Abstract
KLF4 is a transcription factor that is highly expressed in the gastrointestinal tract. Previously we have demonstrated that KLF4 represses HDC promoter activity in a gastric cell line through both an upstream Sp1 binding GC box and downstream gastrin responsive elements. However, the mechanism by which KLF4 inhibits HDC promoter is not well defined. In the current study, by using yeast two-hybrid screening, Tip60 was identified as a KLF4 interacting protein. Further coimmunoprecipitation and functional reporter assays support the interaction between these two proteins. In addition, Tip60 and HDAC7, previously shown to interact with each other and repress transcription, inhibited HDC promoter activity in a dose-dependent fashion. Consistently, knock down of Tip60 or HDAC7 gene expression by specific shRNA increased endogenous HDC mRNA level. Co-immunoprecipitation assays showed that HDAC7 was pulled down by KLF4 and Tip60, suggesting that these three proteins form a repressive complex. Further chromatin immuno-precipitation indicated that all three proteins associated with HDC promoter. Two-hour gastrin treatment, known to activate HDC gene expression, significantly decreased the association of KLF4, Tip60 and HDAC7 with HDC promoter, suggesting that gastrin activates HDC gene expression at least partly by decreasing the formation of KLF4/Tip60/HDAC7 repressive complexes at the HDC promoter.
INTRODUCTION
Kruppel-like factor 4 (KLF4), previously known as gut-enriched Kruppel-like factor, is a transcriptional factor cloned both in mice (1,2) and in humans (3) that binds GC-rich DNA sequences having a consensus core DNA binding sequence of CACCC (1). It is a member of the Kruppel-like factor (KLF) family, which is highly expressed in the gastrointestinal tract and other epithelial tissues (4). KLF4 both activates and represses transcription (5–9). Under most conditions, it inhibits cell proliferation by triggering cell cycle arrest (5,10). Its role as a cell cycle regulator suggests that it may be a tumor suppressor, a role supported by the recent data showing that specific ablation of KLF4 in the gastric epithelium of mice results in premalignant changes (11). However, elevated KLF4 levels have also been linked to cancer. KLF4 mRNA and protein are overexpressed in most squamous–cell carcinomas of the oropharynx (12) and in up to 70% of mammary carcinomas (13). A role for KLF4 as an oncogene has been further supported by the induction of squamous epithelial dysplasia by ectopic KLF4 expression in mice. This paradox was partially resolved by a recent study showing that p21Cip1 status may be a switch that determines the tumor suppressor or oncoprotein function of KLF4 (14). Animal studies have indicated that KLF4 is required for terminal differentiation of goblet cells in colon (11) and that loss of KLF4 in mice causes altered proliferation and differentiation and precancerous changes in adult stomach (11). Given the importance of KLF4 in regulating cell proliferation and differentiation, it is important to delineate how KLF4 regulates gene expression at the molecular level.
Tip60 (Tat-interactive protein, 60 kDa) was originally identified by yeast two-hybrid screening as a protein that interacts with the activation domain of the HIV-1 transactivator protein Tat (15). A specific physical interaction was further demonstrated by the binding of expressed Tip60 to purified Tat in vitro (16). Tip60 belongs to the HAT family, MYST, which includes MOZ, Ybf2/Sas3, Sas2 and Tip60 because of their close sequence similarities (17). Both in vitro and in vivo HAT activities of Tip60 have been demonstrated, as has the role of Tip60 in DNA repair and apoptosis (18). While Tip60 activates gene transcription through its intrinsic HAT activity, it acts as a corepressor for STAT3 by the recruitment of HDAC7 (19). In addition, Tip60 has been implicated in transcriptional repression through interactions with CREB or the transcriptional repressor ZEB (Zinc Finger E Box-binding protein) (16,20). Currently it remains uncertain how Tip60 functions both as an activator and a repressor of transcription.
Histidine decarboxylase (HDC) is the only known enzyme that converts histidine to histamine (21), a bioamine that plays important roles in many physiological processes, including allergy, inflammation, neurotransmission and gastric acid secretion (22–24). In the adult mammal, HDC is highly expressed in enterochromaffin-like (ECL) cells of the stomach, where the HDC activity is tightly regulated by a gut peptide hormone, gastrin (25). HDC promoter activity is upregulated by a variety of stimuli, including gastrin (1), phorbol 12-myristate 13-acetate (PMA) (1,25–27), oxidative stress (28), thrombopointin (29), Helicobacter pylori infection (30,31) and pituitary adenylate cyclase-activating polypeptide (PACAP) (32). Recently, Ying Yang 1 (YY1), a pleiotropic transcription factor which can both upregulate and downregulate gene expression depending on promoter context and cellular environment (33,34), was found to repress the HDC promoter activity through an upstream GC box (35). In addition, KLF4 has been shown to inhibit the HDC promoter activity through three elements: the same upstream GC box and two downstream gastrin responsive elements (36). These studies have confirmed that the Sp1 binding GC box plays an important role in both basal and stimulated HDC gene expression. However, the precise mechanism of how KLF4 represses HDC gene expression remains unclear.
MATERIALS AND METHODS
Yeast two-hybrid screening
Yeast two-hybrid screening was performed according to the manufacturer's instruction (MATCHMAKER Two-Hybrid System; Clontech). Briefly, DNA sequence of KLF4 mRNA (AF022184) that encodes amino acid residues from 170 to 288 aa with EcoRI and BamHI sites added at the 5′ and 3′ ends were cloned in-frame into a yeast two-hybrid system vector pGBKT7 (Clontech, Cat # K1612-1) digested with EcoRI and BamHI. The reporter plasmid was then transformed into yeast strain AH109 to generate the reporter strain with the TRP-selection medium. Background growth of this reporter strain was ablated by titrating with 30 mM 3-aminotrizole (3-AT) (Sigma). Screening was carried out by transforming human lung cDNA library from Clontech (Catalog number HL4044AH) into the reporter strain. Plasmids were retrieved from the yeast colonies growing in SD/-Ade/-His/-Leu/-Trp/X-α-Gal agar plates with 30 mM 3-AT. Plasmids from these clones were then transformed into DH5α bacterial competent cells (Gibco). The cDNA inserts were sequenced and then blasted with GenBank database.
Construction of plasmids
pHA-KLF4 was cloned by PCR with HindIII site at 5′ primer and BamHI site at 3′ primer to amplify KLF4 sequence from KLF4/HisB (36) and by insertion into pEP7/HA/FL vector (a kind gift of Dr John Flemming) predigested with HindIII and BamHI enzymes. Fragments of different KLF4 N-terminal truncation mutants, KLF4AC/HisB (117–471 aa), KLF4BC/HisB (145–471 aa) and KLF4CC/HisB (170–471 aa), were amplified by PCR using KLF4/HisB as the template and cloned in-frame into pcDNA3.1/HisB (named as HisB) using HindIII and EcoRI sites. All PCR-based constructs were confirmed by DNA sequencing. pFLAG-Tip60 and pCMV2-HDAC7 constructs were provided by Dr He-Jin Lee at Parkinson Institute. GAL4BD-Tip60 constructs (pM vector, pM-Tip60 and pM-Tip60M with defective histone acetyltransferase activity) were kindly provided by Dr Thomas C. Sudhof at University of Texas Southwestern Medical Center. pCMX constructs (pCMX vector, pCMX-HDAC1 and pCMX-HDAC7) were kindly provided by Dr Ronald M Evans at Salk Institute for Biological Studies. Different pM/KLF4 constructs were cloned by PCR. The cloning sites in the pM vector are EcoRI and HindIII. All the constructs have been confirmed by DNA sequencing. pCS2+ and GAL4-VP16/pCS2+ were kindly provided by Dr Scott Frazer at California Institute of technology.
