Abstract
The telomerase ribonucleoprotein copies a short template within its integral RNA moiety onto eukaryotic chromosome ends, compensating for incomplete replication and degradation. Non-template regions of telomerase RNA (TER) are also crucial for telomerase function, yet they are highly divergent in sequence among species and their roles are largely unclear. Using both phylogenetic and mutational analyses, we predicted secondary structures for TERs from Kluyveromyces budding yeast species. A comparison of these secondary structure models with the published model for the Saccharomyces cerevisiae TER reveals a common arrangement into three long arms, a templating domain in the center and several conserved elements in the same positions within the structure. One of them, a three-way junction element, is highly conserved in budding yeast TERs. This element also shows sequence and structure similarity to the critical CR4-CR5 activating domain of vertebrate TERs. Mutational analysis in Kluyveromyces lactis confirmed that this element, and in particular the residues conserved across yeast and vertebrates, is critical for telomerase action both in vivo and in vitro. These findings demonstrate that despite the extreme divergence of TER sequences from different organisms, they do share conserved elements, which presumably carry out common roles in telomerase function.
INTRODUCTION
Telomeres are nucleoprotein structures protecting eukaryotic chromosome ends (1). Telomerase is a ribonucleoprotein (RNP) complex that can add short DNA repeats onto telomeres and thus compensate for losses caused by incomplete replication or degradation (2,3). The essential core components of this specialized enzyme are telomerase RNA (TER) and telomerase reverse transcriptase (TERT; Est2 in budding yeasts), which can repeatedly copy a small portion of TER (termed ‘template’) onto the telomere 3′-end. Other proteins that were shown to associate with the telomerase complex in Saccharomyces cerevisiae include Est1 and Est3 (which are essential for telomerase action in vivo), the Sm proteins and Ku80.
TERs are highly divergent in sequence and length even among closely related species. Phylogenetic covariation was used to predict common secondary structure models for TERs within each group of ciliates (4,5), vertebrates (6) and Saccharomyces sensu stricto species (7–10). However, only limited similarity in the general architecture of these models was observed (6,9). Yeast TERs are relatively large (930–1540 nt) and highly divergent in sequence (11,12). While significant portions of these TERs are dispensable for telomerase function, other non-template regions appear to be important (13–15). Several conserved elements were found to contribute crucial functions such as specifying the template boundary (12,16) and recruiting Est1, Ku80 and the Sm proteins (17–19). A unique pseudoknot element containing a major-grove triple-helix was found to be crucial for telomerase function in the budding yeast Kluyveromyces lactis (20) and human (21) TERs, though its specific function is still unclear. A critical domain in vertebrate TERs, CR4-CR5, was suggested to interact with TERT (22). Within this domain, a small stem-loop, P6.1, is essential for the telomerase catalytic activity. In ciliate TERs, stem IV was suggested to be the functional homolog of the vertebrate P6.1 (23). However, a yeast homolog of this element has not been identified yet.
Here, we report on a secondary structure model of the K. lactis TER. Comparing the models of Kluyveromyces and Saccharomyces TERs revealed a similar organization of three long arms and a templating domain at the center. Functional elements, conserved in sequence and/or structure, were identified in the same corresponding positions within the structure in all species. One of these elements, a novel three-way junction, has been further studied by mutational analysis and found to be critical for telomerase function. Interestingly, it shows conservation in both secondary structure and sequence with the vertebrate CR4-CR5 domain, in particular with p6.1, suggesting that the three-way junction is the yeast homolog of the vertebrate CR4-CR5 activating domain.
MATERIALS AND METHODS
Yeast strains, genomic DNA preparation and Southern hybridization
K. lactis strain yJR27 (ter1Δ ura3-1 his2-2 trp1), based on the parental strain 7B520, was used for the CEN-ARS plasmid shuffling of WT and mutant TER genes as described (13). All yeast strains were grown at 25–28°C. Genomic DNA preparation and Southern analysis of telomere restriction fragments were carried out as described previously (20).
Purification of telomerase extracts and in vitro telomerase assay
Cell extracts were prepared from K. lactis strains and partially purified as described previously (24). Telomerase assay was performed as described (12,24), except for the following modifications: 5′-biotinylated oligonucleotides were used as telomerase substrates, the telomerase reaction (20 μl) was incubated at 30°C for 30 min, then heated at 95°C for 4 min, internal control (5′-end 32P-labeled, 3′-biotinylated, 10-mer oligonucleotide: GTGGTGTACG) was added, and the products were purified with Dynabeads MyOne™ Streptavidin C1 (Invitrogen Dynal AS, Oslo, Norway) as follows. Equal volume of 10 mM Tris HCl pH 7, 1 mM EDTA, 2 M NaCl and 50 μg Dynabeads was added, the beads were incubated for binding at room temperature for 30 min, washed 3 times with 10 mM Tris HCl pH 7, 1 mM EDTA, 1 M NaCl and once with 10 mM Tris HCl pH7, 1 mM EDTA, using Dynal magnet according to the manufacturer's instructions. For elution, the beads were resuspended in 10 µl of 95% formamide loading buffer, incubated at 95°C for 5 min, and electrophoresed on an 8 M urea 15% polyacrylamide gel. TER levels in the fractions were quantified by slot blot analysis using a 32P-labeled TER1 probe.
