Abstract
Undifferentiated transcription factor 1 (UTF1) was identified first in mouse embryonic stem cells and is also expressed in human embryonic and adult stem cells. UTF1 transcription ceases at the onset of differentiation, which clearly distinguishes it from less sensitive pluripotency markers, such as Oct4 or Nanog. We present here two transgenic hESC lines, named ZUN. Each line harbors one copy of the UTF1 promoter/enhancer driving a resistance gene and yielded highly homogeneous cultures under selection pressure, with a larger proportion of Oct4 and Sox2 positive cells. While ZUN cultures, like parental HUES8 cultures, retained the capacity to differentiate into tissues of all three germ layers using a SICD mouse teratoma model, they surprisingly exhibited an increased refractoriness to various differentiation cues in vitro. Together with its small size of only 2.4 kb for the entire cassette, these features render our selection system a powerful novel tool for many stem cell applications and human somatic cell reprogramming strategies.
INTRODUCTION
The transcriptional co-activator ‘undifferentiated transcription factor 1′ (UTF1) is expressed in both mouse and human embryonic stem cells (mESCs and hESCs, respectively) (1–3). UTF1 interacts with ATF-2 and TBP-containing complexes, and seems to be involved in cell proliferation control (4,5). UTF1 is regarded as one of the most important pluripotency marker genes defining the hESC niche (6,7). Importantly, its transcriptional regulation is very sensitive to differentiation cues and switched off faster and with a larger magnitude than either Oct4 or Sox2 during embryoid body formation from both mouse and human ESCs (8–10). A similar response is seen when mouse embryonic carcinoma (EC) and mESCs were triggered to differentiate by retinoic acid (RA) (11,12). In a comparison of hESCs cultured in medium containing either fetal calf serum or defined serum replacement, UTF1 was found to be downregulated to a larger extent in serum medium compared to Oct4 or Sox2, suggesting strongly that it is a very reliable marker for early differentiation (13). Furthermore, RNA interference (RNAi) of Oct4 transcripts in mESCs has been shown to cause downregulation of UTF1 transcripts (14), and more than 90% of all cells in undifferentiated mESC populations stain positively for both these markers (15). These two experiments further allude to the prevalence of UTF1 and Oct4 co-expression in undifferentiated ESCs.
Human embryonic stem cell cultures are always heterogeneous and usually contain a subset of less pluripotent and, hence, more differentiated cells (16–18). This is also true for mouse and medaka ESCs (19,20). It was shown that the presence of minor populations of differentiated cells compromises the pluripotency of entire stem cell cultures (21). These features represent a severe problem for many hESC applications which can be addressed either via fluorescence-activated cell sorting (FACS) in order to isolate pluripotent hESCs or by antibiotic selection as a means to eliminate differentiating cells.
FACS strategies generally utilize transgenes, such as EGFP, under the control of genetic elements of a pluripotency marker. For example, mouse Rex1-EGFP and Oct4-EGFP combinations have been used in hESCs to screen for undifferentiated cells via flow cytometry (17,22,23). A recent improvement in semi-automated FACS and clonal recovery utilized a mouse Oct4-EGFP construct to sort for pluripotent hESCs (24). However, this procedure has to be repeated routinely in order to ensure more homogeneous hESC cultures over longer periods of time.
Antibiotic selection has the advantage of being less intrusive to hESCs. Expression of the neomycin resistance (Neo) gene and G418 antibiotic selection, along with EGFP expression, have been shown to be compatible with hESC pluripotency (23,25). In the mouse, the Oct4-Neo combination has been used to isolate ES and EC cell lines from embryos. (19,21,26). Thus far, only one report utilized the twin markers of EGFP and Neo driven by an Oct4 promoter for the maintenance of pluripotency in hESCs (22). However, the ability to select against partially differentiated cells as well as the derivation of more homogeneous pluripotent hESC cultures was not established in these studies.
Here we demonstrate that UTF1 is an exceptionally sensitive pluripotency marker. We found that M1, a genetic element comprising a conserved octamer sequence important for Nanog expression (27), is also present and functional in the UTF1 3′ enhancer, but not in the UTR of Oct4 or Sox2. As a representative example, we present two stable ZUN hESC lines in which Neo is under the control of UTF1 promoter/enhancer. Application of G418 selection results in ablation of differentiated hESCs and homogeneously pluripotent ZUN cultures, containing a significantly larger number of Oct4 and Sox2 positive cells compared to parental cultures. While ZUN cultures retain the capacity to differentiate into progenitors of all three germ layers in vivo, they surprisingly exhibited an increased refractoriness to differentiation cues under various culture conditions.
MATERIALS AND METHODS
Plasmids
The full-length human UTF1 gene was amplified from HeLa genomic DNA using primers UTF1-F (CCGGAATTCAGCGCCAGGACCGACCCCTTA) and UTF1-R (TGCTCTAGAGGGTTCAGCACTTCTCCTGCCTC) via PCR. The product was cleaved with EcoRI and XbaI, and ligated into phagemid pTZ-18R. The UTF1 coding sequence was subsequently replaced with that of EGFP or Neo in the following way. KpnI and NcoI restriction sites were introduced between the end of the UTF1 promoter and before the ATG codon by amplifying the promoter region of pTZ-UTF1 via PCR with primers UTF1-F and UTF1-MR (CGGGGTACCCCGGGGCTGGGGCGCGG). The resulting fragment was cleaved with EcoRI and NcoI, and ligated to a cleaved pCMV-EGFP which replaces the CMV promoter. The plasmid was then digested with EcoRI and BsrGI to produce an (EcoRI) UTF1promoter-EGFP (BsrGI) sticky-ended fragment. pTZ-UTF1 was used as a PCR template to amplify a segment of the UTF1 3′ UTR region and inserting a BsrGI RE site near the end of UTF1 transcription unit using primers UTF1-MF (AGCTGTACAAGTGAGTCCCGGCTGCGGC) and UTF1-BR (AGAATACTCAAGCTATGCATCCAAC). This produced another BsrGI-BamHI sticky-ended 1 kb fragment. pTZ-UTF1 was then digested with EcoRI and BamHI, and this 3.2 kb fragment was ligated to the above two 1 kb fragments, yielding pTZ-UTF1-EGFP. The Neo gene was amplified with flanking KpnI and BsrGI RE sites via PCR with pPGK-Neo as template and Neo-F (CTGGGTACCCCATGATTGAACAAGATGG) and Neo-R (ACTGTGTACATCAGAAGAACTCGTCAAGAA) primers.
