Abstract
Positive supercoils are introduced in cellular DNA in front of and negative supercoils behind tracking polymerases. Since DNA purified from cells is normally under-wound, most studies addressing the relaxation activity of topoisomerase I have utilized negatively supercoiled plasmids. The present report compares the relaxation activity of human topoisomerase I variants on plasmids containing equal numbers of superhelical twists with opposite handedness. We demonstrate that the wild-type enzyme and mutants lacking amino acids 1–206 or 191–206, or having tryptophane-205 replaced with a glycine relax positive supercoils faster than negative supercoils under both processive and distributive conditions. In contrast to wild-type topoisomerase I, which exhibited camptothecin sensitivity during relaxation of both negative and positive supercoils, the investigated N-terminally mutated variants were sensitive to camptothecin only during removal of positive supercoils. These data suggest different mechanisms of action during removal of supercoils of opposite handedness and are consistent with a recently published simulation study [Sari and Andricioaei (2005) Nucleic Acids Res., 33, 6621–6634] suggesting flexibility in distinct parts of the enzyme during clockwise or counterclockwise strand rotation.
INTRODUCTION
DNA topoisomerases are essential enzymes that exert their important cellular roles by relaxing the superhelical tension that inevitably arises during important DNA metabolic processes (1,2). Due to the double-helical nature of DNA, topological challenges are manifested whenever the two strands are separated to expose the genetic material. During transcription and replication, positive supercoils (over-winding of DNA) are generated in front of and negative supercoils (under-winding) behind the tracking polymerases (3,4). Topoisomerases of the type IB family, including eukaryotic and poxviral topoisomerase I (topo I), relax both positive and negative supercoils by introducing transient single-strand breaks in the DNA duplex (5–8). Using an active site tyrosine as a nucleophile, topo I cleaves the DNA backbone and generates a covalent 3′-phosphotyrosyl linkage and a free 5′-hydroxyl DNA end. This hydroxyl group acts as a nucleophile in the ensuing religation reaction, which releases the covalently bound enzyme and restores the continuity of the DNA strand. Relaxation occurs in the duration of the single-stranded DNA break (9,10).
Human topo I is the cellular target for anti-cancer therapeutics of the camptothecin (CPT) class. These compounds exert their cytotoxic function by inhibiting the ligation step of topo I catalysis, thereby trapping covalent protein–DNA complexes that are converted into permanent DNA damage upon collision with the replication and transcription machineries. In addition to affecting the ligation activity, CPT is known to inhibit the DNA relaxation activity of human topo I. Indeed recent single molecule nanomanipulation studies in combination with in vivo investigations using yeast as a model suggest that CPT at least in part exerts the cytotoxic effects by blocking relaxation of positive supercoils (11).
The DNA relaxation event is difficult to address experimentally. Dynamic details concerning this step of topo I catalysis is available only in two recent single-molecule studies, which demonstrate that type IB topoisomerase-catalyzed DNA relaxation proceeds in a stepwise torque-dependent manner (12) and that CPTs slow down the strand rotation step of human topo I catalysis (11). These results are consistent with the ‘controlled rotation’ mechanism for human topo I as suggested by Champoux and coworkers (13). According to this model, DNA relaxation proceeds by rotation of the free 5′-hydroxyl DNA end around the intact strand in a manner that is restricted by the surrounding protein (13–15). Considering this model the observed CPT inhibition of relaxation can be explained by a drug-induced stalling of the enzyme in a conformation that prevents strand rotation by spatial blockage of the cleaved strand imposed by inflexible enzyme parts (16,17).
The controlled rotation model is in accordance with the ability of human topo I to relax supercoils of both signs (6–8). However, since the direction of rotation will depend on the sign of the supercoils to be removed, the model also implies that different parts of the enzyme may be involved in the control of strand rotation during relaxation of negative versus positive supercoils. At present, experimental data that discriminate between relaxation of supercoils with opposite handedness is available for type II topoisomerases (topo II) (18,19) and at the single molecule level for wild-type topo I (11). A recent computer simulation study by Sari and Andricioaei (20) suggested that relaxation of supercoils of opposite sign by human topo I involves different parts of the enzyme. This issue was not addressed in the published work (11).
To fully appreciate the ‘Sari and Andricioaei model’ (20), it is necessary to understand the 3D buildup of human topo I. The enzyme is composed of four domains defined as the N-terminal (aa 1–206), core (aa 207–635), linker (aa 636–712) and C-terminal domains (aa 713–765) (13–15,21–24) of which the latter contains the active site tyrosine (Tyr-723). Structural data have been obtained only for residues 201–765 and reveals a clamp-like enzyme structure embedding the DNA helix in a central protein pore, with two lobes of the protein each binding their side of the helix. One lobe is formed by aa 215–433 of the core domain (the upper cap), which is connected by a flexible hinge to the ‘lower cap’ composed of aa 434–635, the C-terminal domain and the anti-parallel coiled-coil linker domain (13–15,25).
When bound to DNA, human topo I exhibits a closed conformation in which the two caps bring together two opposable loop regions (‘the lips’) located diametrically opposite to the hinge region. It is evident that this architecture necessitates opening/closing of the protein clamp during binding and release of DNA, which most probably is facilitated by motions within the flexible hinge region (13). As mentioned, the ‘controlled rotation model’, implies that the clamp structure of human topo I presents spatial restrictions to the rotation of the free end of the cleaved DNA strand during the topoisomerization step of catalysis (13,14,24). An important role in this process is thought to be played by charged residues in the linker domain and in the so-called nose cone region of the upper cap, but other regions may be involved as well (13,16,23,24). For instance, the N-terminal domain seems to play a role in the control of strand rotation, although the function of this region is less clear due to the lack of extensive structural data (17,26). However, close interactions of Trp-205 to residues in the flexible hinge have been reported (23) suggesting that Trp-205 may be important for motions within the hinge region, which in turn may be involved in the control of strand rotation. In support of such a mechanism, we previously demonstrated a role of Trp-205 during DNA topoisomerization (17,26). This was manifested by altered relaxation kinetics of N-terminally mutated or deleted enzymes and by the DNA relaxation activity of these mutants being unaffected by CPT.