Cell culture and transient transfections
AGS and AGS-E cells that stably express CCKB receptor (36) were grown in complete medium (DMEM containing 10% FCS and 100 IU/ml penicillin and 100 μg/ml streptomycin) in a humidified atmosphere (5% CO2). Transient transfections were performed using Superfect® (Qiagen) according to the manufacturer's protocol. Cells were seeded to ∼60% confluence in 12-well plates. Each well was transfected with 0.005 μg of TK renilla luciferase expression plasmid as an internal control, 0.5 μg of different HDC reporter constructs and 0.5 μg of either the different expression plasmids or the empty vector. Three hours after transfection, media was replaced with complete medium. For gastrin treatment, cells were cultured with serum-free medium overnight, and then stimulated the following day with gastrin (10−8 M) for indicated time period. If gastrin treatment was not needed, the cells were cultured in regular medium after transfection.
Generation of shRNA expressing cell lines
DNA oligos encoding a shRNA targeting the Tip60 and the HDAC7 genes were designed using the online software from Invitrogen. The oligonucleotide sequences are: HDAC7 (AF239243) top strand: 5′ CACCGCTGAAGACCTGGAGACAGATCGAAATCTGTCTCCAGGTCTTCAGC 3′, bottom strand: 5′ AAAAGCTGAAGACCTGGAGACAGATTTCGATCTGTCTCCAGGTCTTCAGC 3′; Tip60 (NM_006388) top strand: 5′ CACCGGATGAATGGGTGACGCATGACGAATCATGCGTCACCCATTCATCC 3′, bottom strand: 5′ AAAAGGATGAATGGGTGACGCATGATTCGTCATGCGTCACCCATTCATCC 3′. Top strand oligonucleotide and bottom strand oligonucleotide were annealed and cloned into Block-iT™ U6 RNAi Entry vector using the manufacturer's manual (Invitrogen 25-0663). DNA sequences that encode shRNA targeting the Tip60 and the HDAC7 genes were then moved to a vector pLenti6/Block-iTTM-DEST by recombination using the manufacturer's manual (Invitrogen 25-0677). The oligonucleotide sequences in the final constructs were confirmed by DNA sequencing. The final Tip60 and HDAC7 shRNA encoding plasmids were transfected into AGSE cells, and stable clones that were blasticidin (40 µg/ml) resistant were propagated. Expression levels of RNA were then tested by real-time PCR. A plasmid that contained DNA sequence encoding shRNA targeting the LacZ gene was used to generate a control stable cell line.
Total RNA from AGS-E cells and quantitative RT-PCR
Cells were grown to 70% confluence in 6-well plates. Forty-eight hours later, RNA was isolated using Trizol® Reagent (Invitrogen) in accordance with the manufacturer's protocol. After extraction, 5 μg of total RNA was then used as a template to synthesize the complimentary cDNA using First Strand Synthesis Kit (Invitrogen). The cDNA from this synthesis was then used in regular reverse transcript PCR (RT-PCR) and quantitative real-time PCR (quantitative RT-PCR) analysis using the following primer pairs of human origin: KLF4-5RT (5′ CAAGTCCCGCCGCTCCATTACCAA 3′) and KLF4-3RT (5′ CCACAGCCGTCCCAGTCACAGTGG 3′) for KLF4 gene, HDC5 (5′-AAGATCATCAAGCCGCCTCAGC-3′) and HDC3 (5′-AGCGCACCGTCTTCTTCTTAGT-3′) for the HDC gene, Tip60-RT5 (5′ TCTCAGACCTTGGCCTCCTATCCT 3′) and Tip60-RT3 (5′ CTTCCCCCTCTTGCTCCAGTCC 3′) for Tip60 gene, HDAC7-RT5 (5′ CCATGACGACGGCAACTTCTT 3′) and HDAC7-RT3 (5′ TGCTGCGTCATGTATCCAAAAC 3′) for HDAC7 gene, and GAPDH-5RT (5′ GACATCAA GAAGGTGGTGAAGC 3′) and GAPDH-3RT (5′ GTC CACCACCCTGTTGCTGTAG 3′) for GAPDH gene.
Luciferase assays of HDC reporter constructs
After incubation with or without gastrin, cells were washed with PBS and then incubated with 250 μl of 1 X Passive Lysis Buffer (Promega) for 20 min with constant shaking. Ten microliters of cell lysate was then assayed in a Monolight™3010 luminometer (Pharmingen). Light units of each reporter were divided by those of the internal control, Renilla luciferase, to represent the relative promoter activity.
Coimmunoprecipitation and western blotting analysis
Combination of different constructs including pHA-KLF4, pFLAG-Tip60, pCMV2-HDAC7 and the vector controls were cotransfected into AGS cells as described above. Forty-eight hours after transfection, cells were harvested and lysed with buffer containing 10 mM Tris/HCl (pH 8.0), 300 mM NaCl, 1% Triton X-100, plus proteinase cocktail (Roche, Cat. # 10130500). Cells were sonicated briefly, and the supernatant was collected by centrifugation. Five micrograms of rabbit polyclonal antibodies [anti-KLF4 antibody from Dr Chi-Chuan Tseng at Boston University, anti-Flag antibody from Sigma (F3165)] was used to perform immunoprecipitation. Extensively washed immunoprecipitates were then separated by SDS-PAGE and analyzed by western blotting with rat monoclonal anti-HA antibody (1:1000, Roche, Cat # 1867423) to detect KLF4 and with Polyclonal anti-Flag antibody (1:1000, F3165 from Sigma) to detect Tip60 and HDAC7. To detect protein expression after transfection of series of KLF4 truncation mutant plasmids (pcDNA3.1/His-Myc backbone), ∼100 μg of total protein was separated on 4–20% SDS-PAGE, and the proteins were transferred to PVDF membrane. Western blotting analysis was then performed using anti-myc-HRP antibody (1:5000, Invitrogen, Cat. #46-0709) and anti-α-tubulin antibody (1:1000, Oncogene, Cat. #CP06-100UG) followed by enhanced chemiluminiscence (ECL) detection procedure.
CHIPs
AGSE cells in 10 cm dishes with and without 10−8 M gastrin treatment were fixed with 1% formaldehyde at 37°C for 10 min and then harvested and lysed with 500 µl of lysis buffer [1%SDS, 5 mM EDTA, 50 mM Tris/HCl (pH 8.0), plus protease inhibitor]. After brief sonication (3 times at 10 s each), extracts were precleared by 2 µg of sheared salmon sperm DNA, 20 µl of control IgG and protein A-Sepharose for 2 h at 4°C. Twenty microliter of cleared chromatin extract was used as input control. Five microgram of antibodies (anti-KLF4 antibody from Chi-Chuan Tseng at Boston University, anti-Tip60 antibody from Upstate, Cat # 07-038, anti-HDAC7 antibody from Dr Hung-Ying Kao at Case Western University and anti-acetyl histone 4 antibody from Upstate, Cat # 06-866) were added to the precleared chromatin preparation and incubation was carried out overnight at 4°C, followed by addition of 45 µl protein A-Sepharose, 2 µg of salmon sperm DNA and incubated for another 1 h at 4°C. Sepharose beads were extensively washed sequentially for 10 min in 1 ml each TSE I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris/HCl, pH 8.1, 150 mM NaCl), TSE II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris/HCl, pH 8.1, 500 mM NaCl), buffer III (0.25M LiCl, 1% NP40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris/HCl, pH 8.1) and TE buffer. DNA were eluted with 100 µl elution buffer (1%SDS, 0.1 M NaHCO3) at RT for 10 min and elutes were heated at 65°C overnight to reverse the formaldehyde crosslink. DNA samples were then extracted using the Qiagen PCR purification kit followed by PCR using a pair of primers to amplify a fragment of human HDC promoter (Top strand: 5′ GAACTGAGGGCTCTTTTACG 3′, Bottom strand: 5′ CAGTGTGGGCCCTTTATTTA 3′), intron 1 (Top strand: 5′ TCTCCCAGGTTCAAGCGATTCT 3′, Bottom Strand: 5′ GACTGGGCCTGACCGATTTGT 3′), intron 2 (Top strand: 5′ CCTGGGCAACAAGAGCGAAACT 3′, Bottom strand: 5′ GCTTAGGCCTCTCATCCCAACACT 3′). PCR products were analyzed on 2.0% agarose gels containing ethidium bromide (0.25 mg/l).