The pTZ-UTF1-EGFP backbone was digested with KpnI and BsrGI to remove EGFP, and ligated with the cleaved Neo fragment to yield pTZ-UTF1-Neo. The promoter-less version of pTZ-UTF1-EGFP was generated using EcoRI and KpnI to remove the UTF1 promoter. The ends were blunted using Klenow Fragment (NEB) followed by self-ligation. The enhancer-less version of pTZ-UTF1-EGFP was generated using PstI to remove the UTF1 enhancer followed by self-ligation.
pTZ-UTF1-EGFP was used as template to introduce the mutant M1 sequence (GTAGTGGT; original sequence GTCTGGGT) (27). Flanking arms 200 bp from M1 were amplified using two primer pairs, UTF1-M1F (GACCCAGGAGTAGCATG) and UTF1-M1mR (CTAGGCCCACCACTACCAGAGCCAC) (near the 5′ XcmI RE site), and UTF1-M1mF (GTGGCTCTGGTAGTGGTGGGCCTAG) and UTF1-M1R (CATTGTTATGCTAGCGG) (near the 3′ NheI RE site). These were combined via assembly PCR using UTF1-M1F and UTF1-M1R as primers. The resulting 370 bp fragment was digested with XcmI and NheI, and ligated to a similarly cleaved pTZ-UTF1-EGFP in order to replace the original M1 sequence.
Plasmids were amplified in Escherichia coli strain DH5α. We used standard PCR conditions with Pfu polymerase (Promega). Restriction enzymes were from New England Biolabs.
Cell culture, transfection and differentiation
F3 human foreskin fibroblasts (28), A549 and HeLa carcinoma cell lines were cultured according to standard procedures. hESC line hES2 (karyotype: 46XX) was obtained from ES Cell International Pte Ltd. HUES8 (karyotype: 46XY) and HUES9 (karyotype: 46XX, inv9) was provided by the Harvard Stem Cell Institute. hESCs were cultured on G418-resistant MEFs (PMEF-N, Chemicon), as previously described (29,30).
F3, A549 and HeLa cells were transfected with 10 μg of reporter plasmid via electroporation as described previously (31). hESCs were transfected with 2 μg of reporter plasmid and Effectene (Qiagen) as described previously (29).
Differentiation medium was prepared by removing basic fibroblast growth factor and exchanging knockout serum replacement with fetal bovine serum (all three from Gibco). Spontaneous differentiation was conducted on matrigel (BD Biosciences) or gelatin surfaces without MEFs. Induced differentiation on MEFs was conducted either with 10 µM (RA) or 1% (DMSO) (Sigma) which was added to differentiation medium (32). Embryoid body formation was induced by plating hESCs onto low-cell binding plates (Nunc).
SCID mouse teratoma assay
To test the differentiation potential of hESC lines in vivo, serially passaged hESCs were manually harvested and injected as clumps with an approximate cell dose of 2−4 × 106 cells in a volume of 50 µl into the quadriceps of the right hind limb of a male SCID mouse (3 mice/hESC line). Mice were maintained under controlled conditions in accordance with the National Institutes of Health (NIH) and National Advisory Committee for Laboratory Animal Research (NACLAR) guidelines, and with approval of the Biopolis Institutional Animal Care and Use Committee (Biopolis IACUC approval 050008, National University of Singapore Institutional Review Board 05-020). Teratoma formation was monitored visually using a simple grading system that was confirmed by caliper measurements (grade 0—no detectible enlargement, grade 1—enlargement just detectible, grade 2—obvious enlargement, grade 3—enlargement impedes locomotion). When teratomas reached grade 3 (after 6–8 weeks), teratomas weighing 1–2 g were excised, fixed (Bouin's solution), paraffin-embedded, sectioned and histologically analyzed following staining with hematoxylin and eosin. Additionally, teratomas were assessed by a pathologist at the Department of Pathology, National University of Singapore.
Characterization of ZUN hESC lines
ZUN cell lines were generated by transfecting HUES8 cells using Effectene with pTZ-UTF1-Neo. Individual colonies were mechanically expanded in the presence of G418 (500 μg/ml). Cloning efficiency was 10−5, with four stable cell lines generated, of which only two contained the full-length UTF1-Neo cassette. These two lines were analyzed here and labeled as ‘ZUN’. Karyotyping via Giemsa staining was performed by Cytogenetics Lab, KK Women's and Children's Hospital, Singapore. 50 μl of BrdU/Colcemid (50 : 50) mixture was added to an 80% confluent 35 mm dish containing hESCs with 2 ml of medium the night before staining and analysis. Transgenic genomic inserts in both cell lines were sequenced from correctly sized genomic PCR products amplified using standard Taq PCR protocols and UTF1-F and UTF1-R primers. Southern blotting was performed as previously described (33), with 10 µg of isolated genomic DNA digested with EcoRI. Neo was detected using a 32P-labeled Neo probe. WST-1 cell proliferation assays were conducted using manufacturer protocols (Roche). Briefly, 3 × 104 cells were seeded onto each MEF-layered 96-well at day 0. WST-1 reagent was added to the wells for 30 min the next day, and 440 nm readings were recorded using a standard photoplate reader. This was repeated for a total of 4 days with different plates. Each cell line had five replicates per time-point. Alkaline phosphatase staining and TRA-1-60/TRA-1-81 surface marker immunostaining (visualized with FITC) were conducted using manufacturer protocols (Chemicon). Flow cytometry of surface markers, Oct4 and Sox2 was performed as previously described (29), with pre-incubation of primary and FITC-conjugated secondary antibodies (Santa Cruz).