Cloning of K. lactis telomerase protein genes
The genes encoding the putative K. lactis orthologs of Est1, Est2, Est3, Ku70 and Ku80 were identified in the K. lactis genome by their sequence homology to the S. cerevisiae proteins. The genes, including 950–1000 nt upstream and 850–900 nt downstream of the coding sequences, were amplified by PCR and cloned into a SmaI site of pKDU7, a K. lactis 2 μ-based, high copy-number plasmid (25). The cloned sequences were confirmed by sequencing.
RESULTS
Secondary structure models for Kluyveromyces telomerase RNAs
To identify conserved functional TER elements, we attempted to compare secondary structure predictions for distant budding yeast species. We had previously cloned TER genes from six Kluyveromyces budding yeast species and identified several common stems supported by covariation, and several elements conserved in sequence and/or structure (12,18,20,26). However, the overall low sequence conservation has hindered the prediction of a detailed secondary structure model based on phylogenetic covariation. Instead, we used short stems and single-stranded sequences supported by phylogenetic covariation and experimental data, to constrain the free-energy based computer program mfold (27) and predict a secondary structure for klTER, as described in Materials and Methods section and shown in Supplementary Figure S1A. The structures of other Kluyveromyces TERs were also predicted in the same way (data not shown).
Common secondary structure models for TERs from Saccharomyces sensu stricto species have previously been published (7–10), revealing an emerging consensus for TER structure (28). However, some inconsistencies exist among the models, particularly in the structures proposed for the pseudoknot domain and the Sm-binding arm. To compare the structure of klTER with that of S. cerevisiae TER (also termed TLC1, but for simplicity will be referred to as scTER), we followed the approach described by Zappulla and Cech (10), except for using TER sequences from seven, instead of four, Saccharomyces sensu stricto species [S. cerevisiae, S. paradoxus, S. mikatae, S. kudriavzevii, S. bayanus, S. pastorianus and S. cariocanus (8,9)]. We first aligned the seven TER sequences using ClustalX (29) and predicted a common secondary structure by RNAalifold (30). Then, several predicted stems and a pseudoknot model proposed previously (9) were used to constrain mfold and predict a detailed secondary structure model for scTER (Figure S1B). The predicted structure was generally consistent with the previously published secondary structure models (8,10). The structure of the 3′ arm was consistent with that proposed by Zappulla and Cech (10).
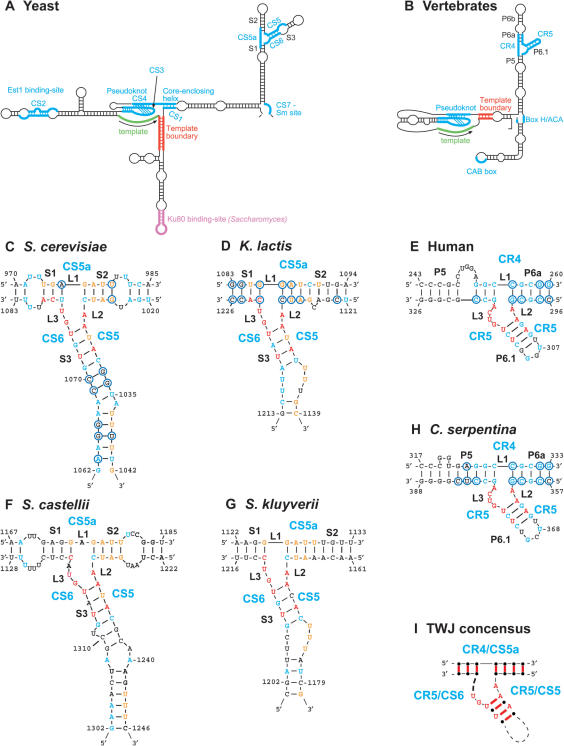
Interestingly, while no sequence identity was apparent between klTER and scTER, the predicted Kluyveromyces TER structures reveal an arrangement of three long arms and a templating domain at the center, as described previously for the Saccharomyces TERs (8–10) and schematically shown in Figure 1A. The lengths of the corresponding arms vary by up to 150 nt between different Kluyveromyces species, and by up to 225 nt between S. cerevisiae and Kluyveromyces TERs (Figure S1 and data not shown). However, seven short sequences (CS1–7) identified previously in the Kluyveromyces species (26) were found in the same structural context in the S. cerevisiae model: CS1, 3 and 4 form the pseudoknot structure adjacent to the template and the core-enclosing helix (9,26); CS2 forms a bulged-stem required for the binding of the telomerase protein Est1 (18) and CS7 forms a binding site for the Sm proteins (19,26). The Ku80 binding site (17) is conserved only in the Saccharomyces but not in the Kluyveromyces species, consistent with the severe effect of Ku80 deletion on telomere length in S. cerevisiae, but not in K. lactis (31). Two other conserved elements, CS5 and CS6, have not been studied previously. In both scTER and klTER structure models, these two sequences are partially paired to each other, and together with an additional, less conserved sequence termed CS5a, form a three-way junction (TWJ; Figures 1C and D, S2A and B).
Figure 1.