RESULTS
Yeast two-hybrid screening to identify KLF4 interacting proteins
To further understand how KLF4 functions at the molecular level in different physiological events such as G1/S cell cycle arrest upon DNA damage and oxidative stress, we identified proteins interacting with KLF4. Yeast two-hybrid screening was utilized with two different regions of full-length KLF4 protein as baits: the N-terminal domain containing the activation domain (aa 1–170) and the middle region containing the repression domain (aa 170–388). A human lung pACT2 cDNA library (from Clontech) was used for screening since KLF4 is expressed in the lung. Because transcriptional factors have intrinsic transcriptional activities, 40 mM 3-AT (3-aminotrizole, a competitive inhibitor of the yeast HIS3 protein used to suppress background growth on medium lacking histidine) was added to minimize background growth during initial screening and the subsequent confirmation step. Among the positive clones obtained from this strategy (Table 1), three were found to contain the in-frame sequence of the Tip60 gene when the middle region of KLF4 was used as the bait. The high frequency of Tip60 from this yeast two-hybrid screening strongly suggests that KLF4 interacts with Tip60.
Table 1.
Positive clones from yeast two-hybrid screening using two fragments of KLF4 as the baits
| Bait | Blast search results from 20 positive colonies (frequency of more than 1 is indicated in parentheses) | Coding region characteristic |
| KLF4A (1–170 aa) | JunD(2), IGKC, Small membrane protein 1, macroglobulin α2, SFTPA2, human upstream binding transcription factor | In-frame |
| c-Jun(2), prothymosin, FKBP 1A, ZFP36, MGAT1, galectin 1, CALM3,DC class II histocompatibility antigen α-chain | Not in-frame | |
| KLF4B (170–388 aa) | Tip60(3), FIM protein, BDR-1, Ubc protein 38197157, PCDH16 | In-frame |
| c-Jun (4), LDLR, SVAP1,DUSP4, APBA3 | Not in-frame | |
Note: in-frame indicates that amino acid coding results of DNA sequence of positive clones are within the same open reading frame of GAL4 activation domain in the cloning vector. Not in-frame is the opposite.
KLF4 interacts with Tip60 by coimmunoprecipitation assay
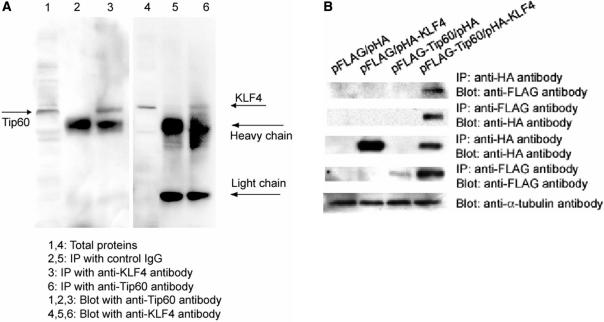
To confirm the interaction between KLF4 and Tip60 coimmunoprecipitation assays were performed. Total proteins from AGS cells, where both KLF4 and Tip60 were expressed, were immunoprecipitated with anti-KLF4 and anti-Tip60 antibodies followed by western blotting analysis using anti-Tip60 and anti-KLF4 antibodies, respectively. As shown in Figure 1A, Tip60 were pulled down by anti-KLF4 antibody (lane 3), and KLF4 was pulled down by anti-Tip60 antibody (lane 6) (a very weak band where KLF4 migrates in lane 5 was observed, but the signal of the band is several magnitudes weaker than that of the band in lane 6). In addition, pHA-KLF4 and pFLAG-Tip60 plasmids were cotransfected into AGS cells. Forty-eight hours after transfection, cells were harvested and the total proteins were extracted. Anti-FLAG and anti-HA antibodies were used to pull down Tip60 and KLF4, respectively. The immunoprecipitates were separated by SDS-PAGE gel followed by western blotting analysis using anti-HA antibody and anti-FLAG antibody. As shown in Figure 1B (first panel), KLF4 successfully pulled down Tip60. Consistently, Tip60 also pulled down KLF4 (Figure 1B, second panel). These coimmunoprecipitation data support the physical interaction between KLF4 and Tip60 from the yeast two-hybrid screening results. To further study the interaction between KLF4 and Tip60, their subcellular localizations were examined by immunostaining, using transient transfection assays in AGS cells. When overexpressed, both KLF4 and Tip60 were predominantly colocalized in the nucleus (Supplementary Figure 1), which is consistent with the co-immunoprecipitation data and raises the possibility that these proteins interact to regulate gene expression.
Figure 1.
KLF4 interacted with Tip60 by coimmunoprecipitation assay. (A) Endogenous KLF4 interacted with endogenous Tip60. Total proteins were immunoprecipitated with control IgG and anti-KLF4 and anti-Tip60 antibodies. The immunoprecipitates were then probed with anti-Tip60 and anti-KLF4 antibodies. Total proteins were loaded as a control (B). KLF4 interacted with Tip60 in vitro. N-terminally HA-tagged KLF4 expression plasmid (pHA-KLF4) and C-terminally FLAG-tagged Tip60 plasmid (pFLAG-Tip60) with the respective vectors were cotransfected in combination into AGS cells. Total protein extracts were prepared 48 h after transfection. Anti-HA antibody or Anti-FLAG antibody conjugated agarose beads were added to protein extracts. After washing, immunoprecipitated proteins were separated on SDS-PAGE gel and then probed with different antibodies to detect interaction between KLF4 and Tip60.