Quantitative real-time PCR
Total RNA was recovered from cell pellets (Qiagen) and cDNA was synthesized via the SSII reverse transcriptase kit, primed with oligo dTs (Invitrogen). SYBR Green qRT-PCR (Qiagen) was conducted using the 7500 real-time PCR System and results analyzed using sequence detection software (SDS) version 1.2.3 (Applied Biosystems). All experiments had at least biological duplicates and assay triplicates, and results were analyzed via the ΔΔCt method using GAPDH as the housekeeping transcript. Primers used were as follows: GAPDH (CCCACTCCTCCACCTTTGAC and CTCCCCTCTTCAAGGGGTCT), Nanog (AGTCCCAAAGGCAAACAACCCACTTC and ATCTGCTGGAGGCTGAGGTATTTCTGTCTC) (6), Oct4 (CTTGCTGCAGAAGTGGGTGGAGGAA and CTGCAGTGTGGGTTTCGGGCA) (1), Sox2 (ATGCACCGCTACGACGTGA and CTTTT GCACCCCTCCCATTT) (1), UTF1 (AGCAGATCCGGAAGCTCATGGG and TCCTCGGGGATGCAGGTG), UTF1-Neo (GGGTACCCCATGATTGAACA and CAGGTCGGTCTTGACAAAAAG), Sox9 (GAAGCTCGCGGACCAGTA and CGTTCTTCACCACTTCCTC), Nestin (CGTTGGAACAGAGGTTGGAG and GAGCGATCTGGCTCTGTAGG), ID2 (CTGGACTCGCATCCCACTAT and CACACAGTGCTTTGCTGTCA), HAND1 (TGCCTGAGAAAGAGAACCAG and ATGGCAGGATGAACAAACAC) (9), IGF2 (TCCTC CCTGGACAATCAGAC and AGAAGCACCAGCAT CGACTT) (9), α1AT (AGACCCTTTGAAGTCAAG GACACCG and CCATTGCTGAAGACCTTAGTG ATGC) (34).
RESULTS
UTF1 is a sensitive pluripotency marker for hESCs
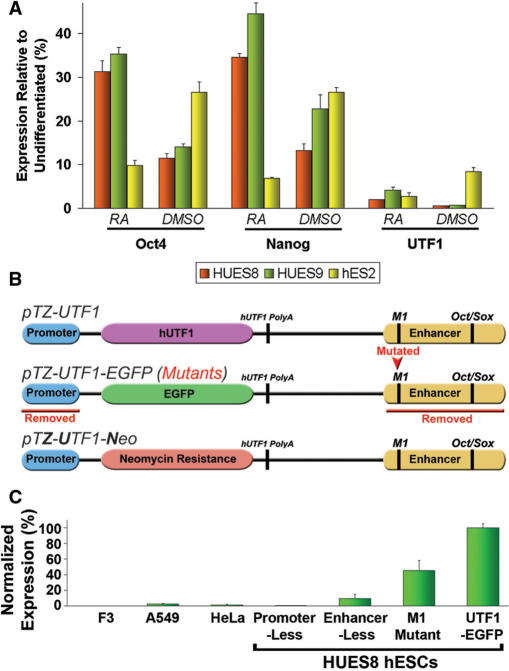
We evaluated first the sensitivity of UTF1 expression with respect to induced differentiation using quantitative real-time PCR (qRT-PCR). Various hESC lines were triggered to differentiate via retinoic acid (RA) or dimethyl sulfoxide (DMSO) treatment. The results show that this almost completely abolished UTF1 expression; a response that was not observed for Nanog and Oct4 (Figure 1A). We next generated the following three UTF1 transgenic constructs: pTZ-UTF1, pTZ-UTF1-EGFP and pTZ-UTF1-Neo (Figure 1B). In addition, three variants of pTZ-UTF1-EGFP were designed: one without the minimal 5′ promoter, one lacking the 3′ enhancer, and one containing an altered M1 sequence. The M1 octamer sequence was found to be conserved in the Nanog promoter of many species (27), and we identified it recently upstream of the Oct4/Sox2 binding site within the 3′ UTF1 enhancer.
Figure 1.
UTF1 expression in hESCs. (A) qRT-PCR Expression analysis of RA- (for 12 days) and DMSO-induced (for 7 days) differentiation in hESCs, normalized to that of respective undifferentiated hESC lines, with two biological replicates for each sample. (B) Schematic drawing of vectors. Full-length UTF1 was cloned into phagemid pTZ-18R yielding pTZ-UTF1. Its coding region was subsequently replaced with enhanced green florescent protein (EGFP) or neomycin (Neo), yielding pTZ-UTF1-EGFP and pTZ-UTF1-Neo, respectively. Red text indicates alterations in pTZ-UTF1-EGFP. (C) Flow cytometry of UTF1-driven EGFP expression in various cell lines following transient transfection of pTZ-UTF1-EGFP. Transfection efficiencies were normalized to those determined in parallel using pCMV-EGFP and are the average of three biological replicates. See Supplementary Figure S7 for representative dot plots.
The results demonstrate hESC-specific expression of the full-length pTZ-UTF1-EGFP after transfection into various human cell lines and hESC line HUES8 (Figure 1C). We next introduced the three pTZ-UTF1-EGFP versions into HUES8 cells and showed that the 5′ promoter and 3′ enhancer regions were essential for EGFP expression (Figure 1C). In addition, the altered M1 element reproducibly led to a significantly smaller number of EGFP expressing hESCs, which is similar in magnitude to its reported effect on Nanog transcription (27). Together, these data confirm that UTF1 transcription is a very sensitive pluripotency marker specific for hESCs and suggests that its regulation in hESCs involves M1-binding factors in addition to Oct4 and Sox2.
Characterization of transgenic pTZ-UTF1-Neo hESC lines
We established stable pTZ-UTF1-Neo cell lines from HUES8 cells via random genomic transgene insertion. Two single-copy lines with normal karyotype, dubbed ZUN1 and ZUN2, were used in subsequent experiments (Supplementary Figure S1). Careful characterization of both lines revealed that they have similar proliferation rates (Supplementary Figure S2), exhibit comparable alkaline phosphatase (AP) staining patterns (Supplementary Figure S3), and express Nanog at a similar level as parental HUES8 cells (Figure 2A). However, an elevated level of Oct4, Sox2 and UTF1 in ZUN cultures was reproducibly detected by qRT-PCR (Figure 2B). This correlation of expression supports the notion that the control of UTF1 involves factors Oct4 and Sox2, but most likely not Nanog (35).
