Abstract
Fifteen years elapsed between the discovery of the double helix (1953) and the first DNA sequencing (1968). Modern DNA sequencing began in 1977, with development of the chemical method of Maxam and Gilbert and the dideoxy method of Sanger, Nicklen and Coulson, and with the first complete DNA sequence (phage ϕX174), which demonstrated that sequence could give profound insights into genetic organization. Incremental improvements allowed sequencing of molecules >200 kb (human cytomegalovirus) leading to an avalanche of data that demanded computational analysis and spawned the field of bioinformatics. The US Human Genome Project spurred sequencing activity. By 1992 the first ‘sequencing factory’ was established, and others soon followed. The first complete cellular genome sequences, from bacteria, appeared in 1995 and other eubacterial, archaebacterial and eukaryotic genomes were soon sequenced. Competition between the public Human Genome Project and Celera Genomics produced working drafts of the human genome sequence, published in 2001, but refinement and analysis of the human genome sequence will continue for the foreseeable future. New ‘massively parallel’ sequencing methods are greatly increasing sequencing capacity, but further innovations are needed to achieve the ‘thousand dollar genome’ that many feel is prerequisite to personalized genomic medicine. These advances will also allow new approaches to a variety of problems in biology, evolution and the environment.
INTRODUCTION
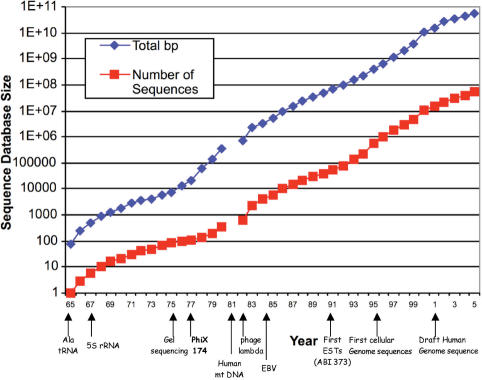
The year 2007 marks the 30th anniversary of the introduction of modern DNA sequencing methods (1,2) and the first complete sequence of a DNA molecule (3,4). These 30 years have seen astounding growth in DNA sequencing capacity and speed. From the first small phage genome, 5386 bases in length, DNA sequencing has advanced to sequence the human genome of ∼3 billion bases (5,6). The total amount of sequence in the databases passed the 100 Gb mark in August 2005 (Figure 1). It is remarkable that such progress has been made using methods that are refinements of the basic ‘dideoxy’ method introduced by Sanger in 1977.
Figure 1.
Growth of the nucleotide sequence database. The number of published nucleotide sequences, and the total number of base pairs of sequence are plotted versus the date of deposition or publication. Data since 1981 are re-plotted from http://www.ncbi.nlm.nih.gov/Genbank/genbankstats.html and data for sequences published before 1981 are from Dayhoff, Nucleic Acid Sequence Database, Vol. 1 (38). The dates of landmark sequences and technological advances are indicated.
THE SEQUENCE CONCEPT IN BIOLOGY
But the story of sequencing began more than a quarter of a century earlier, when Sanger's studies of insulin first demonstrated the importance of sequence in biological macromolecules. That work showed, for the first time, that proteins are composed of linear polypeptides formed by joining amino acid residues in a defined, but apparently arbitrary order (7,8). Summarizing these studies in his 1959 Nobel address (9), Sanger observed: ‘Examination of the sequences of the two chains reveals neither evidence of periodicity of any kind, nor does there seem to be any basic principle which determines the arrangement of the residues. They seem to be put together in an order that is random, but nevertheless unique and most significant, since on it must depend the important physiological action of the hormone.’
Consequently, when the double-helical structure of DNA was proposed soon thereafter (10), it was natural to consider its implications for base sequences in DNA. In their original paper, Watson and Crick pointed out that their structure placed no constraints on the sequence of a DNA molecule. They also observed that it suggested a mechanism for faithful replication of any base sequence. Thus the stage was set for an attack on the coding problem—how is the amino acid sequence of a protein determined by the base sequence of the DNA gene that encodes it? It is interesting that this inferred importance of DNA sequence led to the solution of the coding problem before the experimental determination of any actual DNA sequences, but that is another story.
EARLY DAYS OF DNA SEQUENCING
Fifteen years elapsed between the discovery of the DNA double helix and the first experimental determination of a DNA sequence. This delay was caused by several factors that made the problem intimidating:
The chemical properties of different DNA molecules were so similar that it appeared difficult to separate them.
The chain length of naturally occurring DNA molecules was much greater than for proteins and made complete sequencing seems unapproachable.
The 20 amino acid residues found in proteins have widely varying properties that had proven useful in the separation of peptides. The existence of only four bases in DNA therefore seemed to make sequencing a more difficult problem for DNA than for protein.
No base-specific DNAases were known. Protein sequencing had depended upon proteases that cleave adjacent to certain amino acids.
Some RNA molecules did not share all of these drawbacks with DNA. In particular, transfer RNA molecules were small and individual types could be purified. RNAases with base specificity were known so methods analogous to those used in protein sequencing could be developed. Consequently Escherichia coli alanine tRNA was the first nucleic acid molecule to be sequenced by Holley and coworkers in 1965 (11). Unlike amino acid sequences, which were not interpretable until 3D protein structures were determined by X-ray crystallography, models for the structure of tRNAs could be deduced by assuming base pairing analogous to that found in the DNA double helix.
The first DNA molecule purified to homogeneity was the genome of bacteriophage ϕX174, reported by Sinsheimer in 1959 (12). Equilibrium buoyant density centrifugation of the ϕX virion yielded pure preparations from which the DNA could be easily isolated by phenol extraction. ϕX DNA turned out to be a single-stranded circular molecule that was estimated to be ∼5000 nt in length.
Purification of other viral DNAs was also accomplished soon, in particular phage lambda DNA (13), a linear molecule with cohesive ends that was the subject of the first successful DNA sequencing. Wu and Kaiser (14,15) measured incorporation of radiolabeled nucleotides by E. coli DNA polymerase in reactions that extended the 3′ termini to fill in the complementary cohesive end sequences. They reported a partial sequence in 1968, but completion of the 12 base sequence was not accomplished until 1971 (16). An inroad into DNA sequencing had been made, but the method only applied to short stretches near the ends of lambda and related phage genomes. Wu also provided a means to generalize his approach by introducing the use of oligonucleotide primers in DNA sequencing reactions (17).
The next decisive event, without which DNA sequencing could not have proceeded along the path that it took, was the discovery of type II restriction enzymes by Hamilton Smith and coworkers (18,19). These enzymes recognized and cleaved DNA at specific short nucleotide sequences, typically 4–6 bp in length. It was soon found that enzymes recognizing many different sequences could be identified by screening bacterial strains (20). The restriction enzymes therefore provided a general method for cutting a large DNA molecule into a number of smaller pieces that could be separated by size using gel electrophoresis. These pieces had specific ends that could function as starting points for the sequencing methods that developed over the next few years.