Functional interactions between KLF4 and Tip60
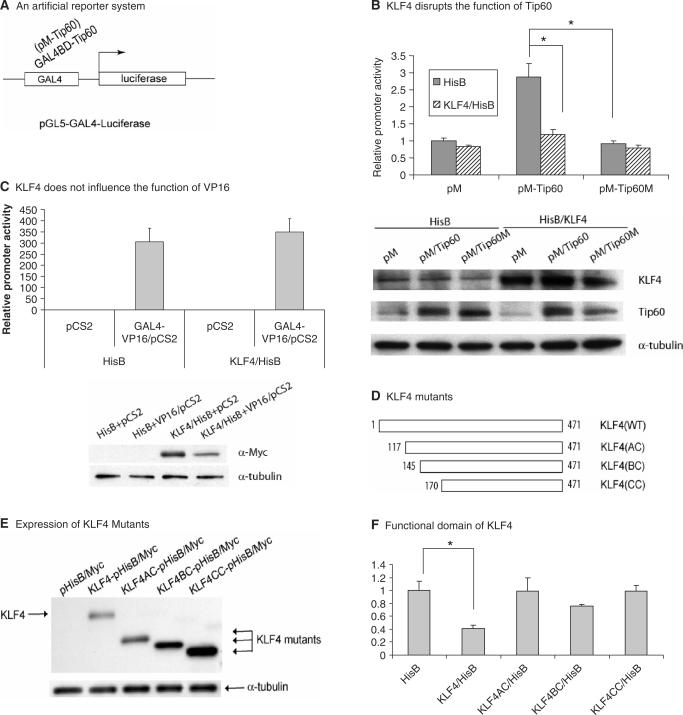
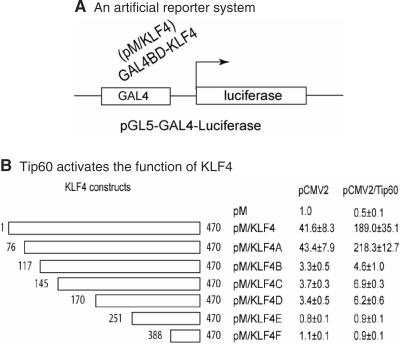
If, as our earlier studies indicated, KLF4 and Tip60 interact, it is possible that KLF4 influences Tip60-mediated transcriptional regulation. To test this hypothesis, an in vitro artificial reporter system was used, in which Tip60 was recruited to the proximal promoter region by the GAL4 DNA binding domain in the construct (Figure 2A). The hypothesis is that because Tip60 has intrinsic transcriptional activity, its recruitment to the proximal promoter should result in the transcriptional regulation. As shown in Figure 2B, Tip60 activated the artificial reporter activity because of its intrinsic histone acetyltransferase (HAT) activity (about 3-fold increase). In the presence of KLF4, Tip60-mediated transcriptional activation was completely abrogated. A mutant Tip60, that does not have the HAT activity, did not activate the transcription in the same assay, and KLF4, as expected, did not have any effect on this mutant. Expression of KLF4 had no effect on Tip60 stability at the protein level (bottom panel of Figure 2B). As a negative control, overexpression of KLF4 did not influence the activation activity of VP16 (Figure 2C) using the same reporter system. These data suggest that KLF4 specifically disrupted Tip60-mediated transcriptional activation. Furthermore, while full-length KLF4 disrupted Tip60-mediated transactivation, KLF4 N-terminal truncation constructs with amino acids 117–470, 145–470 and amino acids 170–470 (Figure 2D–F) did not show significant disruption in the same assay. This suggests that the functional domain in KLF4 that mediated this disruption is located between amino acids 1–117. In a parallel experiment using a similar reporter system where KLF4 was recruited to the proximal promoter (Figure 3A), overexpression of Tip60 activated KLF4-mediated transcriptional activation for ∼4- to 5-folds (Figure 3B). When the N-terminal KLF4 was truncated from 76 to 117 aa, transactivation activity decreased from 43- to 3-folds that is consistent with published data showing a transactivation domain was located between 91 and 117 aa (3). Interestingly, while Tip60 remained its ability to activate KLF4-mediated transactivation (about 5-folds increase) when N-terminal KLF4 was truncated to 76 aa, it had minimal activation (about 1.4-fold) when N-terminal KLF4 was further truncated to 117 aa, suggesting that the region between 76 and 117 aa in KLF4 is required for Tip60 to activated KLF4-mediated transcriptional activation. Expression of Tip60 at a protein level by western blotting analysis did not change in this reporter assay (data not shown).
Figure 2.
KLF4 disrupts Tip60-mediated transcriptional activation. (A) A reporter system (pGL5-GAL4-Luciferase) was shown where Tip60 was recruited to the proximal promoter region by its Gal4 fusion domain to modulate transcription. (B) KLF4 disrupted Tip60-mediated transcriptional activation. Plasmids expressing KLF4 (KLF4/HisB) and GAL4-fused wild-type and mutant Tip60 (pM-Tip60 and pM-Tip60M) with the respective vectors were cotransfected in combination into AGS cells. Cells were lysed 48 h after transfection followed by luciferase assay as described in Materials and Methods section. Means ± SD for three independent experiments were shown. Statistical difference (P < 0.05) of the relative promoter activities of the transfection experiments was indicated by a star (*). The bottom panel showed the protein levels by western blotting analysis in the same transfection setting using anti-KLF4, anti-Tip60 and anti-α-tubulin antibodies. (C) KLF4 did not influence the function of VP16. As described in panel B, similar transfection experiments were performed using KLF4 and VP16 expressing constructs to detect the effect of KLF4 on VP16-mediated transcriptional activation. The bottom panel showed the protein levels of KLF4 (Myc epitope-tagged) and α-tubulin. (D) Different KLF4 truncation mutants were shown with the starting position of amino acids marked with numbers. (E) Expression of different KLF4 truncation mutants was shown. Total proteins were extracted 48 h after transfection followed by western blotting analysis using anti-Myc antibody. (F) Identification of a functional domain in KLF4. A pM-Tip60 construct was cotransfected with different KLF4 mutant constructs as shown in panel D and the reporter into AGS cells. The relative reporter activities with different KLF4 constructs were measured as described above.
Figure 3.
Tip60 activates KLF4-mediated transcriptional activation. (A) As described in Figure 2A, a similar artificial promoter system was used where KLF4 was recruited to the proximal promoter region. (B) Tip60 and vector were cotransfected with different pM/KLF4 constructs to test the function of Tip60 in KLF4-mediated transcriptional regulation. The relative promoter activities were normalized with the control KLF4 plasmid (pM) and with control Tip60 plasmid (pCMV2).
Tip60 and HDAC7 inhibit the HDC promoter activity
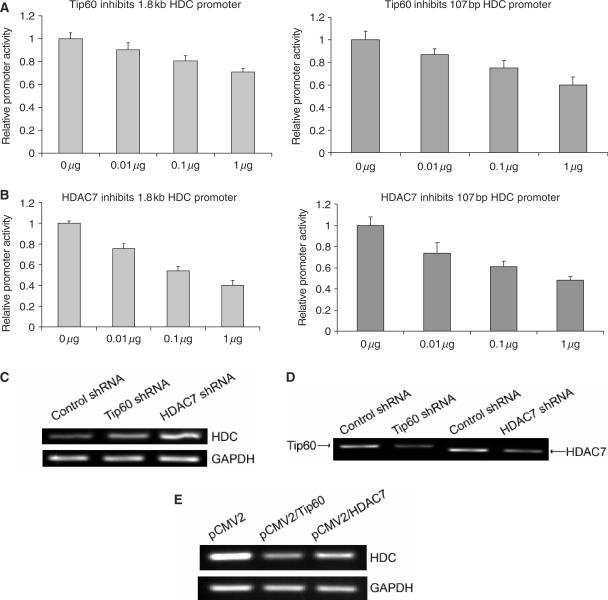
Previously, Tip60 was reported to function as a co-repressor for STAT3 with another protein HDAC7 (19). Tip60 and HDAC7 may therefore function similarly for KLF4 in regulation of HDC promoter. To test this possibility, different amounts of either Tip60 or HDAC7 expressing constructs were cotransfected with either a minimal 107 bp HDC promoter reporter construct or a full length 1.8 kb HDC promoter reporter construct. Relative promoter activities induced by transfection of plasmids expressing Tip60 and HDAC7 were then measured 48 h after transfection. As shown in Figure 4A and B, both Tip60 and HDAC7 dose-dependently inhibited HDC promoter activity by this transient transfection assay, where HDAC7 showed higher potency than did Tip60. To confirm the down-regulation of HDC promoter by Tip60 and HDAC7, stable cell lines that specifically express shRNA against Tip60 and HDAC7 were generated (Figure 4D). Consistent with the in vitro transfection data, endogenous HDC mRNA levels in both Tip60 shRNA and HDAC7 shRNA expressing cell lines were elevated (Figure 4C). In parallel, endogenous HDC mRNA levels were downregulated upon exogenous expression of Tip60 and HDAC7 (Figure 4E). These data indicate that HDC gene expression is repressed by Tip60 and HDAC7.
Figure 4.
Tip60 and HDAC7 inhibited HDC promoter activity. (A) Tip60 dose-dependently inhibited both the full length (1.8 kb, left panel) and minimal (107 bp, right panel) HDC promoter. HDC promoter reporter constructs were cotransfected with increasing amount of Tip60 expressing construct into AGS cells. Forty-eight hours after transfection, cells were lysed, luciferase activities were measured and relative promoter activities were calculated as described in Materials and Methods section. (B) HDAC7 dose-dependently inhibited HDC promoter activity. The experiments were performed similarly as described in panel A. In both A and B, the total amount of plasmids in each transfection was 1 μg. The space besides Tip60 or HDAC7 construct was filled with the empty vector. For all these experiments, means ± SD for three independent experiments were shown. (C) Endogenous HDC mRNA levels were elevated by knocking down Tip60 and HDAC7 gene expression. Total RNA was extracted from a control cell line stably expressing a control shRNA, Tip60 shRNA and HDAC7 shRNA stably expressing cell lines. Reverse transcription was performed to generate cDNA. RT-PCR was then followed to amplify a fragment of HDC cDNA and a fragment of GAPDH cDNA. PCR products were separated on agarose gel and visualized under UV light. (D) Expression of Tip60 and HDAC7 in respective cell lines was shown that stably expressed Tip60 or HDAC7 shRNA. RT-PCR was performed as described in panel C to amplify a fragment of Tip60 and HDAC7 cDNA. (E) Endogenous HDC mRNA levels were downregulated by overexpression of exogenous Tip60 and HDAC7. Total RNA was extracted from AGS cells that transfected with vector (pCMV2), Tip60 (pCMV2/Tip60) and HDAC7 (pCMV2/HDAC7). RT-PCR was performed as described above. One representative of each shRNA cell line (control shRNA, Tip60 shRNA and HDAC7 shRNA), and one representative of Tip60 and HDAC7 overexpression experiments was shown from three independent experiments.