The importance of flexibility within the human topo I clamp structure during topoisomerization is still a matter of debate. Two individual investigations have addressed the effect of covalently closing the protein clamp by sealing the two opposable lips using disulfide bridging. However, they had contradictory results. One study, published by Carey and coworkers (27), showed an unaltered DNA relaxation rate upon closing the protein clamp. In contrast, Woo et al. (28) found that DNA relaxation activity was inhibited within the locked protein clamp structure. It is still uncertain how these conflicting results should be interpreted. The simplest explanation relies on the different positions of the disulfide bridges used to lock the two protein clamps. Woo et al. (28) used a bridge positioned closer to the catalytic active site than did Carey and coworkers (27). Hence, the space left for strand rotation may have been too narrow in the Woo clamp but sufficient to allow relaxation in the Carey clamp.
The molecular dynamics simulation study by Sari and Andricioaei (20) contributed significantly to the discussion concerning the mechanism of human topo I-mediated topoisomerization by proposing that two different mechanisms account for the relaxation activity of the enzyme depending on the handedness of supercoils in the DNA substrate. Their results, which presuppose the validity of the ‘controlled rotation model’ (13), predict that relaxation of positive supercoils requires the loops of the lips to separate by ∼10–14 Å. Relaxation of negative supercoils, on the other hand, is expected to involve an ∼12 Å stretching of the hinge, likely assisted by residues in the N-terminal domain such as Trp-205 (20). The existing experimental data to some extent support the predictions. The two studies that addressed the effect of locking the topo I clamp by sealing the lips were performed on positively supercoiled DNA (27,28). One of these studies demonstrated inhibition of relaxation upon clamp closure as predicted by the computer simulation (28). Our previous investigations suggesting the involvement of N-terminal regions of the enzyme in the control of DNA relaxation, most likely via interactions to the hinge region were performed on negatively supercoiled DNA (17,26).
In this study, we addressed the validity of the ‘Sari and Andricioaei model’ with respect to the putative functions of N-terminal regions in human topo I during topoisomerization. This is accomplished by comparing the activity of three N-terminally mutated human topo I variants, HT(207–765) (lacking residues 1–206), HT▵(191–206) (lacking residues 191–206) and HT(Trp205Gly) (having Trp-205 replaced with Gly), with that of the wild-type enzyme (HT) on positively and negatively supercoiled DNA substrates. We demonstrate that all four topo I variants remove positive supercoils faster than negative supercoils. Moreover, the introduced mutations render the enzymes insensitive towards CPT in relaxation of negative but not positive supercoils. These results support the proposed model and suggest that residues within the N-terminal region are involved in controlling relaxation of negative but not positive supercoils.
MATERIALS AND METHODS
Enzymes and materials
[γ-32P]ATP (7000 Ci/mmol) was from ICN, oligonucleotides were purchased from DNA technology, and Me2SO and CPT were from Sigma-Aldrich. CPT was dissolved in 99.9% Me2SO at 20 mM and stored at –20°C. All other chemicals used were analytical reagent grade.
Wild-type human topo I (HT) as well as the four mutant human topo I enzymes, HT(Trp205Gly), HT▵(191–206), HT(207–765) and HT(Tyr723Phe) were expressed in the Saccharaomyces cerevisiae top1 null strain RS190 (a kind gift from R. Sternglanz, State University of New York, Stony Brook, NY, USA) and purified as described previously (17). Positively supercoiled pBR322 was prepared by incubating negatively supercoiled plasmid with Archaeoglobus fulgidus reverse gyrase as described (18,19). The average number of superhelical twists present in the DNA plasmids was determined by electrophoretic band counting relative to fully relaxed molecules. Positively supercoiled plasmids contained ∼15–17 positive superhelical twists per molecule (σ +0.035 to +0.039) and negatively supercoiled plasmid molecules each contained ∼15–17 negative superhelical twists (σ ∼ −0.035 to –0.039) (18,19).
Assay for human topo I-mediated relaxation of plasmid DNA
DNA relaxation reactions at processive conditions were carried out in 20 μl reaction volumes in the presence of 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 5 mM MgCl2 and 5 mM CaCl2. The enzyme storage buffer additionally provided 15 mM NaCl, 2.5% glycerol, 0.25 mM Tris–HCl (pH 7.5) and 0.025 mM EDTA to the reaction. DNA relaxation activity at distributive conditions was assayed as described for processive conditions, except that additional NaCl was supplied to the reaction buffer, providing a NaCl concentration corresponding to the salt optima of the individual enzymes as reported previously [150 mM for HT, 100 mM for HT(Trp205Gly) and HT▵(191–206),and 75 mM for HT(207–765) (17,26)]. When indicated, a final concentration of 60 μM CPT dissolved in Me2SO [10% (v/v) final concentration] was added to DNA relaxation reactions at processive conditions. Control reactions with no addition of CPT were supplied with Me2SO [10% (v/v) final concentration].
A total of 100 fmol of pBR322 were incubated with purified recombinant enzyme at 37°C for the indicated time intervals. Reactions were terminated by the addition of SDS to a final concentration of 0.2% (w/v). Samples were subjected to proteolytic digestion by 0.5 μg/ml of proteinase K at 37°C for 30 min prior to separation of reaction products in a 1% agarose gel. DNA was visualized by staining the gel with 0.5 μg/ml ethidium bromide and relaxation products were analyzed using the Bio-Rad Gel Doc-2000 system.
Since positively supercoiled plasmids bind ethidium bromide less efficient than negatively supercoiled plasmids, DNA concentrations were determined by spectrophotometric analysis and confirmed by ethidium bromide staining of plasmids linearized with HindIII.
Assay for human topo I-mediated cleavage of plasmid DNA
DNA cleavage reactions were conducted by incubating 100 fmol of positively or negatively supercoiled pBR322 with each of the four enzymes in a reaction buffer containing 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 5 mM MgCl2 and 5 mM CaCl2. Additionally, 15 mM NaCl, 2.5% glycerol, 0.25 mM Tris–HCl (pH 7.5) and 0.025 mM EDTA was supplied by the enzyme storage buffer. Twenty microliter reactions were incubated at 37°C for 2 min and quenched by the addition of 0.2% (w/v) SDS. Samples were treated with proteinase K as described above and analyzed by separation in a 1% agarose gel containing 0.35 μg/ml ethidium bromide.