Early work on DNA sequencing that followed sequencing of the phage lambda cohesive ends used methods similar to those used for RNA sequencing. These methods employed base-specific chemical reactions such as depurination, or enzymes with some degree of specificity, for example E. coli endonuclease IV, to produce fragments that were typically in the range of 10–20 bp in length. The size of the fragments that could be analyzed was limited by the separation methods that were used. These included 2D chromatography, and also electrophoresis in one dimension followed by chromatography in a second. When such a small fragment was incompletely digested from one end by exonuclease then its sequence could be determined by (i) analysis of the terminal nucleotide of each partial digestion product or (ii) by base-specific shifts in the position of the spot following 2D separation of the products.
Although these early methods were not powerful enough to determine complete gene sequences, several important regulatory signals were sequenced using them. Examples are operator sequences from the E. coli lac operon (21) and from phage lambda (22).
GEL-BASED DNA SEQUENCING METHODS
In 1975, Sanger introduced his ‘plus and minus’ method for DNA sequencing (23,24). This was a critical transition technique leading to the modern generation of methods that have completely dominated sequencing over the past 30 years. The key to this advance was the use of polyacrylamide gels to separate the products of primed synthesis by DNA polymerase in order of increasing chain length. The method analyzed the products of DNA polymerase reactions that extended a primer annealed to a single-stranded DNA template, as Wu and Kaiser had done in sequencing the lambda cohesive ends. DNA synthesis to extend the primer was carried out in two sequential DNA polymerase reactions. The first was carried out under conditions where synthesis was slow and asynchronous, resulting in a population of all possible products extending 1,2,3, … up to a few hundred bases. A 32P labeled nucleotide was incorporated at this step. This product was then divided into eight aliquots and used to prime a second round of DNA polymerase reactions. In these reactions, synthesis was terminated in a sequence-specific manner by supplying only one of the four nucleoside triphosphates (‘plus’ reactions), or else three of the four (‘minus’ reactions). The eight reactions were electrophoresed in adjacent lanes of a 12% acrylamide, 8 M urea denaturing gel. Following electrophoresis the gel was placed in contact with X-ray film for a suitable time, typically overnight. When the film was developed, molecules differing by a single nucleotide in length could be resolved as discrete bands on the resulting autoradiograph. This allowed a sequence of ∼50 bases to be deduced in a single experiment. The main problem with the method is the difficulty in determining the length of homopolymer runs. Bands corresponding to the beginning and end of such runs are produced, but no bands are produced for positions internal to runs, so run lengths must be estimated from band spacing in the gel. This becomes unreliable for longer runs. Nevertheless, the plus and minus method was used to produce a complete sequence of the ϕX DNA genome (3). In this work, restriction fragments of double-stranded ϕX replicative form DNA were melted and annealed to single-stranded virion DNA to serve as specific primers for the method.
Maxam and Gilbert (2) developed a DNA sequencing method that was similar to the Sanger and Coulson method in using polyacrylamide gels to resolve bands that terminated at each base throughout the target sequence, but very different in the way that products ending in a specific base were generated. Their method started with a double-stranded DNA restriction fragment radiolabeled at one end with 32P. The fragment was then cleaved by base-specific chemical reactions. One reaction cleaves at both purines (the ‘A + G’ reaction), one preferentially at A (‘A > G’), one at pyrimidines (‘C + T’) and one at cytosines only (‘C’). Unlike the plus and minus method, the chemical method produced bands for every sequence position, including those within homopolymer runs. This advantage led to early widespread adoption of the chemical method following its publication in February 1977 (2)
The problems with the plus and minus method were solved when Sanger developed ‘the dideoxy method’ and published it in December 1977 (1). The underlying concept was to use chain-terminating nucleotide analogs rather than subsets of the four natural dNTPs to cause base-specific termination of primed DNA synthesis. In the original implementation both arabinoside triphosphates and 2′,3′-dideoxy nucleoside triphosphates were tried. These analogs are incorporated in a sequence-specific manner by E. coli Pol I, but the enzyme is unable to further extend the growing DNA strand (in the case of the ddNTPs simply because of the lack of a 3′ hydroxyl group). Synthesis was carried out in the presence of all four dNTPs, one of which was α-32P labeled. Four reactions were set up, each doped with a chain-terminating analog of one of the dNTPs, at an appropriate concentration. If the concentration of ddATP, for example, was adjusted so that it was incorporated in place of the normal dATP ∼1% of the time then a series of chain-terminated products were produced, each ending with an A. Some molecules in the product ended at each of the A residues in the sequence. When such a product was electrophoresed on a denaturing 12% acrylamide gel, a series of bands representing the positions of all A's in the sequence were displayed. Unlike the plus and minus method, bands were produced for each A within runs of consecutive A residues. When the four dideoxy reactions were run in adjacent lanes it was possible to read sequences of ∼100 nt in most cases.
The dideoxy sequencing method as originally described required a single-stranded DNA template. The general applicability of the method was therefore greatly enhanced when Messing and collaborators developed methods for cloning into the single-stranded phage M13 (25–27).
SEQUENCES, SEQUENCES, SEQUENCES
The complete sequence of ϕX determined by the plus and minus method was published in 1977 and then revised slightly in 1978 after resequencing by the dideoxy method. It was a revelation because, to the surprise of many, it turned out to be extremely interesting. Unlike amino acid sequences of proteins, the DNA sequence of the ϕX genome could be interpreted to tell a fascinating story based upon interpretation of the sequence in terms of the genetic code. Analysis of mutations in genes identified by traditional phage genetics, combined with amino acid sequence information for protein components of the ϕX virion, allowed phage genes to be located on the DNA sequence. For the first time translation of a DNA sequence in all possible reading frames identified long open reading frames that could be assigned to genes identified by traditional genetic methods. And, most surprising, it was clear that significant portions of the genome were translated in more than one reading frame to produce two different protein products. These pairs of ‘overlapping genes’ had not been detected by recombination mapping of the ϕX genome but their existence was indisputable when the sequence was analyzed in light of genetic and protein sequence information (28,29).
The sequence of the simian virus SV40 followed quickly in 1978 (30). Sequencing had begun by determining sequences of RNA copies of parts of the genome, but was rapidly completed after publication of the Maxam–Gilbert method.
With the introduction of the gel-based sequencing methods, the rate of DNA sequencing accelerated (Figure 1). Progress in the methodology was incremental and was driven by the selection of sequencing targets of increasing complexity. In the Sanger group, the 16.5 kb human mitochondrial genome (31) was followed by the 48.5 kb complete phage lambda genome (32). Following Sanger's retirement his protégé Bart Barrell led sequencing of the 172 kb Epstein–Barr virus (33) and then the 237 kb human cytomegalovirus genome (34). For 15 years following ϕX, the sequencing group at the MRC Laboratory of Molecular Biology in Cambridge, UK, had continuously held the record for the longest DNA sequence published.
During this period the useful read length of dideoxy sequencing increased from about 100 up to about 400. This improvement was mainly the result of: (i) the use of very thin sequencing gels and (ii) 35S labeling of the DNA, which gives sharper bands than 32P due to the lower energy of the emitted β particles. Sequencing capacity was also increased by the use of gels with narrow lanes, typically 48 lanes on a 20 by 45 cm gel. Sequencing reactions could be done manually in 96-well plates with handheld repetitive pipetting devices. During this period a single person could run 8 gels on a single day, each with 12 sequence ladders, and obtain some 30 kb of primary sequence data. But it was difficult to do this more than about twice a week.