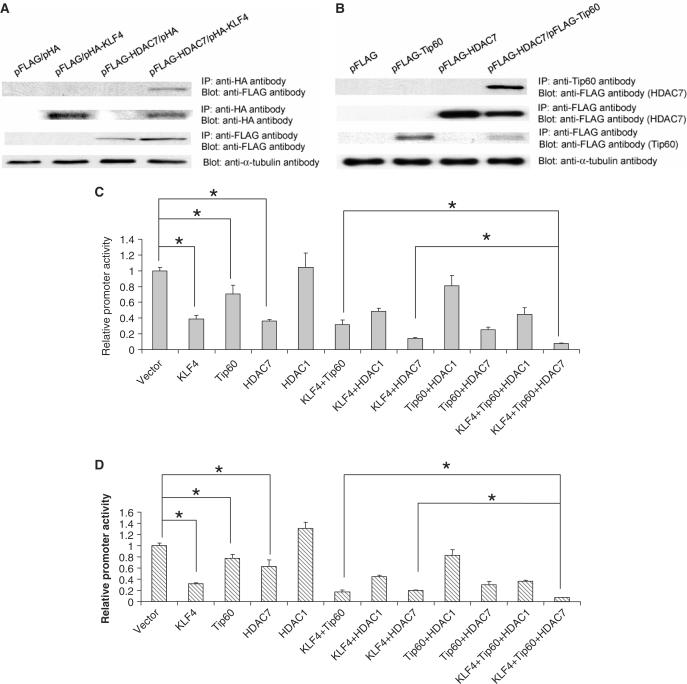
Tip60, KLF4 and HDAC7 form a complex and cooperatively repress the HDC promoter activity
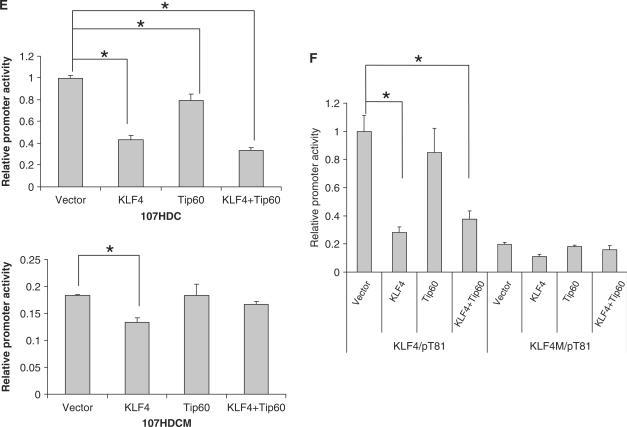
Previously, Tip60 has been reported to form a repressive complex with HDAC7 and function as a corepressor of STAT3. We postulated that Tip60 and HDAC7 may function similarly as corepressors of KLF4 in regulation of HDC gene expression. To test this possibility, physical interactions between KLF4, Tip60 and HDAC7 were examined in an immunoprecipitation assay. As shown in Figure 5, HDAC7 were pulled down by both KLF4 (Figure 5A, top panel) and Tip60 (Figure 5B, top panel). Together with coimmunoprecipitation between KLF4 and Tip60 (Figure 1), the results suggest that these three proteins form a complex. To test the functional importance of this complex, different combinations of KLF4, Tip60 and HDAC7 expression constructs with their respective vectors were cotransfected with the 1.8 kb and the 107-bp human HDC promoter reporter construct into AGS cells to examine their effect on the reporter activity. As seen in Figure 5C and D, while the single or double combination of Tip60, KLF4, and KLF4 showed inhibitory activities in regulating the human HDC reporter, the combination of these three proteins further inhibited the promoter in the same transient cotransfection assays, suggesting that Tip60, KLF4 and HDAC7 cooperatively repress the activity of HDC promoter. In addition, it appears that the major KLF4 response element in HDC promoter (Sp1 binding GC box) is important for Tip60 to inhibit the promoter activity, since the mutation of this site abrogated the inhibitory effect of Tip60 alone and the combination effect of KLF4 and Tip60 (Figure 5E, bottom). As a control, Tip60 alone did not have any inhibitory effect on another KLF4 responsive reporter (KLF4/pT81). Instead, it slightly relieved the inhibitory effect of KLF4 (Figure 5F). These data suggest that KLF4 and Tip60 cooperatively repress transcription of a subset of genes.
Figure 5.
KLF4, Tip60 and HDAC7 formed a complex and cooperatively inhibited HDC promoter activity. (A) HDAC7 was pulled down by KLF4. Coimmunoprecipitation was performed using anti HA-tag antibody after transient transfection of pHA-KLF4 and pFLAG-HDAC7 constructs into AGS cells as described in Methods and Materials section. Anti Flag-tag antibody was used to detect HDAC7 expression in the following western blotting analysis. (B) HDAC7 was pulled down by Tip60. Similar to A, except anti-Tip60 antibody was used for immunoprecipitation. (C) KLF4, Tip60 and HDAC7 cooperatively repressed 1.8 kb HDC promoter activity. KLF4, Tip60, HDAC7 expressing plasmids with their respective vectors were cotransfected into AGS cells in combination of three constructs with the full length 1.8 kb HDC promoter reporter. Forty-eight hours after transfection, cells were lysed, luciferase activities were measured and relative promoter activities were calculated as described in Materials and Methods section. HDAC1 construct was also used as a control for HDAC7. (D) KLF4, Tip60 and HDAC7 cooperatively repressed 107 bp HDC promoter activity. Similar to panel C except the minimal 107 bp human HDC promoter reporter was used instead of the full length 1.8 kb HDC reporter. (E) Tip60 lost its inhibitory effect when a major KLF4 responsive element (Sp1 binding GC box) was mutated in the HDC promoter. Cotransfection experiments were formed similarly as described above with wild-type 107 bp human HDC promoter reporter (top) and mutant reporter (bottom) with mutations in the upstream GC box. (F) Tip60 did not have any effect on a KLF4 responsive artificial promoter. Cotransfection experiments were formed similarly with an artificial reporter KLF4/pT81 with a KLF4 binding site and various combinations of KLF4 and Tip60 overexpression constructs. A mutant reporter (KLF4M/pT81) with mutations in the KLF4 binding site was also used in the experiment. Means ± SD for three independent experiments were shown for reporter assays (panels C–F), and major statistical difference (P < 0.05) of the relative promoter activities of the transfection experiments was indicated by a star (*).