Figure 2.
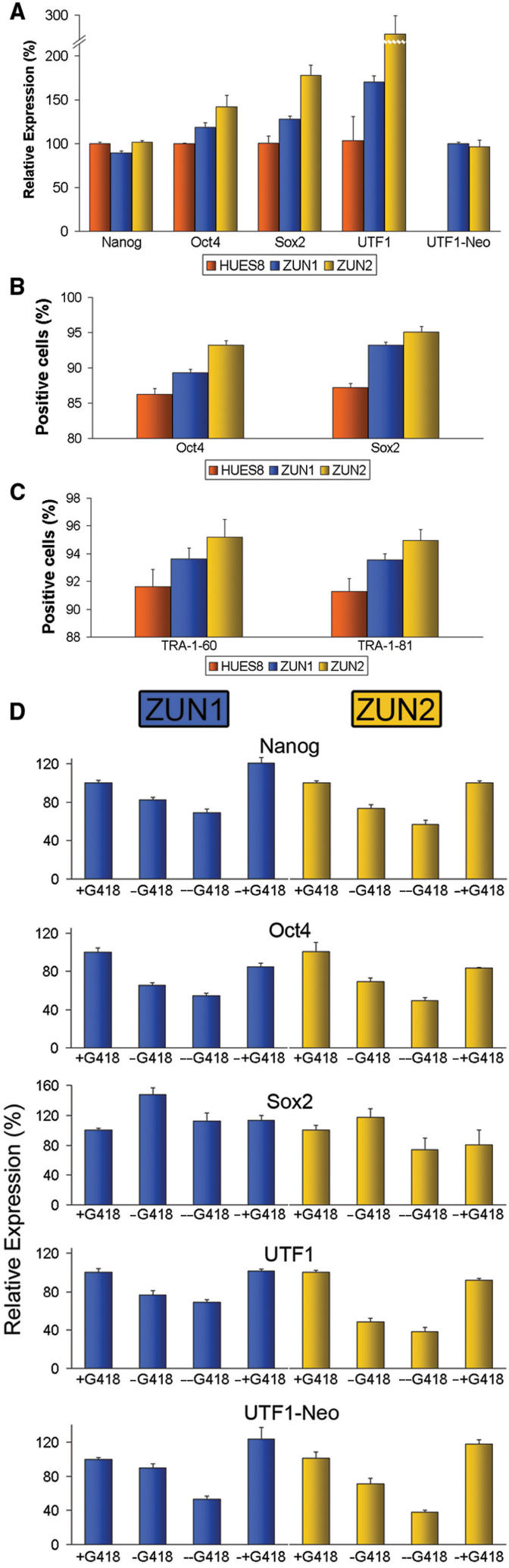
Characterization of ZUN hESC lines. (A) Expression analysis of ZUN hESCs. qRT-PCR comparison of pluripotency markers across the three hESC cultures normalized to HUES8 samples, performed with biological triplicates. (B) Oct4 and Sox2 expression analysis of ZUN hESCs. Percentage of hESCs expressing Oct4 and Sox2. ZUN cultures consist of significantly more Oct4- and Sox2-expressing cells compared to HUES8, based on the two-tailed paired Student's t-test (P-values < 0.05). (C) Surface marker analysis of ZUN hESCs. Percentage of positively stained hESCs for TRA-1-60 and TRA-1-81. Both ZUN lines consist of significantly more positively stained cells compared to parental HUES8, based on the two-tailed paired Student's t-test (P-values < 0.05). (D) qRT-PCR analysis of removal and restoration of G418 selection pressure on ZUN1 and ZUN2 cultures. ZUN lines were cultured in the presence of G418 for 90 days (+G418); G418 removed for 30 (−G418) or 60 (−/−G418) days; G418 removed for 30 days and re-introduced for 30 days (−/+G418). Expression levels were normalized to +G418 values and presented as an average of three experimental repeats.
Flow cytometry revealed that the observed higher level of global Oct4, Sox2 and UTF1 expression was due to the presence of a larger fraction of cells that express these markers. A statistically significant higher percentage of ZUN cells stained positive for Oct4 and Sox2 compared to HUES8 cultures (Figure 2B). We could also demonstrate that ZUN cultures contained more cells which stain positive for surface markers TRA-1-60 and TRA-1-81 (Figure 2C). These results indicate that HUES8 cultures contained more hESCs with a low expression level of pluripotency markers. In contrast, such cells have been ablated in ZUN cultures via G418 selection.
We next analyzed the effect of sequential removal and restoration of G418 selection on both ZUN lines and employed qRT-PCR of pluripotency markers Nanog, Oct4, Sox2 and UTF1 for the detection of the onset of differentiation (9). We also included the transgene UTF1-Neo in this analysis because its regulation should be linked to that of the endogenous UTF1. The results show that spontaneous differentiation can be detected in cultures during 60 days of G418 withdrawal. This is indicated by the progressive reduction in signals obtained from all markers, with the exception of Sox2 (Figure 2D). However, upon re-application of antibiotic selection, original expression levels could be restored in these cultures. Using a transgenic hESC line in which Neo is expressed from the elongation factor 1 alpha (EF-1α) promoter as a control, we confirmed that G418 withdrawal has no effect on the expression of this set of markers which clearly links this effect to the presence of the UTF1-Neo transgene in hESCs (Supplementary Figure S4). Together, these data show that ZUN cultures are more homogeneously pluripotent than the parental cultures and that G418 addition and subsequent ablation of differentiated ZUN cells is required to sustain homogeneous cultures over longer periods of time.