THE BIRTH OF BIOINFORMATICS
Beginning with ϕX, the management and analysis of sequencing data became a major undertaking. The original ϕX data was in the notebooks of nine different workers each concerned with particular portions of the molecule. Michael Smith, on sabbatical in the Sanger group, had a brother-in-law named Duncan McCallum who was a business computer programmer in Cambridge. He wrote the first programs to help with the compilation and analysis of DNA sequence data (in COBOL) (35). We each transcribed our manually deduced sequences onto paper forms, which then were entered in blocks of 60 on punched cards. The programs then (i) compiled and numbered the complete sequence, (ii) allowed the editing of a previously compiled sequence, (iii) searched the sequence for specific short sequences or families of sequences, for example restriction sites and (iv) translated the sequence in all reading frames. Though invaluable, the programs did not produce output suitable for publication, so the original figure displaying the ϕX sequence with its genes and their translation products annotated (3) was hand typed by Peggy Dowding. Roger Staden helped with computer analysis of the original ϕX sequence and quickly wrote the first suite of programs meant to be interactive and ‘designed specifically for use by people with little or no computer experience’ (36). These programs developed into the Staden Package, still in use today (37).
With the proliferation of DNA sequence data, came the need for a DNA sequence database. Margaret Dayhoff was the early pioneer in this area. She had previously established a protein sequence database and published the first collection of nucleotide sequence information in 1981 (38). Shortly thereafter, in 1982 GenBank was created by the NIH to provide a ‘timely, centralized, accessible repository for genetic sequences’ (39).
As the sequence databases grew, methods to compare and align sequences soon became a rate-limiting step in the analysis of sequence data. The development of rapid search programs such as FASTA (40) and BLAST (41) made it practical to identify genes in a new sequence by comparison to all sequences already in the databases.
These are just a few of the early developments in the computer analysis of DNA sequence information. Bioinformatics has developed into a full-blown discipline far beyond the scope of a review such as this. Bioinformatics is central to the interpretation of sequence data and to the generation of testable hypotheses arising from such data.
THE JOHN HENRY SYNDROME
Toward the end of the manual-sequencing era, the first generation of ‘automated sequencers’ appeared. These machines did not initially automate much of the sequencing process. Gels were still prepared manually, and loaded manually. Only the readout of the sequence data was really automated and in the beginning the base-calling algorithms were quite unsatisfactory. Many steeped in the traditions of manual sequencing were doubtful that automation could compete with dedicated graduate student sequencers. The next few years would show how wrong we were.
AUTOMATED SEQUENCING FACTORIES
In 1986 the laboratory of Leroy Hood at Caltech, in collaboration with Applied Biosystems (ABI), published the first report of automation of DNA sequencing (42). This initial report showed that sequencing data could be collected directly to a computer without autoradiography of the sequencing gel. Although the method could, in principle, have been applied to the chemical sequencing method, the dideoxy method was chosen. A sequencing primer was fluorescently end labeled using four different dyes. A differently labeled primer was used in each of the four dideoxy sequencing reactions. The reactions were combined and electrophoresed in a single polyacrylamide tube gel. DNA was observed by fluorescence as it passed a detector near the bottom of the gel and the four dyes were distinguished by their colors. Fluorescence data was continuously recorded and stored by a computer over the course of a typical 13 h run. The sequence could be deduced from the order in which the four different dyes passed the detector. Work at ABI developed programs to automatically interpret the data to produce an actual sequence (43).
The ABI 370A DNA sequencer appeared very shortly, and was first used to determine the sequence of a gene by Craig Venter and colleagues at NIH (44). At NIH, Venter set up a sequencing facility with six automated sequencers and two ABI Catalyst robots. In 1992 Venter established The Institute for Genomic Research (TIGR) to expand his sequencing operation and established a facility with 30 ABI 373A automated sequencers and 17 ABI Catalyst 800 robots (45). The organizational differences between this facility and the manual sequencing projects that preceded it were as important as the use of automation. This was a real factory with teams dedicated to different steps in the sequencing process such as template preparation, gel pouring and sequencer operation. Data analysis was integrated into the process so that problems in earlier steps could be detected and corrected as soon as possible. By contrast, in the CMV project, each participating investigator sequenced a region of the genome single-handedly.
An early demonstration of the power of automated sequencing was the development of the expressed sequence tag (EST) approach to gene discovery. In this approach cDNA copies of messenger RNA were cloned at random and subjected to automated sequencing. In the first report from Venter and colleagues in 1991, 337 new human genes were reported, 48 homologous to genes from other organisms (46). This initial study was expanded to an analysis of 83 million nucleotides of cDNA sequence that identified fragments of more than 87 000 human cDNA sequences, more than 77 000 of which were previously unknown (47). This approach was adopted by many genome projects. Today the EST database contains over 43 million ESTs from over 1300 different organisms.
Another early application of the automated sequencer was the worm genome sequencing project (see below) which was underway by 1992 with the beginning elements of a factory atmosphere as well (48).
In 1993 the Sanger Centre, later renamed the Wellcome Trust Sanger Institute, was established by the Wellcome Trust and the Medical Research Council. This is arguably the most important of the sequencing centers established to participate in the worldwide public effort to sequence the human genome. Located in Hinxton, near Cambridge, ‘the Sanger’ is an outstanding example of a modern sequencing center. The facility has produced ∼3.4 × 109 bases of finished sequence by the 30th anniversary of the dideoxy sequencing method.
CELLULAR GENOMES
Until 1995 the only completely sequenced DNA molecules were viral and organelle genomes. That year Craig Venter's group at TIGR, and their collaborators, reported complete genome sequences of two bacterial species, Haemophilus influenzae (49) and Mycoplasma genitalium (50). The H. influenzae sequence gave the first glimpse of the complete instruction set for a living organism. The M. genitalium sequence showed us an approximation to the minimal set of genes required for cellular life.
The methods used to obtain these sequences were as important for subsequent events as the biological insights they revealed. Sequencing of H. influenzae introduced the whole genome shotgun (WGS) method for sequencing cellular genomes. In this method, genomic DNA is fragmented randomly and cloned to produce a random library in E. coli. Clones are sequenced at random and the results are assembled to produce the complete genome sequence by a computer program that compares all of the sequence reads and aligns matching sequences. Sanger and colleagues used this general strategy to sequence the lambda phage genome (48.5 kb), published in 1982. However, no larger genome was shotgun sequenced until H. influenzae (1.83 Mb). In the interim shotgun sequencing was used extensively, but only to sequence mapped subclones of larger sequences. This was the strategy used for the 230 kb human CMV sequence, the largest sequence finished sequencing before the H. influenzae genome.