Assay for human topo I-mediated DNA binding
Non-covalent DNA binding by HT(Tyr723Phe) to positively and negatively supercoiled plasmid DNA was assessed using a competitive nitrocellulose filter-binding assay. The enzyme was incubated with 2.5 nM of a 5′-radiolabeled 100-mer synthetic DNA duplex substrate in a 20 μl reaction volume containing 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 5 mM MgCl2 and 5 mM CaCl2 and [15 mM NaCl, 2.5% glycerol, 0.25 mM Tris–HCl (pH 7.5) and 0.025 mM EDTA] supplied by the enzyme storage buffer. Binding reactions were carried out at 37°C for 15 min in the absence or presence of indicated amounts of negatively or positively supercoiled competitor pBR322 plasmid ranging from 0 to 20 nM. A nitrocellulose membrane (Protran, Scleicher & Schuell Bioscience) was prepared by treatment with 0.1 μg/ml salmon testis DNA for 1 h at room temperature. Samples were applied to the membrane and filtered in vacuo. Subsequently the membrane was washed four times with a buffer containing 50 mM NaCl, 10 mM Tris–HCl (pH 7.5), 1 mM EDTA 5 mM MgCl2 and 5 mM CaCl2.
The sequence of the synthetic 100-mer DNA substrate is: Top strand: 5′CGA ATT CGC TAT AAT GCC TGC AGG TCG ACT CTA GAG GAT CTA AAA GAC TTA GAA AAA TTT TTG GCT TAA GCA ACA TAT GGT ATC GTC GGA ATT CAA TGA G-3′ and bottom strand: 5′-CTC ATT GAA TTC CGA CGA TAC CAT ATG TTG CTT AAG CCA AAA ATT TTT CTA AGT CTT TTA GAT CCT CTA GAG TCG ACC TGC AGG CAT TAT AGC GAA TTC G-3′. The top strand of the substrate was 5′-radiolabeled prior to hybridization to the bottom strand by applying the T4 polynucleotide kinase reaction using [γ-32P]ATP as the phosphoryl donor. Unreacted ATP was removed by dialysis on a G-50 column. For hybridization 10 pmol of the top strand and 15 pmol of the bottom strand were mixed, heated to 85°C and slowly cooled to room temperature.
The relative amount of radioactive DNA substrate bound by the enzyme was determined as the amount of radioactivity retained on the membrane using a model SF Molecular Dynamics Phosphorimager and quantified using the QuantityOne software (BioRad).
RESULTS
Kinetics of DNA relaxation mediated by HT, HT(207–765), HT▵(191–206), and HT(Trp205Gly) on positively and negatively supercoiled plasmids
At present, only a few studies have addressed the relaxation of positively supercoiled DNA by HT and most of them have been restricted due to limitations posed by the available substrates (6–8). Positively supercoiled plasmids have been generated mainly by the use of intercalating agents or by subjecting a relaxed plasmid to significantly increased temperature. At these conditions, positive supercoils arise as a compensation of local unwinding of the double helix. However, these methods are not readily controlled to produce under- or over-wound DNA with equal numbers of supercoils of opposite sign allowing factors other than topoisomerase action on DNA topology to be ruled out. Due to these limitations, only one recently published report based on single-molecule nanomanipulation (using magnetic tweezers to introduce supercoils of either sign in single DNA fragments) has directly compared the relaxation kinetics of topo I on positively and negatively supercoiled substrates (11).
In the present study, we use enzymatically generated substrates containing equal amounts of positive or negative supercoils to address the influence of supercoil handedness on topo I activity. These substrates are comparable in all parameters other than the sign of the supercoils. Positively supercoiled substrate was prepared by incubating pBR322 with A. fulgidus reverse gyrase. Negatively supercoiled substrate was obtained by purifying pBR322 from Escherichia coli, which naturally maintains all DNA in an under-wound state. The preparation and characterization of both substrates were performed as previously reported by McClendon et al. (19) and will not be described further here. It should be noted, however, that the two substrates each contained 15–17 supercoils of opposite sign.
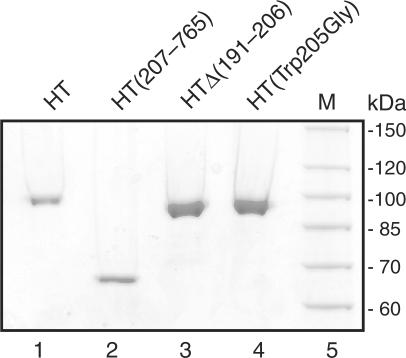
To compare the relaxation of the positively and negatively supercoiled plasmids by HT, HT(207–765), HT▵(191–206), HT(Trp205Gly) each of the enzymes was purified to homogeneity as described previously (17,26). The purified fractions were analyzed in a 10% SDS–polyacrylamide gel and the proteins visualized by Coomassie staining. As evident from Figure 1, only a single band with a mobility corresponding to the expected size of the respective topo I variants could be observed in each of the four enzyme preparations. The identity of the purified proteins as topo I variants were further confirmed by western blot analysis using a human topo I-specific antibody (data not shown).
Figure 1.
Purification of HT, HT▵(191–206), HT(Trp205Gly) and HT(207–765). Purified, recombinant enzymes HT (lane 1), HT(207–765) (lane2), HT▵(191–206) (lane 3) and HT(Trp205Gly) (lane 4) were analyzed by SDS–PAGE and visualized by Coomassie staining. M, protein marker.
The relaxation of positively and negatively supercoiled pBR322 by the four topo I variants was assayed under processive and distributive conditions. Processive conditions were facilitated by a low salt buffer (15 mM NaCl, coming from the enzyme preparations and 5 mM MgCl2) using an enzyme:plasmid ratio of ∼1:1. Under these conditions, each of the four topo I variants have previously been shown to relax negatively supercoiled DNA by completing relaxation of the bound DNA substrate before attacking another substrate (29). Hence, the rate-limiting step of catalysis under these conditions is expected to be mainly the strand rotation while the association/dissociation and cleavage/ligation rates are expected to have less effect on the overall reaction rate. Ideally a molar excess of enzyme compared to plasmid should have been used to limit the effect of association/dissociation on the relaxation rate as much as possible. However, within the technical limitation of this assay, it was impossible to obtain quantitative data on relaxation of positive supercoils using a higher enzyme concentration since under these conditions relaxation of positively supercoiled substrates was completed within <5 s (data not shown).