A common secondary structure model for budding yeast TERs reveals an element similar to the vertebrate CR4-CR5 domain. (A) A schematic representation of a common secondary structure for budding yeast TERs was drawn according to the secondary structure predictions for Kluyveromyces and Saccharomyces TERs shown in Figure S1. (B) A schematic representation of the vertebrate TER structure was drawn according to (41). Indicated are the various functional elements found in telomerase RNA. Secondary structure models for the S. cerevisiae (C), K. lactis (D), human (E), S. castelii (F), S. kluyveri (G) and C. serpentina (H) TWJs were predicted, as described in the Supplementary Data. Indicated in blue are nucleotides conserved within each group [in all six Kluyveromyces, all seven Saccharomyces sensu stricto or in at least 33 of 35 vertebrate TERs (>94%)]. In the S. castellii and S. kluyveri models, indicated in blue are nucleotides conserved with the rest of the Saccharomyces species. Indicated in orange are nucleotides conserved in at least 13 of 15 yeast TERs (>86%). Indicated in red are nine nucleotides conserved across yeast and vertebrates in at least 45 of the 50 TER sequences examined (>90%; see Figure S3). Circled are positions with covariations within the Kluyveromyces, Saccharomyces sensu stricto or the vertebrate groups, supporting the predicted structures. (I) A representation of the consensus TWJ structure for yeast and vertebrate TERs. Indicated in red are conserved base pairs and conserved nucleotides.
The three-way junction is conserved in yeast and vertebrate TERs
Similar structures formed by CS5a, CS5 and CS6 were predicted in all seven Saccharomyces and six Kluyveromyces TERs (Figures 1C and D, S2A and B). In particular, the junction itself is highly conserved, including no unpaired nucleotides between stem 1 and stem 2 (L1 = 0), an A linker between stem 2 and stem 3 (L2 = A), and a GU linker between stem 3 and stem 1 (L3 = GU; with the exception of L3 = GC in K. dobzhanskii). Linkers between the stems of the TWJs were shown to be critical for the topology and the precise angles between the stems (32). In addition to the linkers, paired nucleotides that are close to the junction in stems 1, 2 and especially 3, are highly conserved among all Kluyveromyces and Saccharomyces TERs (indicated in orange and red in Figures 1C and D, S2A and B). All changes found in these nucleotides maintained the predicted secondary structure and therefore considered covariations (indicated by circles). Finally, a stretch of U residues is conserved in the CS5 strand of stem 3 in all Kluyveromyces and Saccharomyces TERs. Altogether, the high conservation of the linkers, the stems and specific nucleotides supported the prediction and suggested that the sequence and topology of the TWJ is conserved among budding yeast TERs. We then searched for the conserved TWJ element in TER sequences of the more distant Saccharomyces sensu lato species S. castellii and S. kluyveri, which were identified in the genbank based on published partial sequences (7). Although these sequences show no apparent similarity to the Kluyveromyces or Saccharomyces sensu stricto TERs, they were also predicted to form similar TWJs with the same junction parameters (except for an extra A in two of the linkers in S. castellii—L1 = A and L3 = GUA), and conserved nucleotides at the corresponding positions (Figure 1F and G).
The position of the TWJ in the secondary structure model appears similar to that of the vertebrate CR4-CR5 domain (Figure 1A and B). Although parts of the CR4-CR5 structure were solved by NMR (33,34), the structure at the junction of P5, P6a and P6.1 remained unsolved. To further study possible conservation between these elements, we aligned the sequences corresponding to the CR4-CR5 domain from 35 vertebrate species (6) using ClustalX (29), and subjected the alignment to RNAalifold (30) for common secondary structure prediction. Interestingly, RNAalifold predicted a structure similar to the yeast TWJ (see Figure 1E and H and sequence alignment in Figure S3). However, the short length of stem P5 between the bulge and the junction (3 bp), and an alternative conformation predicted previously for the human TER [(35) and Figure S2D], raised doubts about the RNAalifold prediction. Therefore, we examined whether the alternative conformation can be formed in all vertebrate TERs and whether phylogenetic covariation supports it. Apparently, the sequence and the length of the bulge (Figure 1E: nt 248–252 in the human TWJ) are not conserved and most variations in the sequence are not compatible with the alternative structure, indicating that the alternative conformation is not likely to form in at least 21 of the 35 vertebrate TERs. In contrast, the stem predicted by RNAalifold, although as short as 3 bp in some species, is conserved in all vertebrate TERs with a one nucleotide change in two species (G-C to G-U), which preserves the structure (encircled in Figure 1E: nt 253–255 and 319–321). In addition, the predicted stem is as short as 3 bp only in the mammalian species (19 of the 20) and in Gallus gallus (Chicken; data not shown). In the rest of the species [the mammalian Dasyurus hallucatus (Quoll), the bird Anodorhynchus hyacinthinus (Macaw), and all the Reptilia, Amphibia and Chondrichthyes species], this stem is longer (4–6 bp) and supported by more covariations (the Chelydra serpentina predicted TWJ is shown as an example in Figure 1H). The higher diversity of the latter group suggests that a longer stem existed in the common evolutionary ancestor of these vertebrates, and its shortening to 3 bp occurred later in the mammalian and avian species. Chemical structure probing of in vitro transcribed hTER (35) is generally consistent with our model: nucleotides UGGA249-52 of the predicted bulge in P5 and GU315-6 in the linker L3 (Figures 1E, S2C and S2D) show moderate to strong accessibility to modifying agents, suggesting that they are likely to be in a single-stranded conformation, while the 3 bp stem was not modified, except for the moderate and weak modification of G253 and G254, respectively. However, chemical probing in vivo showed only moderate modification of AG252-3 and strong modification of G315, while the rest of the P5, P6a and P6.1 nucleotides were only weakly or not at all modified, indicating that a conformational change, additional tertiary interactions or the binding of a protein protected this sequence in vivo. While we cannot exclude the alternative conformation (Figure S2D), the conservation of the RNAalifold-predicted structure in all vertebrates and its similarity to the yeast structure clearly support our model (Figures 1E and S2C). Importantly, stem 2 and the most conserved elements, linker 2 and stem 6.1, are identical in both conformations.