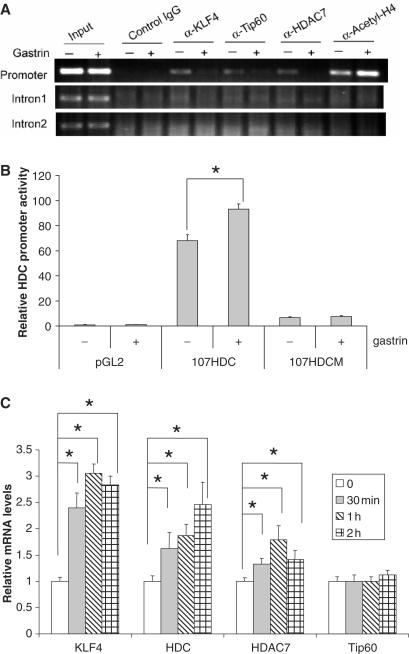
KLF4, Tip60 and HDAC7 bind to HDC promoter by chromatin immunoprecipitation (CHIP) assays
To investigate the in vivo regulation of HDC by KLF4, Tip60 and HDAC7, in vivo binding of these proteins with the promoter were examined by CHIP. Control rabbit IgG and antibodies of KLF4, Tip60, HDAC7 and acetyl-H4 raised in rabbits were used to precipitate DNA–protein complexes after formaldehyde fixation and protein extraction from AGSE cells. DNA samples were extracted from the precipitates and were used as templates in PCR with primer pairs to amplify a fragment of the proximal HDC promoter region (promoter), a fragment in the first intron (Intron 1), and in the second intron (Intron 2) of the HDC gene. As expected, a fragment in the HDC promoter was amplified when anti-KLF4, Tip60 and HDAC7 antibodies but not when control IgG was used, suggesting that all three proteins bind the HDC promoter in vivo (Figure 6A). Two hours of gastrin treatment, which upregulates HDC gene expression (Figure 6B and C), significantly decreased the association of three proteins with the promoter. Consistent with the activation of transcription at the HDC promoter, histone acetylation levels were increased after gastrin treatment as demonstrated by increased binding of acetylated H4 with the promoter. In contrast, binding of KLF4, Tip60, HDAC7 and acetyl-H4 to the intron 1 and 2 was not detected by the same assay, suggesting that all these three proteins bind specifically to the HDC promoter and the binding is regulated by the status of active transcription. Gastrin-induced dissociation of KLF4, Tip60 and HDAC7 with the promoter appears not to occur at the RNA level, since no decreased levels of mRNA of KLF4, Tip60 or HDAC7 were seen by quantitative real-time PCR, instead mRNA levels of KLF4 and HDAC7 were upregulated by gastrin treatment (Figure 6C).
Figure 6.
KLF4, Tip60 and HDAC7 bind to HDC promoter by chromatin immunoprecipitation assay. (A) Overnight starved AGSE cells were treated with 10−8 M gastrin for 2 h. Cells were then fixed with formaldehyde followed by protein extraction as described in Materials and Methods section. Antibodies against KLF4, Tip60, HDAC7, acetyl-H4 plus control IgG were used to precipitate DNA–protein complexes. Purified DNA samples from precipitated DNA–protein complexes were used as template for PCR to amplify a fragment of HDC promoter, intron 1 and intron 2. (B) Gastrin treatment increased 107HDC promoter activity. Vector (pGL2), 107HDC reporter and 107HDCM reporter were transfected into AGSE cells. Before harvesting, cells were treated with 10−8 M gastrin from 2 h. Then, luciferase assays were performed as described. (C) mRNA levels of KLF4, HDC, Tip60 and HDAC7 after 10−8 M gastrin treatment were shown. Total RNA was extracted after gastrin treatment at different time points. Reverse transcription and follow-up quantitative RT-PCR was performed. Relative mRNA level was calculated as described in Materials and Methods section. Means ± SD for three independent experiments were shown, and statistical difference (P < 0.05) of the relative mRNA levels was indicated by a star (*).
DISCUSSION
In this study, we identified in yeast two-hybrid screening Tip60 as a KLF4-interacting protein. Additional cotransfection assays and coimmunoprecipitation analysis suggested that Tip60 and HDAC7 form a complex with KLF4 to regulate HDC promoter activity. The in vivo association of a repressive complex on the HDC promoter comprising KLF4, Tip60 and HDAC7 was further supported by chromatin immunoprecipitation. Finally, gastrin treatment, which activates HDC gene expression, disrupted the association between KLF4, Tip60 and HDAC7 with HDC promoter. Thus, our data provide strong evidence that Tip60 and HDAC7 act as corepressors of KLF4 in regulation of HDC gene expression.
The physical interaction between Tip60 and KLF4 is not surprising given their share roles in DNA damage and apoptosis and the fact that both proteins could be targeted to proteasome-mediated degradation. The KLF4 protein is an unstable protein with a half life of ∼120 min. Its levels in quiescent cells are high, but decrease rapidly after cells are treated with serum. Serum-induced protein degradation is partially mediated by the proteasome, since the application of a proteasome inhibitor partially inhibits the degradation (37). Biologically, KLF4 modulates p53-dependent G1/S cell cycle arrest in response to DNA damage upon γ–irradiation (10). Inhibition of p53 gene expression by KLF4 (14) further strengthens the functional relationship between KLF4 and p53, although the mechanism is not entirely clear. Similarly, Tip60 is targeted by Mdm2 to a proteasome-mediated degradation pathway and also accumulates after UV irradiation (38). Furthermore, it has been established that Tip60 plays a dual role in the p53 pathway. Under normal conditions, Tip60 contributes maintenance of basal levels of p53 by interfering with its degradation; after DNA damage, Tip60 functions as p53 co-activator (39). Our results showing a physical interaction between Tip60 and KLF4 provide additional possibility for how DNA damage pathway could be controlled by different proteins and how these proteins synchronize to protect the cells. It is possible that under normal condition, KLF4 and Tip60 form a complex to regulate p53 stability. This hypothesis is supported by the physical interaction between KLF4 and Tip60 (Figure 1) and between KLF4 and p53 (1), and p53 degradation mediated by Mdm2 and Tip60 (38,39).
Tip60 possesses the intrinsic histone acetyltransferase (HAT) activity, and recruitment of Tip60 to the proximal promoter results in the transcriptional activation of an artificial promoter (Figure 2B). This is consistent with the notion that increased acetylation of histone tails is associated with a transcriptional active state. However, along with our findings, several reports have shown that Tip60 can also repress transcription. Because of the intrinsic HAT activity, it is difficult to understand why Tip60 inhibits transcription. There are several possibilities. First, even though Tip60 can acetylate histone tails with lower efficiency (34), the true substrates of Tip60 may be some other cellular proteins that in turn regulate transcription. For example, Tip60 has been shown to acetylate both ATM (ataxia telangiectasia), a protein kinase that regulates the cell's response to DNA damage (40), and androgen receptor (41). Second, Tip60 may recruit transcriptional corepressors to repress transcription. For example, Tip60 interacts with HDAC7, a protein with histone deacetylases activity that is associated with transcriptional repression, and functions as a corepressor of STAT3 (19). The physical interaction between KLF4 and HDAC7 is consistent with this hypothesis. More importantly, the interacting domain in Tip60 was located in the region that is also required for its HAT activity (19). These data suggest that HAT domain in Tip60 is not used to acetylate any substrates; instead, it is responsible for the recruitment of repressor such as HDAC7 to inhibit transcription. Our data showed that HAT domain of Tip60 is required for its transcriptional activation in an artificial reporter system (Figure 2B). KLF4 completely abrogated Tip60-mediated transcriptional activation to a level that is similar to that mediated by HAT mutant Tip60. This observation raises an intriguing possibility that KLF4 inhibits the HAT activity of Tip60 by protein–protein interaction with this HAT (MYST) domain. Different functions of KLF4 in two artificial systems also appear confusing. In Figure 2B, KLF4 inhibits Tip60-mediated transcriptional activation and in Figure 3B, Tip60 activates KLF4-mediated transactivation. While an artificial promoter and tagged protein constructs were used in both systems that may complicate our interpretation, it is likely that they reflect different roles of KLF4 in transcription: as a transcriptional activator and as a transcriptional repressor, according to the cellular context. For example, KLF4 has been show to inhibit the transcription of p53 in human embryonic kidney cells (Phoenix cells) (14). However, a more recent report indicated that KLF4 activates p53 transcription in vascular smooth muscle cells (42). The fact that the N-terminal domain of KLF4 between aa 76 and 117 is required for Tip60 to activate KLF4-mediated transcriptional activation is also interesting (Figures 2 and 3), since the interaction domain in KLF4 with Tip60 is the middle region between aa 117 and 388 from our initial yeast two-hybrid screening. It is possible that Tip60 physically interacts with KLF4 at the middle region, and activates KLF4-mediated transcriptional activation through the N-terminal domain between aa 76 and 117. Further mapping of interaction domains in Tip60 and KLF4 is needed to understand how KLF4 and Tip60 function coordinately to regulation transcription.