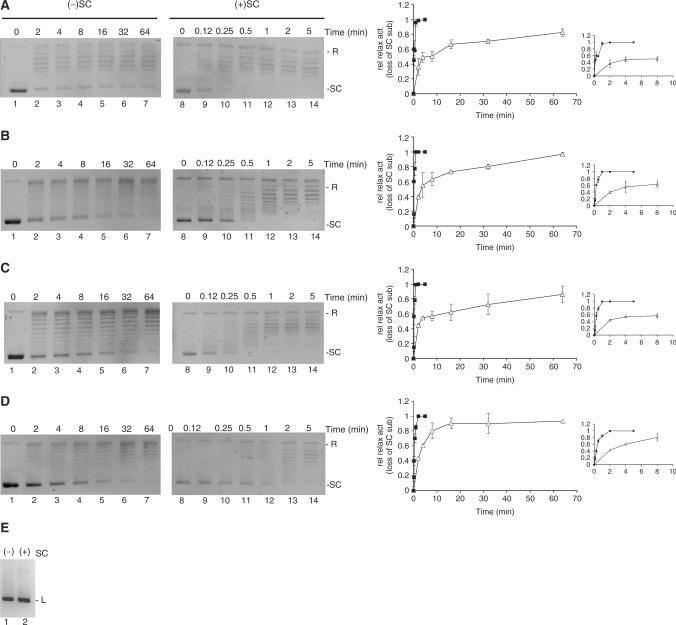
The relative activities of the four topo I variants on negatively supercoiled substrates have been addressed in previously published comparative investigations (17,26). In this study, to ease the comparison of their activities on plasmids with positive relative to negative supercoils, the concentration of each enzyme was adjusted to give similar activity on negatively supercoiled substrates (Figure 2A–D, lanes 1–7). In the experimental setup, 300 ng of either positively or negatively supercoiled pBR322 were incubated with each of the enzymes HT, HT(Trp205Gly), HT▵(191–206) or HT(207–765) for increasing time periods as shown in Figure 2. Reaction products were analyzed in a 1% agarose gel without ethidium bromide. Following electrophoresis, the DNA was visualized by ethidium bromide staining. Note that positively supercoiled plasmids bind ethidium bromide less efficiently than negatively supercoiled plasmids and are, hence, stained less intensively than the corresponding amount of negative supercoiled plasmids. Therefore, to make sure that equal amounts of substrates were used in all experiments, the DNA concentration was measured by spectrophotometric analysis and by ethidium bromide staining of plasmids in the master reaction mixture after linearization by digestion with HindIII (Figure 2E). By comparing lanes 1–7 with lanes 8–14 of Figure 2A–D, it is evident that each of the four topo I variants relaxed the positively supercoiled substrate faster than the negatively supercoiled substrate. Relaxation by all four enzymes proceeded in a processive manner at the investigated assay conditions and no obvious difference in the relaxation mode of the positively and negatively supercoiled substrates was observed for any of the enzymes. Therefore, the relative activities of the enzymes on the two substrates could be estimated rather simply by measuring the amount of fully supercoiled substrate left after the different incubation time periods. Graphical depictions of these measurements are shown in the right panel of Figure 2, where the relaxation activity in each lane is calculated as the loss of fully supercoiled substrate relative to the total amount of plasmids. As evident from the graphs the four human topo I variants HT, HT(Trp205Gly), HT▵(191–206) and HT(207–765) relaxed positively supercoiled plasmids 20–50 times faster than the negatively supercoiled substrates.
Figure 2.
Relaxation activity of HT, HT▵(191–206), HT(Trp205Gly) and HT(207–765) on positively or negatively supercoiled plasmid at processive conditions. Relaxation activities of HT (A), HT(Trp205Gly) (B), HT▵(191–206) (C) or HT(207–765) (D) were assayed at processive conditions by incubating 300 ng negatively (left panels) or positively (right panels) supercoiled pBR322 plasmid with each of the four enzymes at 37°C in an molar enzyme:plasmid ratio of 1:1. Reactions were stopped by addition of 2% (w/v) SDS after indicated periods of time. Following proteinase K digestion, samples were analyzed in a 1% agarose gel stained with ethidium bromide subsequent to electrophoresis. The presented gel picture is a representative of three independent experiments. (E) 300 ng of negatively (lane 1) and positively (lane 2) supercoiled pBR322 plasmid was linearized by HindIII digestion. Graphic depiction of the results are shown in the right panel. Relaxation activity is calculated as the relative loss of supercoiled substrate left after incubation and plotted as a function of incubation time. Filled squares: represent the relaxation of positive supercoils, open triangles: represent relaxation of negative supercoils. SC, supercoiled pBR322. R, relaxed pBR322. L, linearized pBR322.
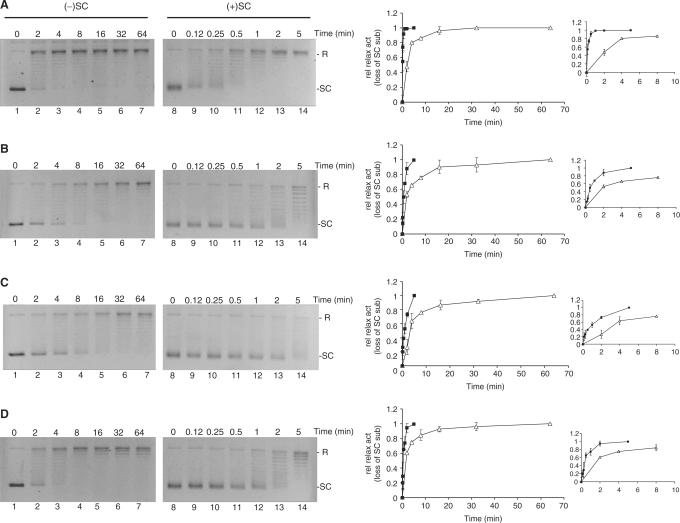
A similar experiment was carried out under distributive conditions which is facilitated by a high salt buffer (corresponding to the optimal salt concentration for each enzyme, see Materials and Methods section) and molar excess of plasmid compared to enzyme. At these conditions the association/dissociation and the cleavage/ligation rates are expected to be rate limiting for DNA relaxation. The results of incubating negatively or positively supercoiled plasmids with each of four topo I variants in a molar ratio of 10:1 under high-salt buffer conditions for increasing time periods are shown in Figure 3. As evident from the graphical depictions of the relaxation activities of HT, HT(Trp205Gly), HT▵(191–206) and HT(207–765) (calculated as described for processive relaxation) all four topo I variants were stimulated 3- to 10-fold by positive supercoils.
Figure 3.