The most conserved features of the yeast TWJ are also conserved in the vertebrate predicted structures. First, the linkers between P5/S1 and P6a/S2 (L1 = 0) and between P6a/S2 and P6.1/S3 (L2 = A) are identical in all 50 vertebrate and yeast TERs, except for L1 = A in S. castellii. The linker between P6.1 and P5 is longer in vertebrates (L3 = GUCA), but it includes the yeast sequence (L3 = GU) with only one exception (L = GAUU in Herpele squalostoma). Furthermore, the additional CA of the vertebrate linker is also conserved in most yeast TERs, although in the yeast predicted structures it is paired in stem 1 (the C is conserved in 31/35 vertebrate and 14/15 yeast TERs, and the A in 34/35 vertebrate and 13/15 yeast TERs). Second, P6.1 shows similarity to stem 3. The first and the third base pairs from the junction, A/U, are absolutely conserved in all 50 yeast and vertebrate TERs. The second and fourth base pairs are conserved within the groups (G/C in all vertebrates, U/A in all Kluyveromyces, C/G in S. kluyveri, U/G and C/G in Saccharomyces sensu stricto and U/A and C/G in S. castellii, for the second and fourth base pairs from the junction, respectively). Finally, a stretch of U residues and a G, which are conserved in the yeast CS5 strand of stem 3 (Figures 1C: nt 1037–1042, 1D: nt 1135–1138, 1F: nt 1243–1245, 1G: nt 1174–1180, S2 and S3), are reminiscent of conserved and critical U and G residue in P6.1 [(Figure 1E: nt 307 and 309; Figure S3; and (36)]. Altogether, the similarity of the yeast TWJ to the vertebrate CR4-CR5 domain, and in particular the linkers and stem 3/P6.1 (Figure 1H), confirms the predictions and suggests that these elements are functional homologs.
The three-way junction is crucial for K. lactis telomerase function in vivo
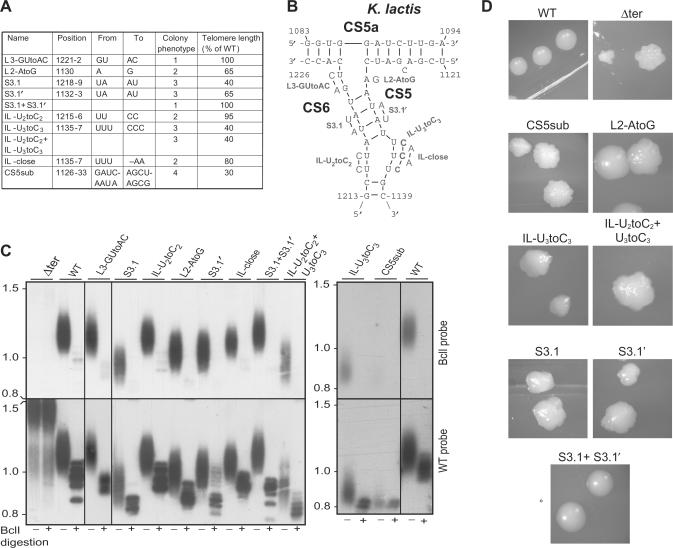
To test whether the conserved features of the TWJ are important for telomerase function, we introduced substitutions into the TER1 gene. We replaced the WT TER1 gene in K. lactis cells with the mutant alleles using a plasmid shuffling system previously described (13), and analyzed telomere length and telomerase activity in vivo. An additional single-nucleotide mutation was introduced into the TER template, resulting in the incorporation of BclI restriction sites into the telomeric repeats. This mutation is otherwise apparently silent, and thus can be used to mark the nascent repeats incorporated by telomerase and analyze its in vivo activity (25). The BclI repeats can be identified by Southern analysis of telomeric fragments hybridized to a BclI-specific telomeric probe, or by BclI restriction endonuclease digestion of the telomeric fragments and hybridization to a WT probe (see scheme in Figure S4).
The first mutation was a transition substitution of 7 nt in CS5 (CS5sub), which was predicted to disrupt stem 2 and stem 3 adjacent to the junction. This mutation severely impaired telomerase function in vivo, as revealed by the rough colony phenotype, a hallmark of impaired telomere maintenance in K. lactis (Figure 2D), the severe shortening of the telomeres to 30% of the WT telomere length and the barely detectable BclI repeat incorporation observed by Southern analysis (Figure 2C). We cloned and sequenced 20 different telomeres from the CS5sub mutant, as described previously (20). In three clones we found interspersed BclI and WT repeats (Figure S5B–D), which were presumably generated by telomeric recombination. This indicates that telomerase was still active and incorporating BclI repeats, but incapable of maintaining stable telomeres. In addition, four telomere clones had misincorporated nucleotides (the most aberrant of them is shown in Figure S5A), which appear to have been synthesized by miscopying of the template. Although this aberrant sequence was found in only one of the 20 clones, we did not observe such a sequence aberration in several hundred other telomere clones of WT and other telomerase mutants. Therefore, we think that the defect caused by the CS5sub mutation is not only in the level, but also in the fidelity, of telomerase activity. Altogether, these results indicate that CS5 is essential for normal telomerase action in vivo.
Figure 2.