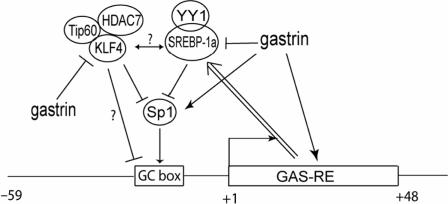
We have previously shown that HDC promoter was inhibited by KLF4 mainly through the upstream Sp1 binding GC box. Our current finding provides an additional mechanism of how KLF4 inhibits HDC promoter activity through the upstream GC box. In general, transcription factors recruit cofactors, either coactivators or corepressors, to the proximal promoter region. These cofactors then modify the structure of local chromatin by changing the status of acetylation, phosphorylation and methylation of histone tails, which resulting in the change of accessibility of transcriptional machinery to the promoter and thus the change of efficiency of transcription. In our system, KLF4 presumably recruits Tip60 and HDAC7 as corepressor to the upstream GC box, since mutation of the site disrupted the inhibitory effect of the complex (Figure 5E bottom panel). Because of the deacetylase activity of HDAC7, which can remove acetyl group from histone tails and is associated with transcriptional inactive state, recruitment of Tip60/HDAC7 complex will repress HDC transcription. Why gastrin decreases binding of the KLF4-Tip60-HDAC7 complex to the HDC promoter (Figure 6A) despite the fact that gastrin increases KLF4 and HDAC7 mRNA levels (6C)? It is likely that gastrin activates Sp1 as reported previously (43). As a result, the accumulation of Sp1 on the HDC promoter would likely lead to decreased binding of KLF4-Tip60-HDAC7 complexes on the HDC promoter, since there is a competition between Sp1 and KLF4 for binding, as we have reported earlier (36). It should be noted that Tip60 does not repress expression of all the genes that was inhibited by KLF4, suggesting that KLF4, Tip60 and HDAC7 cooperatively repress transcription of a subset of genes. The fact that inhibition of HDC promoter activity by YY1 also occurs at the same upstream GC box further suggests the importance of this site in the basal and regulated HDC gene expression (35,44). As previously described, YY1/SREBP-1a complex may displace/compete with the binding of Sp1 with the GC box, resulting in the repression of HDC promoter activity. From our current study, it appears that the KLF4/Tip60/HDAC7 repressive complex function similarly to the YY1/SREBP-1a complex in regulation of HDC promoter activity. However, the relationship between these two complexes remains to be determined (Figure 7).
Figure 7.
A proposed model of transcriptional regulation of HDC is shown by different nuclear factors at the upstream GC box in the promoter. Sp1 has been shown to activate the promoter through this element. Gastrin activates HDC gene expression through both downstream gastrin responsive elements and the upstream GC box. Our published data also suggest that YY1 and SREBP-1a form a complex to compete Sp1 in binding the GC box, resulting in the transcriptional inhibition. The double line with arrow indicates that the downstream GAS-RE is required for this inhibition. Published data and the data presented here suggest that gastrin activates Sp1. Competition between Sp1 and KLF4 on the upstream GC box of the HDC promoter then decreases association of KLF4-Tip60-HDAC7 repressive complexes with the promoter, resulting in the upregulation of HDC gene expression. The relationship between YY1/SREBP-1a complex and KLF4/Tip60/HDAC7 complex remains to be determined.
Given that KLF4 and Tip60 are both involved in many similar biological processes, including DNA damage and apoptosis, the interaction between these two proteins will likely provide more insights in defining the mechanism in these processes. Further studies may focus on the change of interaction between KLF4 and Tip60 in some physiological pathways such as DNA damage response pathway. This will help in the development of KLF4 and/or Tip60 as a target for the treatment of various diseases, including cancers, since DNA damage response has been recently proposed as an early event during tumorigenesis (35,45).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by an NIH grant (RO1 DK-48077) to T.C.W. We also thank Dr Chi-Chuan Tseng at Boston University, Dr Hung-Ying Kao at Case Western Reserve University for kindly providing anti-KLF4 and anti-HDAC7 antibodies and Dr Howard J. Worman at Columbia University of critically reviewing the manuscript. Funding to pay the Open Access publication charges for this article was provided by NIH grant RO1 DK-48077.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhang W, Geiman DE, Shields JM, Dang DT, Mahatan CS, Kaestner KH, Biggs JR, Kraft AS, Yang VW. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J. Biol. Chem. 2000;275:18391–18398. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garrett-Sinha LA, Eberspaecher H, Seldin MF, de Crombrugghe B. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J. Biol. Chem. 1996;271:31384–31390. doi: 10.1074/jbc.271.49.31384. [DOI] [PubMed] [Google Scholar]
- 3.Yet SF, McA'Nulty MM, Folta SC, Yen HW, Yoshizumi M, Hsieh CM, Layne MD, Chin MT, Wang H, et al. Human EZF, a Kruppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J. Biol. Chem. 1998;273:1026–1031. doi: 10.1074/jbc.273.2.1026. [DOI] [PubMed] [Google Scholar]
- 4.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 5.Chen ZY, Shie JL, Tseng CC. Gut-enriched Kruppel-like factor represses ornithine decarboxylase gene expression and functions as checkpoint regulator in colonic cancer cells. J. Biol. Chem. 2002;277:46831–46839. doi: 10.1074/jbc.M204816200. [DOI] [PubMed] [Google Scholar]
- 6.Jenkins TD, Opitz OG, Okano J, Rustgi AK. Transactivation of the human keratin 4 and Epstein-Barr virus ED-L2 promoters by gut-enriched Kruppel-like factor. J. Biol. Chem. 1998;273:10747–10754. doi: 10.1074/jbc.273.17.10747. [DOI] [PubMed] [Google Scholar]
- 7.Higaki Y, Schullery D, Kawata Y, Shnyreva M, Abrass C, Bomsztyk K. Synergistic activation of the rat laminin gamma1 chain promoter by the gut-enriched Kruppel-like factor (GKLF/KLF4) and Sp1. Nucleic Acids Res. 2002;30:2270–2279. doi: 10.1093/nar/30.11.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller KA, Eklund EA, Peddinghaus ML, Cao Z, Fernandes N, Turk PW, Thimmapaya B, Weitzman SA. Kruppel-like factor 4 regulates laminin alpha 3A expression in mammary epithelial cells. J. Biol. Chem. 2001;276:42863–42868. doi: 10.1074/jbc.M108130200. [DOI] [PubMed] [Google Scholar]
- 9.Adam PJ, Regan CP, Hautmann MB, Owens GK. Positive- and negative-acting Kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22alpha in vivo. J. Biol. Chem. 2000;275:37798–37806. doi: 10.1074/jbc.M006323200. [DOI] [PubMed] [Google Scholar]
- 10.Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J. Biol. Chem. 2003;278:2101–2105. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katz JP, Perreault N, Goldstein BG, Actman L, McNally SR, Silberg DG, Furth EE, Kaestner KH. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology. 2005;128:935–945. doi: 10.1053/j.gastro.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 12.Foster KW, Ren S, Louro ID, Lobo-Ruppert SM, McKie-Bell P, Grizzle W, Hayes MR, Broker TR, Chow LT, et al. Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 1999;10:423–434. [PubMed] [Google Scholar]