Relaxation activity of HT, HT▵(191–206), HT(Trp205Gly) and HT(207–765) on positively or negatively supercoiled plasmid at distributive conditions. Relaxation activities of HT (A), HT(Trp205Gly) (B), HT▵(191–206) (C) or HT(207–765) (D) were assayed at distributive conditions by incubating 300 ng negatively (left panels) or positively (right panels) supercoiled pBR322 plasmid with each of the four enzymes at 37°C in an molar enzyme:plasmid ratio of 1:10. Except for these alterations, the experiment was carried out as relaxation under processive conditions shown in Figure 2. Right panel is a graphical depiction of the results, where relaxation activity is calculated as the relative loss of supercoiled substrate left after incubation and plotted as a function of incubation time. Filled squares: represent the relaxation of positive supercoils, open triangles: represent relaxation of negative supercoils. SC, supercoiled pBR322. R, relaxed pBR322. L, linearized pBR322.
Effects of DNA supercoil geometry on DNA cleavage and non-covalent binding
To address whether the increased relaxation rate on positively supercoiled substrates observed in this study could simply be ascribed to enhanced cleavage efficiencies of the four human topo I variants on positively relative to negatively supercoiled substrates, a cleavage assay was performed. For this purpose the amount of cleavage complexes trapped by the addition of 0.2% SDS (w/v) after 2 min of incubation of positive or negative supercoiled plasmids with a surplus of HT, HT(Trp205Gly), HT▵(191–206) or HT(207–765) was compared. This type of experiment is hampered by the fact that cleavage by topo I is rapidly followed by relaxation, which converts the supercoiled plasmids to the relaxed form. In an attempt to circumvent this problem, the experiment was carried out for a short time period under processive conditions in which the enzyme tends to remain bound to the original DNA for an extended period of time. We therefore believe that the detergent-trapped cleavage pattern reflects the initial cleavage of supercoiled plasmids as much as possible.
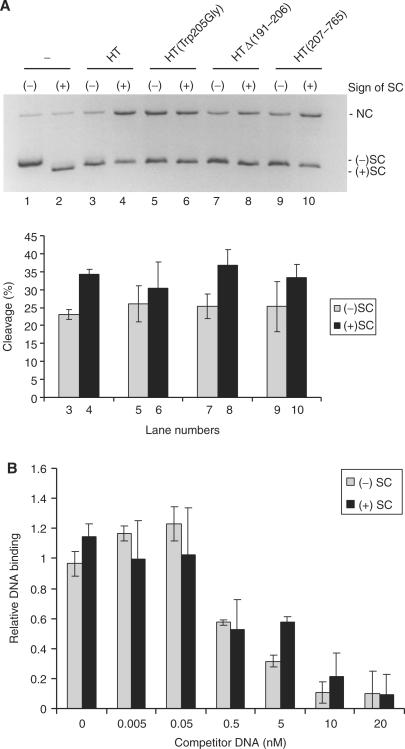
As evident from Figure 4, the four topo I variants maintain a slightly (<2-fold) higher level of cleavage with positively relative to negatively supercoiled substrates (Figure 4A top panel, compare lanes 3 with 4, 5 with 6, 7 with 8 and 9 with 10, for quantifications see lower panel of Figure 4A). This result is consistent (within the scope of expected deviations) with a previous report demonstrating ∼2- to 3-fold stimulation of wild-type topo I cleavage on positively supercoiled substrates (18).
Figure 4.
Cleavage activity of HT, HT▵(191–206), HT(Trp205Gly) and HT(207–765) on positively or negatively supercoiled plasmid. (A) Cleavage activity of the four human topo I variants was assessed by incubating the individual enzyme with negatively (lanes 1,3,5,7 and 9) or positively (lane 2, 4, 6, 8 and 10) supercoiled plasmid at 37°C. After 2 min, reactions were quenched by the addition of 2% (w/v) SDS followed by treatment with proteinase K. Samples were analyzed in a 1% agarose gel containing ethidium bromide. The presented gel picture is chosen as a representative of three independent experiments. Lanes 1 and 2, substrate controls. No enzyme added. Lanes 3 and 4, HT. Lanes 5 and 6, HT(Trp205Gly). Lanes 7 and 8, HT▵(191–206) and lanes 9 and 10, HT(207–765). The lower panel is a graphic depiction showing the cleavage activity calculated as the percentage of the supercoiled substrate converted to the nicked form. (B) Binding of HT(Tyr723Phe) to negatively and positively supercoiled plasmid. The ability of 0–20 nM negatively or positively supercoiled pBR322 to compete with binding of 2.5 nM radiolabeled 100-mer linear substrate DNA by the HT(Tyr723Phe). The amount of bound linear DNA was determined by measuring the radioactivity retained on a nitrocellulose filter. Binding to the radioactive substrate in the presence of each concentration of supercoiled competitor plasmid was normalized relative to binding in the absence of competitor. (–)SC, negatively supercoiled pBR322. (+)SC positively supercoiled pBR322. NC, nicked circular pBR322.
Due to the mentioned limitations of the cleavage assay we also tested the effect of supercoil handedness on non-covalent DNA binding. This was done using a competitive DNA-binding assay (Figure 4B). In this assay, the ability of negatively or positively supercoiled plasmids to compete with a 100-mer radiolabeled synthetic DNA substrate for binding to a human topo I variant lacking the active site tyrosine HT(Tyr723Phe) was monitored on nitrocellulose membranes. HT(Tyr723Phe) was used to avoid cleavage, which is rapidly followed by relaxation and, hence, alteration of the plasmid topology. The enzyme was incubated with radiolabeled substrate and positively or negatively supercoiled plasmid simultaneously for 15 min before the samples were transferred to the membrane that retains protein bound DNA. As evident from Figure 4B, no significant difference in the affinity of HT(Tyr723Phe) for positively or negatively supercoiled substrate was observed.