The three-way junction is crucial for K. lactis telomerase function in vivo. The mutations introduced into the TWJ (A and B) and the resulting mean telomere length, as measured by Southern analysis (C), are summarized in (A). WT and mutant TER1 genes were introduced into K. lactis cells on a CEN-ARS plasmid, replacing the WT TER1 gene. These mutants contain an additional BclI template mutation that is incorporated into telomeres, introducing a BclI restriction site. Otherwise, the BclI mutation is phenotypically silent and can therefore be used to mark the nascent products of the investigated telomerase in vivo (see scheme in Figure S4). Genomic DNA was prepared from the sixth passage (∼90–120 generations), digested with EcoRI or EcoRI+BclI, electrophoresed on a 1% agarose gel, vacuum blotted onto a membrane, and hybridized first with a BclI-specific telomere probe (top) and then with a WT telomeric probe (bottom), as indicated on the right of (C). For simplicity, only a portion of the gel is shown, including 7 of the 12 K. lactis telomeres. (D) Typical colonies of the different strains taken at their forth passage. Impaired telomere maintenance is associated with rough colonies, as opposed to the smooth appearance of WT colonies.
To examine the TWJ in more detail, we introduced specific mutations designed to test the different elements of this structure, and whether their sequence and/or conformation is important (Figure 2). First, we made transition mutations in the linkers between stem 2 and stem 3 (L2-AtoG), and between stem 3 and stem 1 (L3-GUtoAC). The L3-GUtoAC mutation did not have any significant effect. In contrast, the single nucleotide L2-AtoG mutation caused significant telomere shortening to 65% of the WT telomere length. Such an effect for a single nucleotide transition indicates that this linker nucleotide, conserved in all 50 yeast and vertebrate TERs, is critical for telomerase function.
We next tested stem 3, which appears to be conserved between Kluyveromyces, Saccharomyces and vertebrates (P6.1). We introduced 2-nt substitutions in either CS5 or CS6, designed to disrupt the two middle base pairs (S3.1, S3.1′), while the combination of the two (S3.1 + S3. 1′) was expected to restore base pairing. Each of the two-nucleotide mutations resulted in a rough colony phenotype and telomere shortening to 40 and 65% of WT length for S3.1 and 3.1′, respectively (Figure 2), while the combination of the two mutations fully restored normal colony phenotype and telomere length, suggesting that the formation of stem 3 is crucial for telomerase function.
A stretch of U residues is conserved in CS5 in all yeast TERs, reminiscent of the critical U residue conserved in vertebrate TERs (Figures 1, S2 and S3). In the Kluyveromyces and S. kluyveri TERs, these U residues were predicted to form one side of an asymmetrical internal loop, while two other residues in CS6 form the other strand of the loop. In the other Saccharomyces TERs the U residues in CS5 were predicted to pair with G or A residues in CS6, while an A bulge remained in CS5. To test the importance of the sequence and the conformation of this stretch of U residues for telomerase function, we made several mutations in klTER. Transition mutations, replacing the U with C residues in each side of the internal loop (Figure 2A and B), were predicted to preserve the internal loop conformation. Substitution of three U residues conserved in CS5 significantly reduced telomere length to 40% of the WT length (IL-U3toC3). In contrast, the substitution of the two U residues conserved only in Kluyveromyces CS6 only had a slight effect (IL-U2toC2). When we combined these mutations together, changing five U to C residues, there was no further worsening of the phenotype over that of IL-U3toC3. Another mutation was designed to pair the loop by substituting two A residues for the three conserved U residues (IL-close). This mutation only had a moderate effect on telomere length, reducing it to 80% of the WT length, suggesting that these U residues may actually be paired, as predicted for the Saccharomyces TERs. U-U base pairing has been found previously in RNA, e.g. in the human TER (37).
Normal levels of telomerase RNA in three-way junction mutants
CS6 is located ∼30 nt from the Sm site, which is required for the stability of TER. Therefore, we tested whether mutations in the TWJ reduce TER levels rather than impair the function of the enzyme. We measured the TER levels using Northern analysis (data not shown) and real-time PCR (Figure S6). Although some fluctuation in TER levels was observed, no correlation was found between TER levels and the effects of the mutations on telomerase function. The most severe mutant, CS5sub, shows a 30% increase in TER levels, while the apparently normal compensatory mutant S3.1 + S3.1′ shows 40% reduction. These results suggest that the TWJ mutations impair either the assembly of the telomerase RNP complex or its specific activity, rather than reduce TER stability.
The three-way junction is important for the catalytic activity of telomerase in vitro
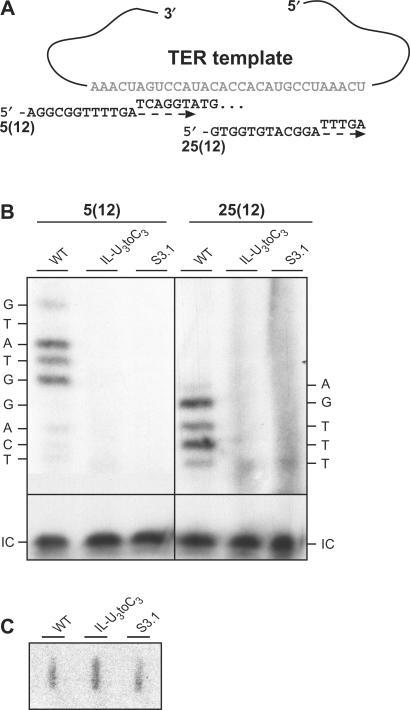
The similarity between the TWJ and the vertebrate CR4-CR5 domain suggests that both elements may have a similar role. The CR4-CR5 domain is critical for telomerase action both in vivo and in vitro (22,36). Therefore, we tested the activity of two TWJ telomerase mutants in vitro: S3.1, a 2-nt substitution in CS6, presumably disrupting stem 3 close to the junction; and IL-U3toC3, a substitution of three cytosine residues for the three conserved uridines in CS5. We partially purified telomerase RNP complexes from cell extracts of the WT and mutant TERs and analyzed in vitro the enzymatic activity of telomerase in the telomerase-enriched fractions. We used two different 12-mer telomerase substrates, which align with their 3′-ends at positions 5 and 25 of the template, as described in Materials and Methods section and illustrated in Figure 3. Strikingly, even though the TWJ appears to be distant from the catalytic core within the secondary structure model, both telomerase mutants were barely active under the in vitro assay conditions (Figure 3). This result indicates that the TWJ structure is crucial for the assembly and/or the action of the telomerase catalytic core, as is the case for the vertebrate CR4-CR5 domain.