ZUN cultures are refractory to various differentiation cues
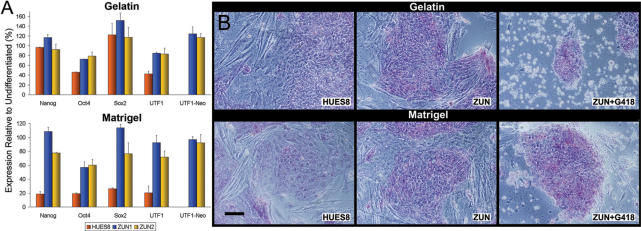
We demonstrated above that ZUN cultures are more homogeneously pluripotent than the parental cultures. We were interested, therefore, in investigating the differentiation behavior of ZUN cells. First, we induced differentiation by plating them on matrigel or gelatin without supporting mouse embryonic fibroblasts (MEFs) and in the absence of G418. In the matrigel protocol, HUES8 cells quickly initiated differentiation as indicated by the 5-fold downregulation of our panel of markers (Figure 3A). However, both ZUN lines maintained significantly higher overall expression levels even after 12 days of culture. Similarly, the gelatin protocol induced a 2-fold downregulation of Oct4 and UTF1 in HUES8, but not in ZUN populations. We observed further that under these conditions, ZUN cultures have a larger number of colonies consisting of cells which display a more compact morphology that stain stronger for alkaline phosphatase (AP), and a reduced occurrence of differentiating cells at colony edges. Upon subsequent addition of G418 for 2 days, only pluripotent cells that stained for AP survived in ZUN cultures, demonstrating again that differentiating ZUN cells are rapidly and efficiently ablated by G418 selection (Figure 3B).
Figure 3.
Differentiation of ZUN hESC lines on Matrigel and Gelatin. (A) Expression analysis of spontaneous differentiation using pluripotency markers. The qRT-PCR of hESCs without MEFs plated on gelatin or matrigel for 12 days was performed with biological triplicates, and presented as expression levels relative to that of respective undifferentiated cell lines. (B) AP staining of hESCs on gelatin and matrigel. Representative phase contrast photomicrographs comparing HUES8 and ZUN cell morphologies on the 12th day and visualization of the ablative effects after 2 days of exposure to G418. Scale bar: 55 µm.
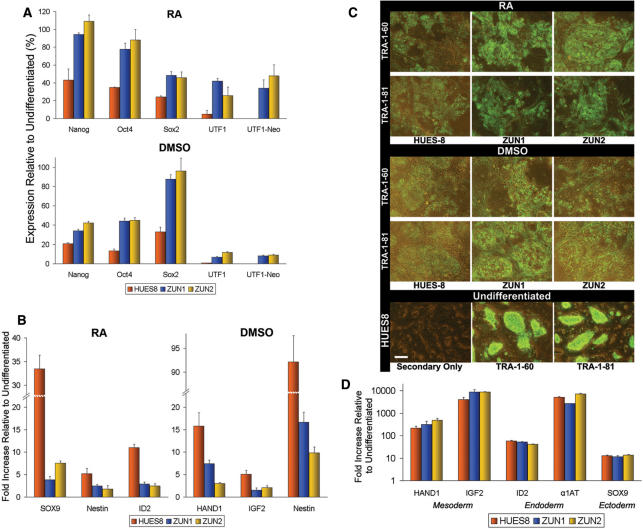
We analyzed next the effects of RA or DMSO treatment on ZUN cultures in the absence of G418 selection. After 12 and 7 days, respectively, qRT-PCR showed that ZUN cultures exhibited a lower rate of differentiation in comparison to HUES8 cultures (Figure 4A). Notably, UTF1 and UTF1-Neo expression diminished by the largest magnitude among the pluripotency markers. Certain differentiation markers also indicated a slower onset of differentiation in ZUN cultures. HUES8 cultures showed a greater (2- to 10-fold) upregulation of Sox9, Nestin and ID2 due to RA treatment, and of HAND1, IGF2 and Nestin after exposure to DMSO (Figure 4B). The expression of surface markers of pluripotency, TRA-1-60 and TRA-1-81, complements these transcript analyses, showing that HUES8 cells exhibited a more muted staining pattern compared to ZUN colonies (Figure 4C). These results confirm our conclusion that under various conditions, ZUN cultures exhibit a more robust phenotype against the onset of differentiation than parental cultures.
Figure 4.
Differentiation of ZUN hESCs induced by RA and DMSO. (A) Expression analysis of induced differentiation using pluripotency markers. qRT-PCR of hESCs on MEFs induced to differentiate via RA (for 12 days) or DMSO (for 7 days) in differentiation medium. (B) Transcript analysis of induced differentiation using differentiation markers. (C) TRA-1-60 and TRA-1-81 surface marker immunostaining for induced differentiation, comparing HUES8 with ZUN cultures. The bottom row shows a control panel of undifferentiated HUES8 cells stained with secondary antibody only, TRA-1-60 or TRA-1-81 antibodies (left to right). Scale bar: 30 μm. (D) Transcript analysis after EB formation. qRT-PCR of EBs from hESCs induced to differentiate for 21 days in suspension cultures. The qRT-PCR experiments were performed with biological duplicates, and presented as expression levels relative to that of respective undifferentiated cell lines.
ZUN cells retain the capacity to differentiate into progenitors of all three germ layers in vitro and in vivo
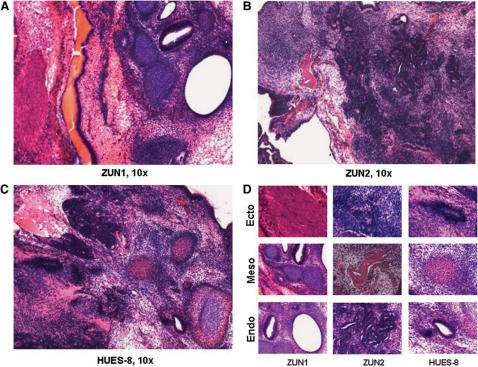
The observed phenotype of increased robustness against the onset of differentiation led us to investigate ZUN cultures under long term differentiation conditions. The results show that ZUN and HUES8 cells cultured under RA and DMSO differentiation protocols for 30 days show similar morphologies and lack of AP staining (Supplementary Figure S5). These cells were completely ablated upon addition of G418, as were those in equally long-term gelatin and matrigel cultures (data not shown). In addition, 21 days of embryoid body formation in suspension cultures caused a significant upregulation of differentiation markers from all three germ layers (Figure 4D). Both ZUN lines and HUES8 show very high expression of HAND1, IGF2 and ID2 as indicated by the large fold increase relative to their undifferentiated counterparts. They also show relatively high expression of α1 anti-trypsin (α1AT) and Sox9. The morphological characteristics and growth rates of HUES8 and ZUN cell-derived EBs were indistinguishable (Supplementary Figure S6). Furthermore, we showed by a teratoma assay that the parental HUES8 line as well as ZUN1 and ZUN2 formed tissue of all three germ layers (i.e. ectodermal structures, mesoderm such as cartilage and bone, as well as mesodermal glandular structures) when transplanted into immune-deficient SCID mice (Figure 5). We conclude, therefore, that the robustness which our ZUN cultures displayed against the onset of differentiation induced by various protocols does not compromise their potential to differentiate, like parental cultures, into all three germ layers in vitro and in vivo.