- 13.Foster KW, Frost AR, McKie-Bell P, Lin CY, Engler JA, Grizzle WE, Ruppert JM. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–6495. [PubMed] [Google Scholar]
- 14.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat. Cell Biol. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 15.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 16.Gavaravarapu S, Kamine J. Tip60 inhibits activation of CREB protein by protein kinase A. Biochem. Biophys. Res. Commun. 2000;269:758–766. doi: 10.1006/bbrc.2000.2358. [DOI] [PubMed] [Google Scholar]
- 17.Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 19.Xiao H, Chung J, Kao HY, Yang YC. Tip60 is a co-repressor for STAT3. J. Biol. Chem. 2003;278:11197–11204. doi: 10.1074/jbc.M210816200. [DOI] [PubMed] [Google Scholar]
- 20.Hlubek F, Lohberg C, Meiler J, Jung A, Kirchner T, Brabletz T. Tip60 is a cell-type-specific transcriptional regulator. J. Biochem. (Tokyo) 2001;129:635–641. doi: 10.1093/oxfordjournals.jbchem.a002901. [DOI] [PubMed] [Google Scholar]
- 21.Medina MA, Quesada AR, Nunez de Castro I, Sanchez-Jimenez F. Histamine, polyamines, and cancer. Biochem. Pharmacol. 1999;57:1341–1344. doi: 10.1016/s0006-2952(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 22.Barocelli E, Ballabeni V. Histamine in the control of gastric acid secretion: a topic review. Pharmacol. Res. 2003;47:299–304. doi: 10.1016/s1043-6618(03)00009-4. [DOI] [PubMed] [Google Scholar]
- 23.Gelfand EW. Role of histamine in the pathophysiology of asthma: immunomodulatory and anti-inflammatory activities of H1-receptor antagonists. Am. J. Med. 2002;113(Suppl. 9A):2S–7S. doi: 10.1016/s0002-9343(02)01431-6. [DOI] [PubMed] [Google Scholar]
- 24.Rangachari PK, Prior T, Bell RA, Huynh T. Histamine potentiation by hydroxylamines: structure-activity relations; inhibition of diamine oxidase. Am. J. Physiol. 1992;263:G632–G641. doi: 10.1152/ajpgi.1992.263.5.G632. [DOI] [PubMed] [Google Scholar]
- 25.Hocker M, Zhang Z, Koh TJ, Wang TC. The regulation of histidine decarboxylase gene expression. Yale J. Biol. Med. 1996;69:21–33. [PMC free article] [PubMed] [Google Scholar]
- 26.Ohgoh M, Yamamoto J, Kawata M, Yamamura I, Fukui T, Ichikawa A. Enhanced expression of the mouse L-histidine decarboxylase gene with a combination of dexamethasone and 12-O-tetradecanoylphorbol-13-acetate. Biochem. Biophys. Res. Commun. 1993;196:1113–1119. doi: 10.1006/bbrc.1993.2366. [DOI] [PubMed] [Google Scholar]
- 27.Hocker M, Zhang Z, Fenstermacher DA, Tagerud S, Chulak M, Joseph D, Wang TC. Rat histidine decarboxylase promoter is regulated by gastrin through a protein kinase C pathway. Am. J. Physiol. 1996;270:G619–G633. doi: 10.1152/ajpgi.1996.270.4.G619. [DOI] [PubMed] [Google Scholar]
- 28.Hocker M, Rosenberg I, Xavier R, Henihan RJ, Wiedenmann B, Rosewicz S, Podolsky DK, Wang TC. Oxidative stress activates the human histidine decarboxylase promoter in AGS gastric cancer cells. J. Biol. Chem. 1998;273:23046–23054. doi: 10.1074/jbc.273.36.23046. [DOI] [PubMed] [Google Scholar]
- 29.Pacilio M, Debili N, Arnould A, Machavoine F, Rolli-Derkinderen M, Bodger M, Arock M, Dumenil D, Dy M, et al. Thrombopoietin induces histidine decarboxylase gene expression in c-mpl transfected UT7 cells. Biochem. Biophys. Res. Commun. 2001;285:1095–1101. doi: 10.1006/bbrc.2001.5296. [DOI] [PubMed] [Google Scholar]
- 30.Wessler S, Rapp UR, Wiedenmann B, Meyer TF, Schoneberg T, Hocker M, Naumann M. B-Raf/Rap1 signaling, but not c-Raf-1/Ras, induces the histidine decarboxylase promoter in Helicobacter pylori infection. FASEB J. 2002;16:417–419. doi: 10.1096/fj.01-0766fje. [DOI] [PubMed] [Google Scholar]
- 31.Wessler S, Hocker M, Fischer W, Wang TC, Rosewicz S, Haas R, Wiedenmann B, Meyer TF, Naumann M. Helicobacter pylori activates the histidine decarboxylase promoter through a mitogen-activated protein kinase pathway independent of pathogenicity island-encoded virulence factors. J. Biol. Chem. 2000;275:3629–3636. doi: 10.1074/jbc.275.5.3629. [DOI] [PubMed] [Google Scholar]
- 32.McLaughlin JT, Ai W, Sinclair NF, Colucci R, Raychowdhury R, Koh TJ, Wang TC. PACAP and gastrin regulate the histidine decarboxylase promoter via distinct mechanisms. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;286:G51–G59. doi: 10.1152/ajpgi.00169.2002. [DOI] [PubMed] [Google Scholar]
- 33.Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto T, Horikoshi M. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J. Biol. Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- 35.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 36.Ai W, Liu Y, Langlois M, Wang TC. Kruppel-like factor 4 (KLF4) represses histidine decarboxylase gene expression through an upstream Sp1 site and downstream gastrin responsive elements. J. Biol. Chem. 2004;279:8684–8693. doi: 10.1074/jbc.M308278200. [DOI] [PubMed] [Google Scholar]
- 37.Chen ZY, Wang X, Zhou Y, Offner G, Tseng CC. Destabilization of Kruppel-like factor 4 protein in response to serum stimulation involves the ubiquitin-proteasome pathway. Cancer Res. 2005;65:10394–10400. doi: 10.1158/0008-5472.CAN-05-2059. [DOI] [PubMed] [Google Scholar]
- 38.Legube G, Linares LK, Lemercier C, Scheffner M, Khochbin S, Trouche D. Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation. EMBO J. 2002;21:1704–1712. doi: 10.1093/emboj/21.7.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Legube G, Linares LK, Tyteca S, Caron C, Scheffner M, Chevillard-Briet M, Trouche D. Role of the histone acetyl transferase Tip60 in the p53 pathway. J. Biol. Chem. 2004;279:44825–44833. doi: 10.1074/jbc.M407478200. [DOI] [PubMed] [Google Scholar]
- 40.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl Acad. Sci. USA. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J. Biol. Chem. 2002;277:25904–25913. doi: 10.1074/jbc.M203423200. [DOI] [PubMed] [Google Scholar]
- 42.Wassmann S, Wassmann K, Jung A, Velten M, Knuefermann P, Petoumenos V, Becher U, Werner C, Mueller C, et al. Induction of p53 by GKLF is essential for inhibition of proliferation of vascular smooth muscle cells. J. Mol. Cell. Cardiol. 2007 doi: 10.1016/j.yjmcc.2007.06.001. (in press) [DOI] [PubMed] [Google Scholar]
- 43.Gerhard M, Neumayer N, Presecan-Siedel E, Zanner R, Lengyel E, Cramer T, Hocker M, Prinz C. Gastrin induces expression and promoter activity of the vesicular monoamine transporter subtype 2. Endocrinology. 2001;142:3663–3672. doi: 10.1210/endo.142.8.8311. [DOI] [PubMed] [Google Scholar]
- 44.Ai W, Liu Y, Wang TC. Yin yang 1 (YY1) represses histidine decarboxylase gene expression with SREBP-1a in part through an upstream Sp1 site. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290:G1096–G1104. doi: 10.1152/ajpgi.00199.2005. [DOI] [PubMed] [Google Scholar]
- 45.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.