Figure 3.
Stem 3 telomerase mutants are barely active in vitro. (A) A scheme showing the two telomerase substrates used, which are each 12 nt long, and which base pair to the telomerase template with their 3′-ends at positions 5 and 25 of the template [5(12) and 25(12), respectively]. A dashed arrow indicates the direction of polymerization, and the nucleotides added are shown above the arrow. (B) Partially purified telomerase fractions were prepared from WT, S3.1 and IL-U3toC3 strains. These fractions (10 μl each) were assayed for telomerase activity as described in Materials and Methods section. The nucleotides added by telomerase to the corresponding products are indicated on the sides of the panel, and the 10-mer oligonucleotide used as internal control is indicated by ‘IC’. (C) Slot-blot analysis with a TER1 probe reveals comparable amounts of TER in the fractions assayed for telomerase activity (10 μl each).
Overexpression of the telomerase proteins Est2 and Est3 partially suppresses the phenotype of a three-way junction mutant
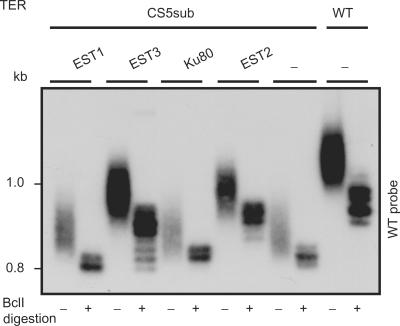
The vertebrate CR4-CR5 domain was shown to interact with TERT (22). If such an interaction occurs between Est2 and the TWJ in K. lactis, the overexpression of Est2 might compensate for a reduced affinity to the TWJ mutant TER and consequently suppress the telomere phenotype, as shown for the interaction of the telomerase protein Est1 with its RNA binding site in scTER (18). We tested whether any of the telomerase proteins can suppress the severe telomere phenotype of the CS5sub when overexpressed. We cloned the K. lactis orthologs of the S. cerevisiae genes Est1, Est2, Est3 and Ku80, known to be associated with the telomerase complex, and expressed them from their endogenous promoters on high copy-number (2 μ) plasmids [based on pKDU7; (25)]. When overexpressed in the WT TER strain, Est2 and Est3 caused some telomere elongation (120 and 107% of WT telomere length, respectively), and Est1 and Ku80 did not affect telomere length (data not shown). While the overexpression of Est1 and Ku80 did not have any apparent effect in the TWJ mutant, both Est2 and Est3 fully restored normal colony phenotype and elongated the telomeres from 30 to 65% of the wild-type length, a more substantial effect than that observed in the WT strain (data not shown and Figure 4). We did not measure higher TER levels in the strains overexpressing Est2 or Est3 (Figure S6), excluding the possibility that the partial suppression of the phenotype was caused by a stabilization effect on TER. Altogether, these results suggest that the TWJ may interact, physically or functionally, with Est2 and/or Est3.
Figure 4.
Overexpression of Est2 and Est3 partially suppresses the CS5sub mutant phenotype. Genes encoding telomerase proteins were cloned into a high copy-number (2 μ) plasmid and introduced into the CS5sub mutant. Genomic DNA was prepared from the strains at their sixth passage, and telomere length and BclI-repeat incorporation were analyzed by Southern, as described in Figure 2 legend. Only WT hybridization is shown.
DISCUSSION
Telomerase RNA (TER) not only provides a template for telomeric repeat synthesis by the telomerase reverse transcriptase, but also has various other roles in the function of the telomerase RNP (38). However, the extreme divergence of TERs has hindered the functional analysis of this non-coding RNA. Unlike the highly conserved and structured ribosomal RNA, telomerase RNA appears to include small elements conserved in sequence and/or structure, which are located within much larger non-conserved regions, as illustrated by the ‘beads on a string’ model (15). Several such elements were found to be conserved within each group of ciliate, vertebrate and yeast TERs (38). The first element found to be conserved across yeast and vertebrates is a major-grove triple-helix within a pseudoknot structure that is crucial for telomerase function (20,21). Here we report on another such element, which shows conservation in structural parameters, sequence and location within the TER secondary structure, across distant yeast and vertebrate species (Figure 1), suggesting that it contributes a conserved telomerase role.