Effects of CPT on the relaxation of positively or negatively supercoiled substrates by HT, HT(207–765), HT▵(191–206) and HT(Trp205Gly)
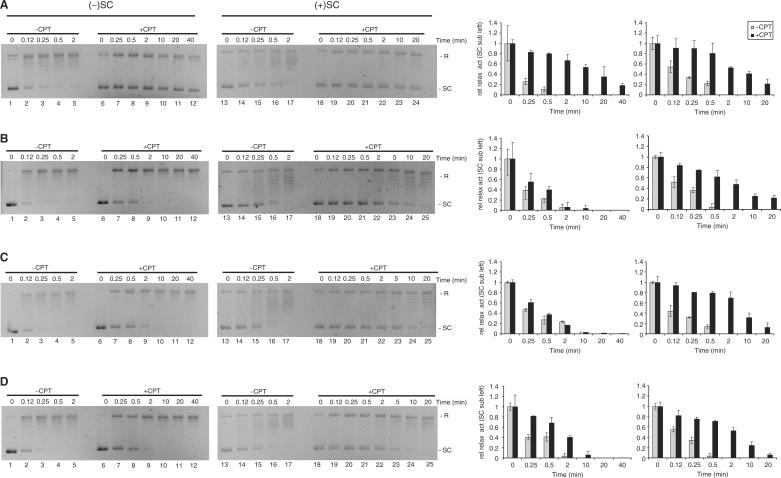
We have previously shown that although the DNA religation activities of HT(Trp205Gly), HT▵(191–206) and HT(207–765) display wild-type sensitivity to CPT, their ability to relax negative supercoils is unaffected by the drug. In contrast, DNA relaxation activity of wild-type human topo I on negatively supercoiled DNA is severely inhibited by CPT (26). To address how CPT influences the relaxation of supercoils with different handedness by the four enzyme variants, we compared the effects of CPT on the relaxation of positively and negatively supercoiled substrates mediated by HT, HT(Trp205Gly), HT▵(191–206) or HT(207–765). First, it was confirmed that DNA ligation by all four enzyme preparations was inhibited by CPT to a comparable level (data not shown). Next, 300 ng of either negatively or positively supercoiled pBR322 were incubated with each of the human topo I variants in the absence or presence of 60 μM CPT for increasing time periods as stated in Figure 5. For comparison, 10% Me2SO was added to reactions performed without CPT to match the conditions in samples containing the drug that was dissolved in Me2SO. Note that in this experiment, enzyme concentrations were adjusted to allow the relaxation rates of either positive or negative supercoils with or without added CPT to be compared. Due to the very different relaxation rates on positively versus negatively supercoiled substrates and the limitations of incubation times posed by drug and enzyme stability, the effects of CPT on the relaxation of either substrate could be allowed only by using ∼10-fold more enzyme for relaxation of negative (enzyme:plasmid ratio 1:1) than positive supercoils (enzyme:plasmid ratio 1:10). Hence, the experiments are not designed to allow relaxation of the two substrates to be compared directly, but only to address the relative effects of CPT.
Figure 5.
Sensitivity of HT, HT▵(191–206), HT(Trp205Gly) and HT(207–765) towards CPT during the relaxation of positive and negative supercoils. Relaxation activity of HT, HT▵(191–206), HT(Trp205Gly) or HT(207–765) was assayed in the absence or presence of 60 μM CPT. (A) Relaxation activity of HT. Lanes 1–5, samples containing 300 ng of negatively supercoiled pBR322 were incubated with HT at 37°C for 0, 0.12, 0.25, 0.5 or 2 min in the presence of 10% Me2SO. Lanes 6–12, same as lanes 1–5 except that 60 μM CPT was added to the reaction mixture and incubation was performed for 0, 0.25, 0.5, 3, 10, 20 or 40 min. Lanes 13–17, same as lanes 1–5 except that the utilized substrate was positively supercoiled. Lanes 18–24, same as lanes 6–11 except that the substrate was positively supercoiled. (B) Same as (A) except that HT was replaced with HT(Trp205Gly). (C) Same as (A) except that HT▵(191–206) was used instead of HT. (D) Same as (A) except that relaxation by HT(207–765) was assayed. All reactions were quenched at the time points indicated in the figure. The right panel is bar charts of the results represented as the relative amounts of supercoiled substrate left after incubation for the indicated time intervals. The presented data is representative of three independent experiments. SC, supercoiled pBR322. R, relaxed pBR322.
Relaxation of negatively supercoiled plasmid DNA by HT was inhibited ∼150 times by the presence of CPT when estimated from the amount of fully supercoiled substrate remaining after incubation for 0.25 or 40 min in the absence or presence of drug, respectively (Figure 5A, compare lane 2 with lane 12, see also right panel for quantifications). HT(Trp205Gly), HT▵(191–206) and HT(207–765) were largely unaffected by CPT when relaxing negative supercoils (Figure 5B–D, lanes 1–12). These results are consistent with our previously published observations (26).
When positively supercoiled plasmid was used as the substrate for relaxation activity the situation changed dramatically. On this substrate all four topo I variants were inhibited ∼40- to 60-fold upon addition of CPT to the reaction mixtures (Figure 5A, compare lanes 16 and 24, and Figure 5B–D, compare lanes 16 and 25). As will be discussed, these results support a model where the regions of the enzyme engaged in control of strand rotation depend on the sign of the supercoils to be relaxed.
DISCUSSION
The present study was prompted by the ‘Sari and Andricioaei model’ for HT, which implies different mechanisms of relaxation depending on the handedness of the supercoils to be removed (20). To address this possibility experimentally, we have compared the abilities of human topo I and variants mutated or deleted in the N-terminal domain to relax plasmid substrates with an equal number of either positive or negative supercoils. As a result of these studies, we find that HT, HT(Trp205Gly), HT▵(191–206) and HT(207–765) all relax positively supercoiled plasmids ∼20–50 times faster than negatively supercoiled plasmid DNA under processive conditions (molar enzyme:plasmid ratio of ∼1:1, low salt buffer) and 3–10 times faster under distributive conditions (molar enzyme:plasmid ratio of ∼1:10, high salt buffer). Cleavage activity was slightly (<2-fold) higher on positively supercoiled compared to negatively supercoiled substrates whereas no difference in binding affinity between the two substrates was observed. The relatively modest stimulation of relaxation by positive supercoils under distributive conditions may be explained by the modest stimulation of cleavage. The more drastic stimulation of relaxation under processive conditions by positive supercoils is unlikely to be accounted for solely by an increased cleavage rate. In surplus of enzyme, the relaxation rate under processive conditions is generally accepted to depend mainly on the speed of strand rotation. Due to technical limitations of our assay, we were not able to perform quantitative analyses using surplus of enzyme for the relaxation of both positive and negative supercoils. However, although not being able to determine the fold stimulation (relaxation of positive supercoils was completed too fast) we did observe significantly faster relaxation of positive versus negative supercoils also when using a molar enzyme:plasmid ratio of 5:1 (data not shown). This argues in favor of strand rotation being faster during relaxation of positively supercoiled DNA. In indirect support of this model is the observation that positive supercoils were relaxed relatively faster than negative supercoils under processive compared to distributive conditions. If increased binding/cleavage rates were the main determinant of the stimulated relaxation by positive supercoils, one would expect a more pronounced stimulation of the reaction under distributive rather than under processive conditions, which contradicts our observations. Based on these arguments we believe that strand rotation is faster during uncoiling of positive relative to negative supercoils. A recently published single-molecule nanomanipulation study where relaxation was monitored on a single DNA fragment in which supercoils were introduced by magnetic tweezers did not reveal a significant difference in the rate of positive or negative supercoil removal by human topo I (11). However, a bias towards a higher velocity in the uncoiling of positive compared to negative supercoils was observed even at the single DNA molecule level and within the second scale time frame of the experiment (11). Since diminutive effects at the single molecule level will be accumulative in our experimental setup performed on a population of substrate and enzyme molecules in a minute scale time frame, we believe that the results obtained by the single molecule technology do not contradict the observations reported here.