Venter and colleagues introduced critical improvements that made it feasible, for the first time, to shotgun sequence complete cellular genomes. Perhaps most important was adoption of the ‘paired ends’ strategy (51,52). The automated sequencing procedure used in the H. influenzae project used melted double-stranded DNA as template, whereas the HCMV project had used single-stranded M13 vectors. With double-stranded templates it was convenient to sequence each clone from both ends. Because the randomly sheared DNA was carefully sized before cloning, the distance between the reads from the ends of each clone could be estimated. The assembly program used this information to construct ‘scaffolds’ from the blocks of completely overlapped sequence (‘contigs’). When two contigs contained sequences from opposite ends of a single clone, then the two contigs could be linked, although a ‘sequence gap’ was said to exist between them. Sequence gaps remaining at the end of the shotgun phase of sequencing could be closed by sequencing from a primer for a site internal to a clone bridging the gap. Gaps between scaffolds are ‘physical gaps’ that contain sequences, which do not occur within any of the sequenced clones. Other measures, such as PCR between the ends of scaffolds using a genomic DNA template, were used to close physical gaps.
Another critical factor in the application of shotgun sequencing to cellular genomes was the TIGR assembler. Previous assembly programs were not designed to handle thousands of sequence reads involved in even the smallest cellular genome projects. However, the TIGR assembler that had been designed to assemble vast amounts of EST data was adequate for the job.
Once these initial sequences were reported the floodgates were open and a steady stream of completed genome sequences has been appearing ever since. It is only possible here to touch on a few of the most significant. Because of the large communities of scientists actively engaged in studies that would benefit from the availability of a genome sequence I have chosen to mention the bacteria E. coli and Bacillus subtilis, the yeast Saccharomyces cerevisiae, the nematode Caenorhabditis elegans, the fruit fly Drosophila melanogaster and humans.
Because of its position as the pre-eminent model organism of molecular biology, sequencing of the genome of E. coli (4.6 Mb) was proposed by Blattner as early as 1983 (53). Sequencing proceeded as sequencing technology improved, starting with manual methods and finishing in 1997 with automated sequencers (54). Early sequences covering ∼1.9 Mb, were deposited starting in 1992, and were obtained from an overlapping set of cosmid clones. The final ∼2.5 Mb was obtained by shotgun sequencing of ∼250 Kb I-Sce I fragments. This E. coli genome sequence, along with several other strains sequenced subsequently has yielded a wealth of information about bacterial evolution and pathogenicity (55,56).
Meanwhile, another model for large-scale genome sequencing projects had emerged; the international consortium. The first genome sequence to be completed by this approach was the yeast S. cerevisiae (12.0 Mb) (57), in late 1996. This was the also the first eukaryotic organism to be sequenced. The project involved about 600 scientists in Europe, North America and Japan. The participants included both academic laboratories and large sequencing centers.
The next success of the consortium approach was the genome of the bacterium B. subtilis (4.2 Mb) (58), in 1997. The project began in 1990 with the participation of five European laboratories. The project finally became a consortium of 25 laboratories in six European countries coordinated at the Institut Pasteur by Frank Kunst (coordinator) and Antoine Danchin. A consortium of seven Japanese laboratories, coordinated by Naotake Ogasawara and Hiroshi Yoshikawa at the Nara Institute of Science and Technology, Japan, also participated, as well as one Korean and two US laboratories.
The first animal genome sequenced was that of ‘the worm’ C. elegans (97 Mb) (59), in 1998. The authorship of this work was simply ‘The C. elegans Sequencing Consortium’, which was a collaboration between the Washington University Genome Sequencing Center in the United States and the Sanger Centre in UK.
In 1996, ABI introduced the first commercial DNA sequencer that used capillary electrophoresis rather than a slab gel (the ABI Prism 310), and in 1998 the ABI Prism 3700 with 96 capillaries was announced. For the first time DNA sequencing was truly automated. The considerable labor of pouring slab gels was replaced with automated reloading of the capillaries with polymer matrix. Samples for electrophoresis were automatically loaded from 96-well plates rather than manually loaded as the previous generation of sequencers had been. Celera Genomics was found by Applera Corporation (the parent company of ABI) and Craig Venter in May 1998 to exploit these new machines by applying Venter's methods for WGS sequencing to the human genome, in direct competition with the publicly funded Human Genome Project. Celera acquired 300 of the machines, each capable of producing 1.6 × 105 bases of sequence data per day, for a total theoretical capacity of ∼5 × 107 bases of raw sequence data per day.
Celera chose the D. melanogaster genome to test the applicability of the WGS approach to a complex eukaryotic genome (60). This involved a scientific collaboration between the scientists at Celera and those of the Berkeley and European Drosophila Genome Projects. These projects finished 29 Mb of the 120 Mb of euchromatic portion of the genome. (About one-third of the 180 Mb Drosophila genome is centromeric heterochromatin.) Using the WGS approach, data was collected over a 4-month period that provided more than 12× coverage of the euchromatic portion of the genome. The results validated the data produced by the ABI 3700s, the applicability of the WGS approach to eukaryotic genomes, and the assembly methods developed at Celera (61). This was a nearly ideal test case because the WGS data could be analyzed separately and then portions of it could be compared with finished sequence already produced by the Drosophila Genome Projects. At the same time the sequence information provided a valuable resource for Drosophila genetics. More than 40 scientists at an ‘Annotation Jamboree’ did initial annotation of the sequence. These scientists, mainly drawn from the Drosophila research community, met at Celera for a 2-week period to identify genes, predict functions, and begin a global synthesis of the genome sequence information.
SEQUENCING OF THE HUMAN GENOME
Eventual sequencing of the human genome became an imaginable goal at the outset of the sequencing era 30 years ago. Formal discussions of the idea began in 1985 when Robert Sinsheimer organized a meeting on human genome sequencing at the University of California, Santa Cruz (62). That same year Charles DeLisi and David A. Smith commissioned the first Santa Fe conference, funded by the DOE, to study the feasibility of a Human Genome Initiative. Discussions continued and in 1988 reports recommending a concerted genome research program were issued by committees of the congressional Office of Technology Assessment and the National Research Council. In 1990 the DOE and NIH presented a joint 5-year US Human Genome Project plan to Congress. It was estimated that the project would take 15 years and cost ∼3 billion US$.
The US Human Genome Project established goals of mapping, and in some cases sequencing, several model organisms as well as humans. These included E. coli, yeast (S. cerevisiae), the worm (C. elegans), drosophila (D. melanogaster) and mouse (laboratory strains of Mus domesticus). So, several of the sequences discussed above received funding from the Human Genome Project. The publicly funded effort became an international collaboration between a number of sequencing centers in the United States, Europe and Japan. Each center focused sequencing efforts on particular regions of the genome, necessitating detailed mapping as a first step. In 1994, a detailed genetic map of the human genome was published including 5840 mapped loci with a mean spacing of 0.7 cM (1 centiMorgan = ∼106 bp) (63). In 1998 the public project, now in a race with Celera, also adopted the new ABI Prism 3700 capillary sequencers. In 1999 the Human Genome Project celebrated passing the billion base-pair mark, and the first complete sequence of a human chromosome was reported [chromosome 22 (64)].
Meanwhile at Celera, human genome sequencing was underway using the WGS strategy. Human genome sequencing began in September 1999 and continued until June 2000, when data collection was completed and an initial assembly was achieved. The Celera data provided approximately 5-fold coverage of the genome. An additional 3-fold coverage of unordered and unoriented BAC sequences from the public effort was included in the assembly. The power of the WGS strategy was amply demonstrated.