Using free energy calculations constrained by phylogenetic covariation and experimental data, we predicted secondary structure models for Kluyveromyces TERs (Figure S1A and data not shown). Comparing the Kluyveromyces and Saccharomyces TER models revealed a similar architecture of three long arms and a templating domain at the center, as described previously for the Saccharomyces TERs (8–10) (Figures 1A, S1A and B). The lengths of the corresponding arms vary by up to 225 nt between different TERs, suggesting that long insertions and deletions occurred in these arms during evolution, while preserving the overall secondary structure. No sequence similarity is observed between the Saccharomyces and Kluyveromyces TERs except for seven short sequences (CS1–7) (26), which are located in the same structural context within the secondary structure models of both groups of yeasts. Three conserved elements, CS5a, CS5 and CS6, form a three-way junction (TWJ) not characterized previously (Figure 1). Although a different structure was previously proposed for this arm of the Saccharomyces TERs (8), the remarkable conservation of the CS5a, CS5 and CS6 sequences among budding yeast TERs, as well as phylogenetic covariation among species, support the TWJ prediction (Figures 1, S2 and S3).
Since the position of the TWJ in yeast appeared similar to that of the vertebrate CR4-CR5 domain (Figure 1A and B), we examined the sequence and structure of the CR4-CR5 domain using multiple sequence alignment of 35 vertebrate TERs (6) and covariation-based secondary structure prediction by RNAalifold (30). Strikingly, the CR4-CR5 domain was predicted to fold into a TWJ with similar conformational parameters to those in yeast (Figure 1E and H). Within the structure, 6 nt are absolutely conserved in the corresponding positions of the linkers L1 and L2, and stem 3/P6.1, across all 50 yeast and vertebrate species examined (Figure 1I). Three additional nucleotides are conserved in at least 45 of the species (>90%), and a stretch of U residues and a conserved G in all yeasts are reminiscent of the critical U and G residues conserved in all vertebrates [Figures 1C–H, S2 and S3 and (36)].
To test the importance of the TWJ, we introduced a transition substitution into CS5 of klTER, predicted to disrupt the TWJ structure. Indeed, this mutant telomerase was barely active (Figure 2); while some BclI repeats were incorporated, this activity was apparently not sufficient to maintain stable telomere length, as revealed by telomeric recombination (Figure S5). This indicates that the TWJ is critical for telomerase function in vivo. Next, we examined the various conserved elements in the TWJ by introducing small substitutions into the sequence. The linker between stem 2 and stem 3 (L2 = A), the stretch of U residues, and stem 3 (Figures 1C–H, S2 and S3) were all found to be important for telomerase function, consistent with their high conservation (see Figure 2 and Results section). On the other hand, a substitution mutation in the less conserved linker 3 (L3 = GU) did not affect telomere length. A compensatory mutation in stem 3 restored normal telomere length, confirming the structure prediction. Similarly, in vertebrates, 4-nt mutations in either strand of P6.1 abolished telomerase activity, while the compensatory mutation restored it (36). Closing the internal U loop by replacing the three U residues in CS5 with two A residues only had a moderate effect (Figure 2), suggesting that these U residues may actually be paired, as predicted for the Saccharomyces TERs. In the Kluyveromyces TWJ, they would form U-U base pairs instead of the U-G and U-A predicted in Saccharomyces. Interestingly, the closing base pair in P6.1 is a wobble U/G in 15 vertebrate species (Figure 1E; nt 306 and 310), but is predicted to be a U/U pair in 12 other species (Figure 1H; nt 367 and 371). In other species, other non-conventional base pairs may form (see Figure S3).
We then examined the nature of the defect in the TWJ mutants. We did not observe any correlation between telomere shortening and reduced TER levels (Figure S6), excluding the possibility that the mutations affected the function of the nearby Sm site. Instead, we saw a severe defect in the specific activity of telomerase in vitro (Figure 3), suggesting that the defect lies in the action of the catalytic core rather than in a regulatory function required for the in vivo activity only. We found some sequence aberrations in individual telomeres cloned from the CS5sub mutant (Figure S5), suggesting that in addition to the clearly reduced specific activity, the fidelity of the enzyme may have also been impaired. Finally, we found that overexpression of Est2 or Est3, but not Est1 or Ku80, partially suppressed the severe telomere phenotype of CS5sub. While we have no other indication for the interaction of these proteins with the TWJ, the ability of the catalytic reverse transcriptase Est2 to partially suppress the phenotype is consistent with the possibility that it interacts with the TWJ domain, similarly to the interaction of the vertebrate TERT with P6.1 (39). The specific function of Est3 is still unclear, but its ability to partially suppress the phenotype of CS5sub may provide an opportunity to study its possible role in telomerase assembly or activation.
Here, we describe a critical domain of yeast TER that has features conserved with the vertebrate CR4-CR5 activating domain. In ciliates, stem IV was suggested to have structural and functional roles similar to those of the vertebrate CR4-CR5 domain (23,40). In addition to the previously reported triplex-containing pseudoknot (20,21,39), the TWJ demonstrates that while TERs are highly divergent, they do share conserved elements presumably providing conserved telomerase functions. These elements are embedded in non-conserved RNA fragments, as suggested by the ‘beads on a string’ model (15). The comparative analyses of these functional elements in different organisms will contribute to the understanding of their specific roles in telomerase function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Elizabeth Blackurn, Noa Gil, Ruth Sperling and Nikolai Ulyanov for critical reading of the manuscript, and the members of the Tzfati laboratory for stimulating conversations. This work was supported by the United States-Israel Binational Science Foundation (2005088); and in part by the Israel Science Foundation (676/02) and the German-Israeli Foundation (I-849-253.13/2004). Funding to pay the Open Access publication charges for this article was provided by the United States-Israel Binational Science Foundation (2005088).
Conflict of interest statement. None declared.