Positive supercoils are primarily generated in cellular DNA ahead of DNA tracking processes such as replication and transcription (3,30). During recent years it has been a matter of debate which of the eukaryotic topoisomerases topo I, topo IIα or IIβ are mainly responsible of removing such supercoils (19,31,32). Although necessary for ongoing replication and transcription, topoisomerase action in front of DNA tracking processes also poses a cellular risk, since collision with polymerases convert accidentally trapped cleavage complexes to permanent DNA damage (33–35). Hence, a potent relaxation activity associated with the lowest possible occurrence of cleavage complexes at any given time may be favorable to cells. This consideration leads to the recent suggestion of topo IIα being a safe key player in the removal of topological stress ahead of DNA tracking polymerases since this enzyme shows a >10–fold stimulated relaxation activity and a decreased cleavage activity on positively compared to negatively supercoiled plasmids (18,19). The stimulation of human topo I relaxation by positive supercoils associated with only a slightly enhanced cleavage activity reported here also argues in favor of human topo I being an important cellular factor in the relaxation of positive supercoils. This notion is supported by the recent finding that positive supercoils accumulate in yeast cells upon CPT-induced inhibition of topo I relaxation activity (11).
We have shown previously that the relaxation activities of HT(207–765), HT▵(191–206) and HT(Trp205Gly) on negatively supercoiled plasmid DNA are unaffected by CPT (26). This result was confirmed in the present study. More interestingly, we found that relaxation of positive supercoils by all four enzymes [(HT, HT(Trp205Gly), HT▵(191–206) and HT(207–765)) were inhibited 40- to 60-fold by CPT. In our hands the inhibition of positive supercoil relaxation by HT was less pronounced than was the inhibition of negative supercoil relaxation (150-fold inhibition). This result contradicts recently published data obtained by single DNA molecule and in vivo investigations suggesting a more pronounced drug inhibition of positive supercoil than of negative supercoil removal by human topo I. The reason for this difference is not clear. It may in part reflect different timescales and drug concentrations used in the two experimental setups. For the aim of the present study, however, the important thing to note is the comparable drug inhibition level of all four human topo I variants when relaxing positive supercoils as opposed to the lack of inhibition of HT(Trp205Gly), HT▵(191–206) and HT(207–765) during relaxation of negative supercoils.The inhibitory effect of CPT on DNA relaxation by human topo I is believed to result from a drug induced stalling of the enzyme in a rather rigid conformation that hinders rotation of the cleaved DNA strand around the intact strand due to collision with the ‘frozen’ enzyme (16,17). In favor of this theory the flexible linker domain has only been crystallized in protein–DNA complexes bound by a CPT derivative suggesting that drug binding renders the linker region inflexible (25,36). In further support of the notion that drug inhibition of DNA relaxation is mediated by spatial restrictions posed by the bound enzyme is that various deletions and/or mutations (including the mutations described in this study) of human topo I leave the enzyme insensitive towards CPT in relaxation of negative supercoils without affecting the drug-induced inhibition of ligation (16,17,26). Based on this model for CPT inhibition of relaxation, we believe the different responses of the mutated enzymes towards CPT when relaxing positive versus negative supercoils reflect different mechanisms of strand rotation being engaged in the removal of supercoils of opposite sign. More specifically, the sensitivity of the N-terminally mutated enzymes HT(Trp205Gly), HT▵(191–206) and HT(207–765) towards CPT during relaxation of positively but not negatively supercoiled plasmids suggest a role of Trp-205 and possibly surrounding residues in the control of strand rotation during the removal of negative but not positive supercoils.
The recently published computer simulations of controlled strand rotation by human topo I performed by Sari and Andricioaei suggest stretching of the hinge region in topo I to be prerequisite during relaxation of negative supercoils, and separation of the lips to be required when positive supercoils are removed (20). The experimental data presented here support the simulation model in terms of the involvement of the hinge region in the removal of negative supercoils since both structural and biochemical evidence suggest that Trp-205 and surrounding residues controls motions within the hinge. Hence, based on the available evidence, we suggest that the absence of important N-terminal residues leads to a constitutively stretched conformation of the hinge region allowing a rather uncontrolled clockwise strand rotation during the removal of negative supercoils as implied by the ‘Sari and Andricioaei model’ (20). As suggested by the same authors, stretching of the hinge region may have little or no effect on the conformation of the lips, which they believe to be involved in the control of counterclockwise strand rotation during relaxation of positive supercoils. This in turn is consistent with the CPT sensitivity of HT(Trp205Gly), HT▵(191–206) and HT(207–765) in the relaxation of positive supercoils presented here.
ACKNOWLEDGEMENTS
We thank Maria Vinter for excellent technical assistance. The Carlsberg Foundation; the Danish Research Councils; the Danish Cancer Society; the Novo Nordisk Foundation; the Augustinus Foundation; the Hartmann Foundation; the Aase and Ejnar Danielsen Foundation; the Harboe Foundation; National Institutes of Health (GM33944 to N.O., 5 T32 CA09582 to A.K.M and A.C.G.). Funding to pay the Open Access publication charges for this article was provided by the Danish Research Councils.
Conflict of interest statement. None declared.