On 25 June 2000 at the White House, President Clinton with Prime Minister Tony Blair publicly announced draft versions of the human genome sequence from both the publicly funded project and from Celera. In February 2001 the Celera (5) and the public (6) draft human genome sequences were published the same week in Science and Nature. The race was officially a tie, but it was clear to all that the entry of Celera had speeded the process by several years. Both projects ended up needing the other to make the progress that was made. The Celera assembly benefited from data produced in the public project and the public project quickly adopted some of Celera's methods, in particular the paired-end strategy. Celera's basic methods have now been adopted by all publicly funded genome projects.
Sequencing of the human genome captured public attention in a way that is extremely rare for a scientific topic. Several books for the general public have centered around the ‘race’ for the human genome sequence (65,66). Leaders of the public and the private projects have even published books describing events from their own personal perspectives (67,68).
BEYOND THE HUMAN GENOME
The major purpose of this review is to commemorate the beginning of the DNA sequencing era with a discussion of the early history of sequencing. But a brief summary of current directions and future horizons is useful to allow an appreciation of the long-term implications of those historical events. It has been said that a full understanding of the human genome sequence may take the better part of the 21st century. Perhaps ironically, the major tool currently available for achieving this understanding is DNA sequencing, on a scale much larger than in the past. Also, we have entered an era where very large-scale DNA sequencing provides a feasible approach to a multitude of problems concerning biology, disease and the environment. This review will conclude with an overview of these developments.
NEXT GENERATION SEQUENCING TECHNOLOGY
In the last few years methods have emerged, which for the first time challenge the supremacy of the dideoxy method. The common feature of these methods is that they are ‘massively parallel’, meaning that the number of sequence reads from a single experiment is vastly greater than the 96 obtained with modern capillary electrophoresis-based Sanger sequencers. At present this very high throughput is achieved with substantial sacrifices in length and accuracy of the individual reads when compared to Sanger sequencing. Nonetheless, assemblies of such data can be highly accurate because of the high degree of sequence coverage obtainable. The methods are designed for projects that employ the WGS approach. They are most readily applied to resequencing, in which sequence data is aligned with a reference genome sequence in order to look for differences from that reference. A few examples of specific instruments that employ massively parallel strategies are discussed below. Other technologies are under development (69–75), and all of these methods will undoubtedly continue to improve.
The first of the massively parallel methods to become commercially available was developed by 454 Life Sciences (76) and is based on the ‘pyrosequencing’ technique (77,78). This system allows shotgun sequencing of whole genomes without cloning in E. coli or any host cell. First DNA is randomly sheared and ligated to linker sequences that permit individual molecules captured on the surface of a bead to be amplified while isolated within an emulsion droplet (79). A very large collection of such beads is arrayed in the 1.6 million wells of a fiber-optic slide. As with the Sanger method, sequencing is carried out using primed synthesis by DNA polymerase. The array is presented with each of the four dNTPs, sequentially, and the amount of incorporation is monitored by luminometric detection of the pyrophosphate released (hence the name ‘pyrosequencing’). A CCD imager coupled to the fiber-optic array collects the data. In sequencing across a homopolymer run, the run length is estimated from the amount of pyrophosphate released, which is proportional to the number of residues incorporated. Errors that result from misjudging the length of homopolymer runs result in single-base insertions and deletions (indels). These constitute the major source of errors in 454 data. The ‘plus and minus’ method of Sanger and Coulson had this same difficulty. Nonetheless, the second generation 454 Genome Sequencer FLX is reportedly able to produce 100 Mb of sequence with 99.5% accuracy for individual reads averaging read over 250 bases in length.
Another promising technique is the Solexa technology (80,81). A key difference between this method and the 454 is that it uses chain-terminating nucleotides. The fluorescent label on the terminating base can be removed to leave an unblocked 3′ terminus, making chain termination a reversible process. The method reads each base in a homopolymer run in a separate step and therefore does not produce as many indels within such runs as the 454. Because the reversible dye terminator nucleotides are not incorporated efficiently, the read length of the Solexa method is less than for 454. Also more base-substitution errors are observed due to the use of modified polymerase and dye terminator nucleotides. The method sequences clusters of DNA molecules amplified from individual fragments attached randomly on the surface of a flow cell. Because of the very high densities of clusters that can be analyzed, the machine can reportedly produce 1 billion bases (1 Gb) of 30–40 base sequence reads in a single run.
Applied Biosystems is also developing a massively parallel sequencer, its Supported Oligonucleotide Ligation and Detection system (SOLiD). The technology is based on a hybridization-ligation chemistry (73). The sample preparation aspect of this technology including library preparation, clonal amplification of the target DNA by emulsion PCR on beads is very similar to the 454 processes in principle. However, the size of the beads used for emPCR (1 µm versus 26 µm) and the array format (random versus ordered) are different. These differences afford the SOLiD technology the potential of generating a significantly higher density sequencing array (potentially over a few hundred fold higher), as well as more flexibility in terms of sample input format. The sequence interrogation is done through the repeated cycles of hybridization of a mixture of sequencing primers and fluorescently labeled probes, followed by ligation of the sequencing primers and the probes, then the detection of the fluorescent signals on the probes which encode the bases that are being interrogated. Although it has a short read length of about 25–35, it can generate ∼2–3 Gb of sequence per run.
During the development of these new technologies it may be useful to combine sequence data obtained using different techniques. Combination of 454 data with a smaller amount of capillary sequencing data has been shown to be cost-effective in certain situations (82). Similarly, the combination of Solexa data with a smaller amount of 454 data could be effective. The 454 data would provide longer read lengths and a method for obtaining paired end sequences from long DNA fragments to greatly aid assembly (83).
Massively parallel methods of the type described above are likely to dominate high-throughput sequencing applications for the next few years. However, a variety of other approaches are being investigated that may eventually develop into practical methods. Several of these approaches, for example ‘nanopore sequencing’, have been reviewed (71,84).
GENOMIC MEDICINE
All disease has a genetic basis, whether in genes inherited by the affected individual, environmentally induced genetic changes that produce a cancer, or the genes of a pathogen and their interaction with those of the infected individual. This emerging field of genomic medicine cannot be considered in detail here, but is the subject of several reviews (85–87). Sequencing of the human genome as well as all major pathogens is beginning to have a major impact on the diagnosis, treatment and prevention of diseases. Genome sequences have provided potential targets for drug therapy (88,89) as well as vaccine candidates (90,91).
An era of personalized medicine, informed by information concerning the patient's genotype, has been widely predicted. The Holy Grail in this field has become ‘the $1000 genome’ sequence. To stimulate work in this area the X Prize Foundation has established the $10 million Archon X Prize for Genomics (92) (http://genomics.xprize.org/).
METAGENOMICS
Only a very small fraction of the microbes found in nature have been grown in pure culture. Consequently we lack a comprehensive view of the genetic diversity to be found on Earth. An approach to this problem has emerged called ‘metagenomics’ or ‘environmental genomics’ (93,94). DNA is isolated directly from environmental samples and sequenced, without attempting to culture the organisms from which it comes.