Figure 5.
Teratoma formation SCID mice studies. The hESC lines ZUN1 (A), ZUN2 (B) and the parental line HUES8 (C) were injected into the hind leg of male SCID mice, and the teratoma formation was followed by a simple grading system (see Materials and Methods section). When teratomas were full developed (1–2 g tissue weight, 6–8 weeks), they were harvested and processed by routine histological procedures, followed by heamatoxilin/eosin staining. (D) Detailed histology of ectodermal (epithelium, neuroectoderm), mesodermal (bone in red, cartilage in blue) and endodermal (glandular structures) structures are shown for each hESC line.
DISCUSSION
Expression of UTF1 is controlled by a 5′ TATA-less promoter consisting of four GC boxes. The 3′ enhancer element harbors a twin octamer sequence where the synergistic binding of Oct4 and Sox2 is essential for UTF1 expression in both mouse and human ESCs (8,35). Sp1-like transcription factors that bind to GC boxes present in the UTF1 promoter are most likely involved in the regulation of UTF1, since they are regarded as regulators of embryonic development in vertebrates (36) and shown to be involved in the transcriptional control of both Oct4 and Nanog (37,38). We showed here that UTF1 is indeed expressed in hESCs, but not in human primary fibroblasts and carcinoma cell lines. Further, both 5′ promoter and 3′ enhancer are needed for expression. A novel finding is that an octamer sequence (M1) which is conserved in the Nanog promoter of many species (27) is also present upstream of the analogous human Oct4/Sox2 cognate sequence in the 3′ UTF1 enhancer. The fact that M1 mutations resulted in equally muted expression levels of UTF1 and Nanog, strongly suggests that similar M1-binding factor(s) are involved in the control of both pluripotency marker genes. Our results also confirm previous reports showing that endogenous UTF1 is downregulated faster than Oct4 or Nanog at the onset of differentiation to nearly undetectable levels. In addition, detailed analysis of SymAtlas data from the Genomics Institute of the Novartis Research Foundation showed that both UTF1 and Nanog expression are greatly diminished at day 8.5 of mouse embryo formation, unlike Oct4 or Sox2 (http://symatlas.gnf.org). Together, these results firmly establish UTF1 as a sensitive and reliable pluripotency marker for hESCs.
Recent reports revealed that UTF1 is also an important marker expressed in other stem cell types. For example, spermatogonial stem cells isolated from adult mouse testis were shown to exhibit features reminiscent of pluripotent ESCs. These cells express Oct4, Nanog, Rex1 and UTF1 and, interestingly, also downregulate UTF1 significantly faster than these other markers upon embryoid body formation (39). Further, pluripotent stem cells derived from re-programmed primary mouse fibroblasts through co-expression of transgenes encoding Oct4, Sox2, c-Myc and Klf4 also express UTF1. It is noteworthy that the expression level of UTF1 in pluripotent stem cells derived from different sources is always more tightly linked with that of the key marker Oct4, and not with other markers like Cripto or Nanog (40). These data, in conjunction with results presented here, indicate that our UTF1-based selectivity tool should be widely applicable to select for pluripotent human stem cell lines derived from different sources.
We demonstrated that the control of both endogenous and exogenous UTF1 in ZUN cells is linked, as expected, and that UTF1-driven Neo expression in hESCs in conjunction with G418 selection can be used to efficiently ablate differentiating cells in standard culture conditions. This resulted in ZUN cultures which display a higher global expression level of key pluripotency markers. We showed that this elevated level is most likely due to an increase in the fraction of Oct4 and Sox2 positive hESCs. Furthermore, the number of cells staining positive for surface markers TRA-1-60 and TRA-1-81 was also elevated which, if taken together, indicates that ZUN cultures are more homogeneously pluripotent than the parental cultures (Figure 2A–C). Upon removal of selection pressure from ZUN cultures, the overall expression level of these markers declined, which indicates loss of pluripotency and, consequently, more heterogeneous hESC cultures. Interestingly, the absence of selection pressure for 60 days did not cause widespread silencing of the exogenous UTF1-Neo. This enabled us to add G418 only periodically in order to select against differentiating hESCs in ZUN cultures. Hence, the UTF1 promoter/enhancer combination could be a generally applicable control element for sustained expression of transgenes in pluripotent hESCs, even in the absence of selection.
We showed that the generation of ZUN lines harnessed the sensitivity of UTF1 as a pluripotency marker which resulted in rapid and efficient ablation of differentiating cells. An unexpected finding was the refractoriness of ZUN cultures to the onset of differentiation. We attribute this phenotype to the increased fraction of pluripotent hESCs in homogeneous ZUN cultures compared to typical, more heterogeneous hESC cultures. Given the propensity for heterogeneous cultures to spontaneously differentiate (21), the increased stability of human stem cell cultures with respect to the uniformity in pluripotency combined with efficient ablation of differentiating cells offered by the ZUN system could significantly improve large scale growth for many future hESCs applications. In addition, ZUN cultures show a higher expression of pluripotency markers when compared to HUES8 cultures following thawing after standard slow cooling cryopreservation procedures (data not shown), thereby counteracting a reported negative effect associated with cryopreservation (41).
An intrinsic advantage of the UTF1-based selection system over other pluripotency marker-based strategies is its enhanced sensitivity towards many differentiation pathways. Although the well-characterized pluripotency marker Oct4 has been a component in strategies for both mouse and human ESCs (21,22), the use of Oct4 in this context can be problematic. Apart from being less sensitive to the onset of differentiation, its expression level is also very tightly controlled. A less than 2-fold increase in Oct4 expression alone leads to differentiation into primitive endodermal and mesodermal lineages in mouse ESCs (42). This implies that the Oct4-Neo system is potentially less sensitive to these differentiation pathways, and, thus, will not be able to efficiently ablate cells that are undergoing differentiation into these lineages. In addition, the <2 kb length of the entire human UTF1 promoter/enhancer element represents a significant advantage for genetic manipulation of hESCs, or other human cell types, if one compares this to an unwieldy 8 kb of the Oct4 control element.