REFERENCES
- 1.Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326:414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- 2.Brill SJ, Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988;54:403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- 3.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 5.Shuman S. Vaccinia virus DNA topoisomerase: a model eukaryotic type IB enzyme. Biochim. Biophys. Acta. 1998;1400:321–337. doi: 10.1016/s0167-4781(98)00144-4. [DOI] [PubMed] [Google Scholar]
- 6.Champoux JJ, Dulbecco R. An activity from mammalian cells that untwists superhelical DNA – a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay) Proc. Natl Acad. Sci. USA. 1972;69:143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camilloni G, Di Martino E, Caserta M, di Mauro E. Eukaryotic DNA topoisomerase I reaction is topology dependent. Nucleic Acids Res. 1988;16:7071–7085. doi: 10.1093/nar/16.14.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camilloni G, Di Martino E, Di Mauro E, Caserta M. Regulation of the function of eukaryotic DNA topoisomerase I: topological conditions for inactivity. Proc. Natl Acad. Sci. USA. 1989;86:3080–3084. doi: 10.1073/pnas.86.9.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leppard JB, Champoux JJ. Human DNA topoisomerase I: relaxation, roles, and damage control. Chromosoma. 2005;114:75–85. doi: 10.1007/s00412-005-0345-5. [DOI] [PubMed] [Google Scholar]
- 10.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 11.Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 12.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 13.Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 14.Redinbo MR, Champoux JJ, Hol WG. Structural insights into the function of type IB topoisomerases. Curr. Opin. Struct. Biol. 1999;9:29–36. doi: 10.1016/s0959-440x(99)80005-0. [DOI] [PubMed] [Google Scholar]
- 15.Redinbo MR, Stewart L, Kuhn P, Champoux JJ, Hol WG. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 16.Stewart L, Ireton GC, Champoux JJ. A functional linker in human topoisomerase I is required for maximum sensitivity to camptothecin in a DNA relaxation assay. J. Biol. Chem. 1999;274:32950–32960. doi: 10.1074/jbc.274.46.32950. [DOI] [PubMed] [Google Scholar]
- 17.Lisby M, Olesen JR, Skouboe C, Krogh BO, Straub T, Boege F, Velmurugan S, Martensen PM, Andersen AH, et al. Residues within the N-terminal domain of human topoisomerase I play a direct role in relaxation. J. Biol. Chem. 2001;276:20220–20227. doi: 10.1074/jbc.M010991200. [DOI] [PubMed] [Google Scholar]
- 18.McClendon AK, Osheroff N. The geometry of DNA supercoils modulates topoisomerase-mediated DNA cleavage and enzyme response to anticancer drugs. Biochemistry. 2006;45:3040–3050. doi: 10.1021/bi051987q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIalpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J. Biol. Chem. 2005;280:39337–39345. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 20.Sari L, Andricioaei I. Rotation of DNA around intact strand in human topoisomerase I implies distinct mechanisms for positive and negative supercoil relaxation. Nucleic Acids Res. 2005;33:6621–6634. doi: 10.1093/nar/gki935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart L, Ireton GC, Champoux JJ. The domain organization of human topoisomerase I. J. Biol. Chem. 1996;271:7602–7608. doi: 10.1074/jbc.271.13.7602. [DOI] [PubMed] [Google Scholar]
- 22.Stewart L, Ireton GC, Parker LH, Madden KR, Champoux JJ. Biochemical and biophysical analyses of recombinant forms of human topoisomerase I. J. Biol. Chem. 1996;271:7593–7601. doi: 10.1074/jbc.271.13.7593. [DOI] [PubMed] [Google Scholar]
- 23.Redinbo MR, Champoux JJ, Hol WG. Novel insights into catalytic mechanism from a crystal structure of human topoisomerase I in complex with DNA. Biochemistry. 2000;39:6832–6840. doi: 10.1021/bi992690t. [DOI] [PubMed] [Google Scholar]
- 24.Redinbo MR, Stewart L, Champoux JJ, Hol WG. Structural flexibility in human topoisomerase I revealed in multiple non-isomorphous crystal structures. J. Mol. Biol. 1999;292:685–696. doi: 10.1006/jmbi.1999.3065. [DOI] [PubMed] [Google Scholar]
- 25.Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB, Jr, Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl Acad. Sci. USA. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frohlich RF, Andersen FF, Westergaard O, Andersen AH, Knudsen BR. Regions within the N-terminal domain of human topoisomerase I exert important functions during strand rotation and DNA binding. J. Mol. Biol. 2004;336:93–103. doi: 10.1016/j.jmb.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Carey JF, Schultz SJ, Sisson L, Fazzio TG, Champoux JJ. DNA relaxation by human topoisomerase I occurs in the closed clamp conformation of the protein. Proc. Natl Acad. Sci. USA. 2003;100:5640–5645. doi: 10.1073/pnas.1031537100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woo MH, Losasso C, Guo H, Pattarello L, Benedetti P, Bjornsti MA. Locking the DNA topoisomerase I protein clamp inhibits DNA rotation and induces cell lethality. Proc. Natl Acad. Sci. USA. 2003;100:13767–13772. doi: 10.1073/pnas.2235886100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McConaughy BL, Young LS, Champoux JJ. The effect of salt on the binding of the eucaryotic DNA nicking-closing enzyme to DNA and chromatin. Biochim. Biophys. Acta. 1981;655:1–8. doi: 10.1016/0005-2787(81)90059-9. [DOI] [PubMed] [Google Scholar]
- 30.Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: conformations at the fork. Proc. Natl Acad. Sci. USA. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mondal N, Zhang Y, Jonsson Z, Dhar SK, Kannapiran M, Parvin JD. Elongation by RNA polymerase II on chromatin templates requires topoisomerase activity. Nucleic Acids Res. 2003;31:5016–5024. doi: 10.1093/nar/gkg705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salceda J, Fernandez X, Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25:2575–2583. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsiang YH, Lihou MG, Liu LF. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- 34.Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, Antony S, Meng L, Liao Z, et al. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat. Res. 2003;532:173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 35.Strumberg D, Pilon AA, Smith M, Hickey R, Malkas L, Pommier Y. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 2000;20:3977–3987. doi: 10.1128/mcb.20.11.3977-3987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staker BL, Feese MD, Cushman M, Pommier Y, Zembower D, Stewart L, Burgin AB. Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex. J. Med. Chem. 2005;48:2336–2345. doi: 10.1021/jm049146p. [DOI] [PubMed] [Google Scholar]