/G imaging shows that at least some siRNA molecules end up hybridized in the cytosol after import into and export from the nucleus, suggesting that no helicase is involved. The observed strong influx and subsequent export raises questions concerning their significance in gene silencing. Indeed, while some triple-strand forming antisense agents act by inhibiting transcription in the nucleus (27), antisense research including siRNA will likely focus primarily on the major silencing pathways acting on mRNAs in the cytoplasm (28). The export of siRNA (among other RNAs) from the nucleus was shown to be dependent on exportin-5 (20). It is thus possible that factors governing the activity of exportin-5 are affected by interventions such as microinjection or transfection, and that our observations reflect a certain trauma on the physiological level of the cell. If so, observation of this phenomenon may be useful in the search of optimized transfection procedures.
Finally, beyond tracing the intact siRNAs after transfection, our method is suited to quantify them. Our first estimates show that a fraction of only ∼1% of the applied amount, corresponding to ∼106 molecules is left intact in the cell 48 h after a standard transfection protocol. Our imaging tool presented here may prove useful in determining the fate of this still huge reservoir of potentially active siRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
M.H. gratefully acknowledges the Heidelberg Rotary Club for funding, Heike Einberger for technical assistance, and the Nikon Imaging Center at the University of Heidelberg. We thank Andres Jäschke for generous support. Funding to pay the Open Access publication charges for this article was provided by the University of Heidelberg.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 3.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 4.Regelin AE, Fankhaenel S, Gurtesch L, Prinz C, von Kiedrowski G, Massing U. Biophysical and lipofection studies of DOTAP analogs. Biochim. Biophys. Acta. 2000;1464:151–164. doi: 10.1016/s0005-2736(00)00126-7. [DOI] [PubMed] [Google Scholar]
- 5.Dallas A, Vlassov AV. RNAi: a novel antisense technology and its therapeutic potential. Med. Sci. Monit. 2006;12:RA67–74. [PubMed] [Google Scholar]
- 6.Spagnou S, Miller AD, Keller M. Lipidic carriers of siRNA: differences in the formulation, cellular uptake, and delivery with plasmid DNA. Biochemistry. 2004;43:13348–13356. doi: 10.1021/bi048950a. [DOI] [PubMed] [Google Scholar]
- 7.Sioud M, Sorensen DR. Cationic liposome-mediated delivery of siRNAs in adult mice. Biochem. Biophys. Res. Commun. 2003;312:1220–1225. doi: 10.1016/j.bbrc.2003.11.057. [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 9.Holen T, Amarzguioui M, Babaie E, Prydz H. Similar behaviour of single-strand and double-strand siRNAs suggests they act through a common RNAi pathway. Nucleic Acids Res. 2003;31:2401–2407. doi: 10.1093/nar/gkg338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uhler SA, Cai D, Man Y, Figge C, Walter NG. RNA degradation in cell extracts: real-time monitoring by fluorescence resonance energy transfer. J. Am. Chem. Soc. 2003;125:14230–14231. doi: 10.1021/ja036854b. [DOI] [PubMed] [Google Scholar]
- 11.Chiu YL, Dinesh CU, Chu CY, Ali A, Brown KM, Cao H, Rana TM. Dissecting RNA-interference pathway with small molecules. Chem. Biol. 2005;12:643–648. doi: 10.1016/j.chembiol.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 12.Raemdonck K, Remaut K, Lucas B, Sanders NN, Demeester J, De Smedt SC. In situ analysis of single-stranded and duplex siRNA integrity in living cells. Biochemistry. 2006;45:10614–10623. doi: 10.1021/bi060351b. [DOI] [PubMed] [Google Scholar]
- 13.Harborth J, Elbashir SM, Vandenburgh K, Manninga H, Scaringe SA, Weber K, Tuschl T. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 14.Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 15.Campbell TN, Choy FY. Knockdown of chimeric glucocerebrosidase by green fluorescent protein-directed small interfering RNA. Genet. Mol. Res. 2004;3:282–287. [PubMed] [Google Scholar]
- 16.Wu P, Brand L. Resonance energy transfer: methods and applications. Anal. Biochem. 1994;218:1–13. doi: 10.1006/abio.1994.1134. [DOI] [PubMed] [Google Scholar]
- 17.Massing U. Manufacture of lipid-based nanoparticles using a dual asymmetric centrifuge. 2005 Patent application EP 05823526.8; PCT/EP2005/057157. [Google Scholar]
- 18.Roux F, Durieu-Trautmann O, Chaverot N, Claire M, Mailly P, Bourre JM, Strosberg AD, Couraud PO. Regulation of gamma-glutamyl transpeptidase and alkaline phosphatase activities in immortalized rat brain microvessel endothelial cells. J. Cell Physiol. 1994;159:101–113. doi: 10.1002/jcp.1041590114. [DOI] [PubMed] [Google Scholar]
- 19.Elbashir SM, Harborth J, Weber K, Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- 20.Ohrt T, Merkle D, Birkenfeld K, Echeverri CJ, Schwille P. In situ fluorescence analysis demonstrates active siRNA exclusion from the nucleus by Exportin 5. Nucleic Acids Res. 2006;34:1369–1380. doi: 10.1093/nar/gkl001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiu YL, Rana TM. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol. Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- 22.Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen TM, Hansen C, Martin F, Redemann C, Yau-Young A. Liposomes containing synthetic lipid derivatives of poly(ethylene glycol) show prolonged circulation half-lives in vivo. Biochim. Biophys. Acta. 1991;1066:29–36. doi: 10.1016/0005-2736(91)90246-5. [DOI] [PubMed] [Google Scholar]
- 24.Larson JM. The Nikon C1si combines high spectral resolution, high sensitivity, and high acquisition speed. Cytometry A. 2006;69:825–834. doi: 10.1002/cyto.a.20305. [DOI] [PubMed] [Google Scholar]
- 25.Grunweller A, Gillen C, Erdmann VA, Kurreck J. Cellular uptake and localization of a Cy3-labeled siRNA specific for the serine/threonine kinase Pim-1. Oligonucleotides. 2003;13:345–352. doi: 10.1089/154545703322617023. [DOI] [PubMed] [Google Scholar]
- 26.Leonetti JP, Mechti N, Degols G, Gagnor C, Lebleu B. Intracellular distribution of microinjected antisense oligonucleotides. Proc. Natl Acad. Sci. USA. 1991;88:2702–2706. doi: 10.1073/pnas.88.7.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Besch R, Giovannangeli C, Degitz K. Triplex-forming oligonucleotides—sequence-specific DNA ligands as tools for gene inhibition and for modulation of DNA-associated functions. Curr. Drug Targets. 2004;5:691–703. doi: 10.2174/1389450043345100. [DOI] [PubMed] [Google Scholar]
- 28.Zhang C, Tang N, Liu X, Liang W, Xu W, Torchilin VP. siRNA-containing liposomes modified with polyarginine effectively silence the targeted gene. J. Control Release. 2006;112:229–239. doi: 10.1016/j.jconrel.2006.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.