REFERENCES
- 1.Bertuch AA, Lundblad V. The maintenance and masking of chromosome termini. Curr. Opin. Cell. Biol. 2006;18:247–253. doi: 10.1016/j.ceb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 3.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 4.Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 5.Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- 6.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 7.Chappell AS, Lundblad V. Structural elements required for association of the Saccharomyces cerevisiae telomerase RNA with the Est2 reverse transcriptase. Mol. Cell. Biol. 2004;24:7720–7736. doi: 10.1128/MCB.24.17.7720-7736.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dandjinou AT, Levesque N, Larose S, Lucier JF, Abou Elela S, Wellinger RJ. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 9.Lin J, Ly H, Hussain A, Abraham M, Pearl S, Tzfati Y, Parslow TG, Blackburn EH. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc. Natl Acad. Sci. USA. 2004;101:14713–14718. doi: 10.1073/pnas.0405879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl Acad. Sci. USA. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu H, McEachern MJ, Dandjinou AT, Tzfati Y, Orr E, Blackburn EH, Lue NF. Telomerase core components protect Candida telomeres from aberrant overhang accumulation. Proc. Natl Acad. Sci. USA. 2007;104:11682–11687. doi: 10.1073/pnas.0700327104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tzfati Y, Fulton TB, Roy J, Blackburn EH. Template boundary in a yeast telomerase specified by RNA structure. Science. 2000;288:863–867. doi: 10.1126/science.288.5467.863. [DOI] [PubMed] [Google Scholar]
- 13.Roy J, Fulton TB, Blackburn EH. Specific telomerase RNA residues distant from the template are essential for telomerase function. Genes Dev. 1998;12:3286–3300. doi: 10.1101/gad.12.20.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livengood AJ, Zaug AJ, Cech TR. Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol. Cell. Biol. 2002;22:2366–2374. doi: 10.1128/MCB.22.7.2366-2374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zappulla DC, Goodrich K, Cech TR. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat. Struct. Mol. Biol. 2005;12:1072–1077. doi: 10.1038/nsmb1019. [DOI] [PubMed] [Google Scholar]
- 16.Seto AG, Umansky K, Tzfati Y, Zaug AJ, Blackburn EH, Cech TR. A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. RNA. 2003;9:1323–1332. doi: 10.1261/rna.5570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- 18.Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 2002;16:2800–2812. doi: 10.1101/gad.1029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 20.Shefer K, Brown Y, Gorkovoy V, Nussbaum T, Ulyanov NB, Tzfati Y. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol. Cell. Biol. 2007;27:2130–2143. doi: 10.1128/MCB.01826-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell JR, Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell. 2000;6:361–371. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 23.Mason DX, Goneska E, Greider CW. Stem-loop IV of tetrahymena telomerase RNA stimulates processivity in trans. Mol. Cell. Biol. 2003;23:5606–5613. doi: 10.1128/MCB.23.16.5606-5613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fulton TB, Blackburn EH. Identification of Kluyveromyces lactis telomerase: discontinuous synthesis along the 30-nucleotide-long templating domain. Mol. Cell. Biol. 1998;18:4961–4970. doi: 10.1128/mcb.18.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartkevi, cbreve, umacr, te D, Siek, sbreve, tele R, Sasnauskas K. Heterologous expression of the Kluyveromyces marxianus endopolygalacturonase gene (EPG1) using versatile autonomously replicating vector for a wide range of host. Enzyme Microb. Technol. 2000;26:653–656. doi: 10.1016/s0141-0229(00)00155-1. [DOI] [PubMed] [Google Scholar]
- 26.Tzfati Y, Knight Z, Roy J, Blackburn EH. A novel pseudoknot element is essential for the action of a yeast telomerase. Genes Dev. 2003;17:1779–1788. doi: 10.1101/gad.1099403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen JL, Greider CW. An emerging consensus for telomerase RNA structure. Proc. Natl Acad. Sci. USA. 2004;101:14683–14684. doi: 10.1073/pnas.0406204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 31.Carter SD, Iyer S, Xu J, McEachern MJ, Astrom SU. The role of nonhomologous end-joining components in telomere metabolism in Kluyveromyces lactis. Genetics. 2007;175:1035–1045. doi: 10.1534/genetics.106.067447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leeper T, Leulliot N, Varani G. The solution structure of an essential stem-loop of human telomerase RNA. Nucleic Acids Res. 2003;31:2614–2621. doi: 10.1093/nar/gkg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leeper TC, Varani G. The structure of an enzyme-activating fragment of human telomerase RNA. RNA. 2005;11:394–403. doi: 10.1261/rna.7222505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antal M, Boros E, Solymosy F, Kiss T. Analysis of the structure of human telomerase RNA in vivo. Nucleic Acids Res. 2002;30:912–920. doi: 10.1093/nar/30.4.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen JL, Opperman KK, Greider CW. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic Acids Res. 2002;30:592–597. doi: 10.1093/nar/30.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theimer CA, Finger LD, Trantirek L, Feigon J. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc. Natl Acad. Sci. USA. 2003;100:449–454. doi: 10.1073/pnas.242720799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr. Opin. Struct. Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Bachand F, Triki I, Autexier C. Human telomerase RNA-protein interactions. Nucleic Acids Res. 2001;29:3385–3393. doi: 10.1093/nar/29.16.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Fender J, Legassie JD, Jarstfer MB, Bryan TM, Varani G. Structure of stem-loop IV of Tetrahymena telomerase RNA. EMBO J. 2006;25:3156–3166. doi: 10.1038/sj.emboj.7601195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen JL, Greider CW. Telomerase RNA structure and function: implications for dyskeratosis congenita. Trends Biochem. Sci. 2004;29:183–192. doi: 10.1016/j.tibs.2004.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.