Our ZUN system has a large range of potential applications. In its current context as a stable transgene in hESCs, it can be used for the maintenance of homogeneously pluripotent cultures. In combination with gene targeting technologies, this application can be upscaled to meet the larger cell quantities ultimately needed for stem cell-based therapies. ZUN lines can also be employed to screen and characterize synthetic factors, media components or surface matrices that permit maintenance of pluripotency, and it can be used to test 3D scaffolds or aid in the derivation of new hESC lines.
As indicated above, the ZUN system shows potential in selection protocols in order to isolate adult stem cells from primary cultures. It can also be used in hESCs in conjunction with cell differentiation strategies as a component of positive-negative selection. The system can also function as a tool in somatic cell reprogramming experiments to identify successfully reprogrammed pluripotent cells. It can also be used to improve future human somatic cell nuclear transfer protocols. With our first demonstration of its utility here and its many possible applications, we envision that our ZUN system will become a valuable selectivity tool for many facets of stem cell research.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank R.J. Zhou for help with the Southern blot analysis, C.-T. Lee for the construction of pTZ-UTF1-EGFP(M1) and K. Neef for hESC line EPN14. Special thanks go to C.A. Davey for critical comments on the manuscript. Funding to pay the Open Access publication charges for this article was provided by NTU special investigator grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ginis I, Luo Y, Miura T, Thies S, Brandenberger S, Gerecht-Nir S, Amit M, Hoke A, Carpenter MK, et al. Differences between human and mouse embryonic stem cells. Dev. Biol. 2003;269:360–380. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 2.Brimble SN, Zeng X, Weiler DA, Luo Y, Liu Y, Lyons IG, Freed WJ, Robins AJ, Rao MS, et al. Karyotypic stability, genotyping, differentiation, feeder-free maintenance, and gene expression sampling in three human embryonic stem cell lines derived prior to August 9, 2001. Stem Cells Dev. 2004;13:585–597. doi: 10.1089/scd.2004.13.585. [DOI] [PubMed] [Google Scholar]
- 3.Sun Y, Li H, Liu Y, Shin S, Mattson MP, Rao MS, Zhan M. Cross-species transcriptional profiles establish a functional portrait of embryonic stem cells. Genomics. 2007;89:22–35. doi: 10.1016/j.ygeno.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukushima A, Okuda A, Nishimoto M, Seki N, Hori TA, Muramatsu M. Characterization of functional domains of an embryonic stem cell coactivator UTF1 which are conserved and essential for potentiation of ATF-2 activity. J. Biol. Chem. 1998;273:25840–25849. doi: 10.1074/jbc.273.40.25840. [DOI] [PubMed] [Google Scholar]
- 5.Nishimoto M, Miyagi S, Yamagishi T, Sakaguchi T, Niwa H, Muramatsu M, Okuda A. Oct-3/4 maintains the proliferative embryonic stem cell state via specific binding to a variant octamer sequence in the regulatory region of the UTF1 locus. Mol. Cell. Biol. 2005;25:5084–5094. doi: 10.1128/MCB.25.12.5084-5094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharya B, Miura T, Brandenberger R, Mejido J, Luo Y, Yang AX, Joshi BH, Ginis I, Thies RS, et al. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood. 2004;103:2956–2964. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- 7.Trounson A. The production and directed differentiation of human embryonic stem cells. Endocr. Rev. 2006;27:208–219. doi: 10.1210/er.2005-0016. [DOI] [PubMed] [Google Scholar]
- 8.Nishimoto M, Fukushima A, Okuda A, Muramatsu M. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol. 1999;19:5453–5465. doi: 10.1128/mcb.19.8.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai J, Chen J, Liu Y, Miura T, Luo Y, Loring JF, Freed WJ, Rao MS, Zeng X. Assessing self-renewal and differentiation in human embryonic stem cell lines. Stem Cells. 2006;24:516–530. doi: 10.1634/stemcells.2005-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei CL, Miura T, Robson P, Lim SK, Xu XQ, Lee MY, Gupta S, Stanton L, Luo Y, et al. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem. Cells. 2005;23:166–185. doi: 10.1634/stemcells.2004-0162. [DOI] [PubMed] [Google Scholar]
- 11.Eifert C, Sangster-Guity N, Yu LM, Chittur SV, Perez AV, Tine JA, McCormick PJ. Global gene expression profiles associated with retinoic acid-induced differentiation of embryonal carcinoma cells. Mol. Reprod. Dev. 2006;73:796–824. doi: 10.1002/mrd.20444. [DOI] [PubMed] [Google Scholar]
- 12.Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 13.Skottman H, Stromberg A-M, Matilainen E, Inzunza J, Hovatta O, Lahesmaa R. Unique gene expression signature by human embryonic stem cells cultured under serum-free conditions correlates with their enhanced and prolonged growth in an undifferentiated stage. Stem Cells. 2005;24:151–167. doi: 10.1634/stemcells.2004-0189. [DOI] [PubMed] [Google Scholar]
- 14.Sen G, Wehrman TS, Myers JW, Blau HM. Restriction enzyme-generated siRNA (REGS) vectors and libraries. Nat. Genet. 2004;36:183–189. doi: 10.1038/ng1288. [DOI] [PubMed] [Google Scholar]