Early studies using manual DNA sequencing methods focused on 16S rRNA genes as a gauge of phylogenetic diversity (95). High-throughput sequencing methods make shotgun sequencing of the whole metagenome from environmental samples informative. A study of the DNA isolated from an acid-mine biofilm yielded nearly complete genome sequences for two organisms and partial sequences for three others from just 76.2 Mb of data (96). Studies of the more complex environment of the oceans have shed new light on the diversity of life on Earth. A total of more than 1.6 Gb of sequence from Sargasso Sea samples yielded 1.2 million previously unknown gene sequences, only about one-third of which were recognizable homologs of previously known genes (97). Before analysis of the Sargasso Sea data the NCBI non-redundant amino acid (nraa) dataset contained some 1.5 million peptides, about 630 000 of which were classified as bacterial. In an expanded study, ocean samples were collected during an expedition from the North Atlantic through the Panama Canal and into the South Pacific. Shotgun sequencing of these samples produced a dataset of 7.7 million sequence reads yielding 6.3 Gb of sequence (98–101). More than 6 million proteins are predicted from this global ocean sampling (GOS) data. These include members of almost all previously known prokaryotic protein families. When these GOS data were clustered with all previously known protein sequences, about 4000 out of a total of 17 000, medium and large clusters contained only GOS sequences (101). The metagenomic approach is being applied to study microbial populations in many environments, for example the human gut (102).
A difficulty with current metagenomic sequence datasets from complex microbial communities is that the vast majority of the data cannot be assembled. This is mainly due to the fact that the cost of acquiring enough sequencing data for the full assembly of all or most of the organisms present in any given sample is still prohibitively high using the state-of-the-art Sanger-based sequencing technology. The development of strategies to apply the new massively parallel sequencing methods may solve this problem if the sequence read length and the accuracy of these technologies can be dramatically improved. In the meantime, another approach is the sequencing of individual cells isolated from the environment. Substantial progress toward this goal has been achieved (103,104). It is clear that the metagenomic approach is becoming a major new tool for understanding our environment. It will also provide a vast array of new genes for application in the emerging field of synthetic biology.
WHAT NEXT?
The amount of nucleotide sequence in the databases has increased logarithmically by nine orders of magnitude over the 40-year period from 1965 to 2005 (Figure 1). This amounts to an average doubling time of about 16 months. Upon closer inspection, the logarithmic rate of increase is not quite constant over time. Inflection in the curve (Figure 1) appear to correspond to technical innovations, such as the development of the gel-based sequencing techniques 30 years ago, and the introduction of automation. This suggests that we are about to see another inflection in the curve resulting from the next generation of massively parallel sequencers.
In 1977 it was possible to imagine a benchtop machine that could sequence the E. coli genome in a few days (although it could only be seen as a black box). The sequencing factory would have been almost inconceivable because it was at odds with the whole culture of biological research. But the sequencing factory did come to pass, and with it came vast amounts of data ideally suited for computational analysis. Taken at face value, the 16-month doubling time for the sequence database is substantially faster than the 24-month doubling in computer power predicted by Moore's law (105) (http://www.intel.com/technology/mooreslaw/). Even if these trends continue it seems unlikely that this difference could begin to limit sequence data collection in the near term. However, some common problems in the analysis of sequence data are already rate-limiting steps in research. A prime example is all-against-all sequence comparison, which requires a computation time that increases with the square of the amount of sequence data. It appears possible that methods for collecting sequence data could soon outstrip our capacity to adequately analyze that data. At present a sizable bioinformatics core is an essential part of a sequencing center. The currently popular vision that an investigator with a single benchtop machine could replace a large sequencing center can only be realized with increases in the productivity of computers and bioinformaticians even more dramatic than that expected for sequencers. It appears that for our individual $1000 genome sequences to be truly useful, fundamental advances in computation and bioinformatics will be essential. The obvious importance of computational analysis of sequence data has led to a greater overall appreciation of the role of theory in biology. A relationship between theory and experiment, not unlike that found in 20th century physics, seems to be taking shape.
ACKNOWLEDGEMENTS
I want to thank Fred Sanger for the privilege of spending a sabbatical year in his laboratory (1975–1976), and for the chance to participate in sequencing the ϕX174 genome. I am grateful for the sabbatical year I spent in Bart Barrell's laboratory (1987–1988) working on the human CMV sequence. I also thank Craig Venter and Ham Smith for our ongoing long-term collaboration, which began in 1995 with the sequencing of the M. genitalium genome. These experiences have allowed me to view much of the history of sequencing at close quarters, and may obviously have affected my perspective on a number of, sometimes controversial, issues. I thank Ham Smith for helpful discussions during the writing of this article, concerning matters ranging from the early days of Maxam and Gilbert sequencing to the human genome and beyond. Yu-Hui Rogers has generously shared insights into the new massively parallel sequencing methods. Funding to pay the Open Access publication charges for this article was waived by the editors for this Summary and Survey article.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maxam AM, Gilbert W. A new method for sequencing DNA. Proc. Natl Acad. Sci. USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- 4.Sanger F, Coulson AR, Friedmann T, Air GM, Barrell BG, Brown NL, Fiddes JC, Hutchison CA, III, Slocombe PM, et al. The nucleotide sequence of bacteriophage phiX174. J. Mol. Biol. 1978;125:225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- 5.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 6.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Sanger F. The terminal peptides of insulin. Biochem. J. 1949;45:563–574. doi: 10.1042/bj0450563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanger F, Tuppy H. The amino-acid sequence in the phenylalanyl chain of insulin. 2. The investigation of peptides from enzymic hydrolysates. Biochem. J. 1951;49:481–490. doi: 10.1042/bj0490481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanger F. Chemistry of insulin: determination of the structure of insulin opens the way to greater understanding of life processes. Science. 1959;129:1340–1344. doi: 10.1126/science.129.3359.1340. [DOI] [PubMed] [Google Scholar]
- 10.Watson JD, Crick FH. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 11.Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, Merrill SH, Penswick JR, Zamir A. Structure of a ribonucleic acid. Science. 1965;147:1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- 12.Sinsheimer RL. A single-stranded DNA from bacteriophage phi X174. J. Mol. Biol. 1959;1:43. [PubMed] [Google Scholar]
- 13.Kaiser AD, Hogness DS. The transformation of Escherichia coli with deoxyribonucleic acid isolated from bacteriophage lambda-dg. J. Mol. Biol. 1960;2:392–415. doi: 10.1016/s0022-2836(60)80050-2. [DOI] [PubMed] [Google Scholar]
- 14.Kaiser AD, Wu R. Structure and function of DNA cohesive ends. Cold Spring Harb. Symp. Quant. Biol. 1968;33:729–734. doi: 10.1101/sqb.1968.033.01.083. [DOI] [PubMed] [Google Scholar]
- 15.Wu R, Kaiser AD. Structure and base sequence in the cohesive ends of bacteriophage lambda DNA. J. Mol. Biol. 1968;35:523–537. doi: 10.1016/s0022-2836(68)80012-9. [DOI] [PubMed] [Google Scholar]
- 16.Wu R, Taylor E. Nucleotide sequence analysis of DNA. II. Complete nucleotide sequence of the cohesive ends of bacteriophage lambda DNA. J. Mol. Biol. 1971;57:491–511. doi: 10.1016/0022-2836(71)90105-7. [DOI] [PubMed] [Google Scholar]
- 17.Padmanabhan R, Wu R. Nucleotide sequence analysis of DNA. IX. Use of oligonucleotides of defined sequence as primers in DNA sequence analysis. Biochem. Biophys. Res. Commun. 1972;48:1295–1302. doi: 10.1016/0006-291x(72)90852-2. [DOI] [PubMed] [Google Scholar]
- 18.Smith HO, Wilcox KW. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J. Mol. Biol. 1970;51:379–391. doi: 10.1016/0022-2836(70)90149-x. [DOI] [PubMed] [Google Scholar]
- 19.Kelly TJ, Jr, Smith HO. A restriction enzyme from Hemophilus influenzae. II. J. Mol. Biol. 1970;51:393–409. doi: 10.1016/0022-2836(70)90150-6. [DOI] [PubMed] [Google Scholar]
- 20.Middleton JH, Edgell MH, Hutchison CA., III Specific fragments of phi X174 deoxyribonucleic acid produced by a restriction enzyme from Haemophilus aegyptius, endonuclease Z. J. Virol. 1972;10:42–50. doi: 10.1128/jvi.10.1.42-50.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert W, Maxam A. The nucleotide sequence of the lac operator. Proc. Natl Acad. Sci. USA. 1973;70:3581–3584. doi: 10.1073/pnas.70.12.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maniatis T, Ptashne M, Barrell BG, Donelson J. Sequence of a repressor-binding site in the DNA of bacteriophage lamda. Nature. 1974;250:394–397. doi: 10.1038/250394a0. [DOI] [PubMed] [Google Scholar]
- 23.Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 24.Sanger F. The Croonian Lecture, 1975: Nucleotide sequences in DNA. Proc. R. Soc. Lond. B. Biol. Sci. 1975;191:317–333. doi: 10.1098/rspb.1975.0131. [DOI] [PubMed] [Google Scholar]
- 25.Messing J, Gronenborn B, Muller-Hill B, Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc. Natl Acad. Sci. USA. 1977;74:3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gronenborn B, Messing J. Methylation of single-stranded DNA in vitro introduces new restriction endonuclease cleavage sites. Nature. 1978;272:375–377. doi: 10.1038/272375a0. [DOI] [PubMed] [Google Scholar]
- 27.Heidecker G, Messing J, Gronenborn B. A versatile primer for DNA sequencing in the M13mp2 cloning system. Gene. 1980;10:69–73. doi: 10.1016/0378-1119(80)90145-6. [DOI] [PubMed] [Google Scholar]
- 28.Barrell BG, Air GM, Hutchison CA., III Overlapping genes in bacteriophage phiX174. Nature. 1976;264:34–41. doi: 10.1038/264034a0. [DOI] [PubMed] [Google Scholar]
- 29.Smith M, Brown NL, Air GM, Barrell BG, Coulson AR, Hutchison CA, III, Sanger F. DNA sequence at the C termini of the overlapping genes A and B in bacteriophage phi X174. Nature. 1977;265:702–705. doi: 10.1038/265702a0. [DOI] [PubMed] [Google Scholar]
- 30.Fiers W, Contreras R, Haegemann G, Rogiers R, Van de Voorde A, Van Heuverswyn H, Van Herreweghe J, Volckaert G, Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978;273:113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- 31.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 32.Sanger F, Coulson AR, Hong GF, Hill DF, Petersen GB. Nucleotide sequence of bacteriophage lambda DNA. J. Mol. Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 33.Baer R, Bankier AT, Biggin MD, Deininger PL, Farrell PJ, Gibson TJ, Hatfull G, Hudson GS, Satchwell SC, et al. DNA sequence and expression of the B95-8 Epstein–Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 34.Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, Chee MS, Hutchison CA, III, Kouzarides T, Martignetti JA, et al. The DNA sequence of the human cytomegalovirus genome. DNA Seq. 1991;2:1–12. doi: 10.3109/10425179109008433. [DOI] [PubMed] [Google Scholar]
- 35.McCallum D, Smith M. Computer processing of DNA sequence data. J. Mol. Biol. 1977;116:29–30. doi: 10.1016/0022-2836(77)90116-4. [DOI] [PubMed] [Google Scholar]
- 36.Staden R. Sequence data handling by computer. Nucleic Acids Res. 1977;4:4037–4051. doi: 10.1093/nar/4.11.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol. Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 38.Dayhoff MO, Schwartz RM, Chen HR, Hunt LT, Barker WC, Orcutt BC. Nucleic acid sequence database. 1981;1 doi: 10.1089/dna.1.1981.1.51. National Biomedical Research Foundation, Georgetown University Medical Center, Washington D.C. 20001. [DOI] [PubMed] [Google Scholar]
- 39.Bilofsky HS, Burks C, Fickett JW, Goad WB, Lewitter FI, Rindone WP, Swindell CD, Tung CS. The GenBank genetic sequence databank. Nucleic Acids Res. 1986;14:1–4. doi: 10.1093/nar/14.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 42.Smith LM, Sanders JZ, Kaiser RJ, Hughes P, Dodd C, Connell CR, Heiner C, Kent SB, Hood LE. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 43.Connell C, Fung S, Heiner C, Bridgham J, Chakerian V, Heron E, Jones B, Menchen S, Mprdan W, et al. Automated DNA-Sequence analysis. Biotechniques. 1987;5:342–348. [Google Scholar]
- 44.Gocayne J, Robinson DA, FitzGerald MG, Chung FZ, Kerlavage AR, Lentes KU, Lai J, Wang CD, Fraser CM, et al. Primary structure of rat cardiac beta-adrenergic and muscarinic cholinergic receptors obtained by automated DNA sequence analysis: further evidence for a multigene family. Proc. Natl Acad. Sci. USA. 1987;84:8296–8300. doi: 10.1073/pnas.84.23.8296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams MD, Kerlavage AR, Kelley JM, Gocayne JD, Fields C, Fraser CM, Venter JC. A model for high-throughput automated DNA sequencing and analysis core facilities. Nature. 1994;368:474–475. doi: 10.1038/368474a0. [DOI] [PubMed] [Google Scholar]
- 46.Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 47.Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, et al. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 1995;377:3–174. [PubMed] [Google Scholar]
- 48.Sulston J, Du Z, Thomas K, Wilson R, Hillier L, Staden R, Halloran N, Green P, Thierry-Mieg J, et al. The C. elegans genome sequencing project: a beginning. Nature. 1992;356:37–41. doi: 10.1038/356037a0. [DOI] [PubMed] [Google Scholar]
- 49.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 50.Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 51.Edwards A, Voss H, Rice P, Civitello A, Stegemann J, Schwager C, Zimmermann J, Erfle H, Caskey CT, et al. Automated DNA sequencing of the human HPRT locus. Genomics. 1990;6:593–608. doi: 10.1016/0888-7543(90)90493-e. [DOI] [PubMed] [Google Scholar]
- 52.Roach JC, Boysen C, Wang K, Hood L. Pairwise end sequencing: a unified approach to genomic mapping and sequencing. Genomics. 1995;26:345–353. doi: 10.1016/0888-7543(95)80219-c. [DOI] [PubMed] [Google Scholar]