- 15.Hoof DV, Passier R, Oostwaard DW-V, Pinkse MWH, Heck AJR, Mummery CL, Krijgsveld J. A quest for human and mouse embryonic stem cell-specific proteins. Mol. Cell Proteomics. 2006;5:261–273. doi: 10.1074/mcp.M500405-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, Itskovitz-Eldor J, Thomson JA. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev. Biol. 2000;227:271–278. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- 17.Eiges R, Schuldiner M, Drukker M, Yanuka O, Itskovitz-Eldor J, Benvenisty N. Establishment of human embryonic stem cell-transfected clones carrying a marker for undifferentiated cells. Curr. Biol. 2001;11:514–518. doi: 10.1016/s0960-9822(01)00144-0. [DOI] [PubMed] [Google Scholar]
- 18.Stewart MH, Bosse M, Chadwick K, Menendez P, Bendall SC, Bhatia M. Clonal isolation of hESCs reveals heterogeneity within the pluripotent stem cell compartment. Nat. Methods. 2006;3:807–815. doi: 10.1038/nmeth939. [DOI] [PubMed] [Google Scholar]
- 19.McWhir J, Schnieke AE, Ansell R, Wallace H, Colman A, Scott AR, Kind AJ. Selective ablation of differentiated cells permits isolation of embryonic stem cell lines from murine embryos with a non-permissive genetic background. Nat. Genet. 1996;14:223–226. doi: 10.1038/ng1096-223. [DOI] [PubMed] [Google Scholar]
- 20.Hong Y, Winkler C, Schartl M. Pluripotency and differentiation of embryonic stem cell lines from the medakafish (Oryzias latipes) Mech. Dev. 1996;60:33–44. doi: 10.1016/s0925-4773(96)00596-5. [DOI] [PubMed] [Google Scholar]
- 21.Mountford P, Nichols J, Zevnik B, O'Brien C, Smith A. Maintenance of pluripotential embryonic stem cells by stem cell selection. Reprod. Fertil. Dev. 1998;10:527–533. doi: 10.1071/rd98087. [DOI] [PubMed] [Google Scholar]
- 22.Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat. Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- 23.Gerrard L, Zhao D, Clark AJ, Cui W. Stably transfected human embryonic stem cell clones express OCT4-specific green fluorescent protein and maintain self-renewal and pluripotency. Stem Cells. 2005;23:124–133. doi: 10.1634/stemcells.2004-0102. [DOI] [PubMed] [Google Scholar]
- 24.Hewitt Z, Forsyth NR, Waterfall M, Wojtacha D, Thomson AJ, McWhir J. Fluorescence-activated single cell sorting of human embryonic stem cells. Cloning Stem Cells. 2006;8:225–234. doi: 10.1089/clo.2006.8.225. [DOI] [PubMed] [Google Scholar]
- 25.Liu YP, Dovzhenko OV, Garthwaite MA, Dambaeva SV, Durning M, Pollastrini LM, Golos TG. Maintenance of pluripotency in human embryonic stem cells stably over-expressing enhanced green fluorescent protein. Stem Cells Dev. 2004;13:636–645. doi: 10.1089/scd.2004.13.636. [DOI] [PubMed] [Google Scholar]
- 26.Gallagher EJ, Lodge P, Ansell R, McWhir J. Isolation of murine embryonic stem and embryonic germ cells by selective ablation. Transgenic Res. 2003;12:451–460. doi: 10.1023/a:1024225225302. [DOI] [PubMed] [Google Scholar]
- 27.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 28.Amit M, Margulets V, Segev H, Shariki K, Laevsky I, Coleman R, Itskovitz-Eldor J. Human feeder layers for human embryonic stem cells. Biol. Reprod. 2003;68:2150–2156. doi: 10.1095/biolreprod.102.012583. [DOI] [PubMed] [Google Scholar]
- 29.Tan SM, Dröge P. Comparative analysis of sequence-specific DNA recombination systems in human embryonic stem cells. Stem Cells. 2005;23:868–873. doi: 10.1634/stemcells.2005-0044. [DOI] [PubMed] [Google Scholar]
- 30.Cowan CA, Klimanskaya I, McMahon J, Atienza J, Witmyer J, Zucker JP, Wang S, Morton CC, McMahon AP, et al. Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 31.Corona T, Bao Q, Christ N, Schwartz T, Li J, Dröge P. Activation of site-specific DNA integration in human cells by a single chain integration host factor. Nucleic Acids Res. 2003;31:5140–5148. doi: 10.1093/nar/gkg711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandenberger R, Wei H, Zhang S, Lei S, Murage J, Fisk GJ, Li Y, Xu C, Fang R, et al. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat. Biotechnol. 2004;22:707–716. doi: 10.1038/nbt971. [DOI] [PubMed] [Google Scholar]
- 33.Christ N, Dröge P. Genetic manipulation of mouse embryonic stem cells by mutant λ integrase. Genesis. 2002;32:203–208. doi: 10.1002/gene.10031. [DOI] [PubMed] [Google Scholar]
- 34.Schuldiner M, Yanuka O, Itskovitz-Eldor J, Melton DA, Benvenisty N. Effects of eight growth factors on the differentiation of cells derived from human embryonic stem cells. Proc. Natl Acad. Sci. USA. 2000;97:11307–11312. doi: 10.1073/pnas.97.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao C, Meng A. Sp1-like transcription factors are regulators of embryonic development in vertebrates Develop. Growth Differ. 2005;47:201–211. doi: 10.1111/j.1440-169X.2005.00797.x. [DOI] [PubMed] [Google Scholar]
- 37.Yang HM, Do HJ, Oh JH, Kim JH, Choi SY, Cha KY, Chung HM, Kim JH. Characterization of putative cis-regulatory elements that control the transcriptional activity of the human Oct4 promoter. J. Cell Biochem. 2005;96:821–830. doi: 10.1002/jcb.20588. [DOI] [PubMed] [Google Scholar]
- 38.Wu DY, Yao Z. Functional analysis of two Sp1/Sp3 binding sites in murine Nanog gene promoter. Cell Res. 2006;16:319–322. doi: 10.1038/sj.cr.7310040. [DOI] [PubMed] [Google Scholar]
- 39.Guan K, Nayernia K, Maier LS, Wagner S, Dressel R, Lee JH, Nolte J, Wolf F, Li M, et al. Pluripotency of spermatogonial stem cells from adult mouse testis. Nature. 2006;440:1199–1203. doi: 10.1038/nature04697. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 41.Katkov II, Kim MS, Bajpai R, Altman YS, Mercola M, Loring JF, Terskikh AV, Snyder EY, Levine F. Cryopreservation by slow cooling with DMSO diminished production of Oct-4 pluripotency marker in human embryonic stem cells. Cryobiology. 2006;53:194–205. doi: 10.1016/j.cryobiol.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.