- 53.Blattner FR. Biological Frontiers. Science. 1983;222:2. doi: 10.1126/science.222.4625.719. [DOI] [PubMed] [Google Scholar]
- 54.Blattner FR, Plunkett G., III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 55.Perna NT, Plunkett G., III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 56.Welch RA, Burland V, Plunkett G., III, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, et al. Life with 6000 genes. Science. 1996;274:546, 563–547. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 58.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 59.The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 60.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 61.Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 62.Sinsheimer RL. To reveal the genomes. Am. J. Hum. Genet. 2006;79:194–196. doi: 10.1086/505887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murray JC, Buetow KH, Weber JL, Ludwigsen S, Scherpbier-Heddema T, Manion F, Quillen J, Sheffield VC, Sunden S, et al. A comprehensive human linkage map with centimorgan density. Cooperative Human Linkage Center (CHLC) Science. 1994;265:2049–2054. doi: 10.1126/science.8091227. [DOI] [PubMed] [Google Scholar]
- 64.Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- 65.Wickelgren I. 1st. New York: Times Books/Henry Holt and Co.; 2002. The Gene Masters: How a New Breed of Scientific Entrepreneurs Raced for the Biggest Prize in Biology. [Google Scholar]
- 66.Shreeve J. The Genome War: How Craig Venter Tried to Capture the Code of Life and Save the World. New York: Alfred A. Knopf; 2004. [PubMed] [Google Scholar]
- 67.Collins FS. The Language of God: A Scientist Presents Evidence for Belief. New York: Free Press; 2006. [Google Scholar]
- 68.Venter JC. A Life Decoded. New York: Viking Press; 2007. in the press. [Google Scholar]
- 69.Metzker ML. Emerging technologies in DNA sequencing. Genome Res. 2005;15:1767–1776. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- 70.Rogers YH, Venter JC. Genomics: massively parallel sequencing. Nature. 2005;437:326–327. doi: 10.1038/437326a. [DOI] [PubMed] [Google Scholar]
- 71.Chan EY. Advances in sequencing technology. Mutat. Res. 2005;573:13–40. doi: 10.1016/j.mrfmmm.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 72.Braslavsky I, Hebert B, Kartalov E, Quake SR. Sequence information can be obtained from single DNA molecules. Proc. Natl Acad. Sci. USA. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 74.Blazej RG, Kumaresan P, Mathies RA. Microfabricated bioprocessor for integrated nanoliter-scale Sanger DNA sequencing. Proc. Natl Acad. Sci. USA. 2006;103:7240–7245. doi: 10.1073/pnas.0602476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rich A. The rise of single-molecule DNA biochemistry. Proc. Natl Acad. Sci. USA. 1998;95:13999–14000. doi: 10.1073/pnas.95.24.13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nyren P, Pettersson B, Uhlen M. Solid phase DNA minisequencing by an enzymatic luminometric inorganic pyrophosphate detection assay. Anal. Biochem. 1993;208:171–175. doi: 10.1006/abio.1993.1024. [DOI] [PubMed] [Google Scholar]
- 78.Ronaghi M, Karamohamed S, Pettersson B, Uhlen M, Nyren P. Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- 79.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl Acad. Sci. USA. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 81.Bennett ST, Barnes C, Cox A, Davies L, Brown C. Toward the 1,000 dollars human genome. Pharmacogenomics. 2005;6:373–382. doi: 10.1517/14622416.6.4.373. [DOI] [PubMed] [Google Scholar]
- 82.Goldberg SM, Johnson J, Busam D, Feldblyum T, Ferriera S, Friedman R, Halpern A, Khouri H, Kravitz SA, et al. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proc. Natl Acad. Sci. USA. 2006;103:11240–11245. doi: 10.1073/pnas.0604351103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ng P, Tan JJ, Ooi HS, Lee YL, Chiu KP, Fullwood MJ, Srinivasan KG, Perbost C, Du L, et al. Multiplex sequencing of paired-end ditags (MS-PET): a strategy for the ultra-high-throughput analysis of transcriptomes and genomes. Nucleic Acids Res. 2006;34:e84. doi: 10.1093/nar/gkl444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deamer DW, Akeson M. Nanopores and nucleic acids: prospects for ultrarapid sequencing. Trends Biotechnol. 2000;18:147–151. doi: 10.1016/s0167-7799(00)01426-8. [DOI] [PubMed] [Google Scholar]
- 85.Guttmacher AE, Collins FS. Genomic medicine–a primer. N. Engl. J. Med. 2002;347:1512–1520. doi: 10.1056/NEJMra012240. [DOI] [PubMed] [Google Scholar]
- 86.Peltonen L, McKusick VA. Genomics and medicine. Dissecting human disease in the postgenomic era. Science. 2001;291:1224–1229. doi: 10.1126/science.291.5507.1224. [DOI] [PubMed] [Google Scholar]
- 87.Khoury MJ, McCabe LL, McCabe ER. Population screening in the age of genomic medicine. N. Engl. J. Med. 2003;348:50–58. doi: 10.1056/NEJMra013182. [DOI] [PubMed] [Google Scholar]
- 88.Drews J. Drug discovery: a historical perspective. Science. 2000;287:1960–1964. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- 89.Hopkins AL, Groom CR. The druggable genome. Nat. Rev. Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 90.Wizemann TM, Heinrichs JH, Adamou JE, Erwin AL, Kunsch C, Choi GH, Barash SC, Rosen CA, Masure HR, et al. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun. 2001;69:1593–1598. doi: 10.1128/IAI.69.3.1593-1598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Adu-Bobie J, Capecchi B, Serruto D, Rappuoli R, Pizza M. Two years into reverse vaccinology. Vaccine. 2003;21:605–610. doi: 10.1016/s0264-410x(02)00566-2. [DOI] [PubMed] [Google Scholar]
- 92.Pennisi E. Genomics. On your mark. Get set. Sequence! Science. 2006;314:232. doi: 10.1126/science.314.5797.232. [DOI] [PubMed] [Google Scholar]
- 93.Tringe SG, Rubin EM. Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet. 2005;6:805–814. doi: 10.1038/nrg1709. [DOI] [PubMed] [Google Scholar]
- 94.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 95.Schmidt TM, DeLong EF, Pace NR. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J. Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 97.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 98.Yutin N, Suzuki MT, Teeling H, Weber M, Venter JC, Rusch DB, Beja O. Assessing diversity and biogeography of aerobic anoxygenic phototrophic bacteria in surface waters of the Atlantic and Pacific Oceans using the Global Ocean Sampling expedition metagenomes. Environ. Microbiol. 2007;9:1464–1475. doi: 10.1111/j.1462-2920.2007.01265.x. [DOI] [PubMed] [Google Scholar]
- 99.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, et al. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS. Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. Structural and Functional Diversity of the Microbial Kinome. PLoS. Biol. 2007;5:e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, et al. The Sorcerer II Global Ocean Sampling Expedition: expanding the Universe of protein families. PLoS. Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang K, Martiny AC, Reppas NB, Barry KW, Malek J, Chisholm SW, Church GM. Sequencing genomes from single cells by polymerase cloning. Nat. Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- 104.Hutchison CA, III, Venter JC. Single-cell genomics. Nat. Biotechnol. 2006;24:657–658. doi: 10.1038/nbt0606-657. [DOI] [PubMed] [Google Scholar]
- 105.Schaller RR. Moore's law: past, present and future. IEEE Spectrum. 1997;34:8. [Google Scholar]