Abstract
Pyrosequencing technology allows us to characterize microbial communities using 16S ribosomal RNA (rRNA) sequences orders of magnitude faster and more cheaply than has previously been possible. However, results from different studies using pyrosequencing and traditional sequencing are often difficult to compare, because amplicons covering different regions of the rRNA might yield different conclusions. We used sequences from over 200 globally dispersed environments to test whether studies that used similar primers clustered together mistakenly, without regard to environment. We then tested whether primer choice affects sequence-based community analyses using UniFrac, our recently-developed method for comparing microbial communities. We performed three tests of primer effects. We tested whether different simulated amplicons generated the same UniFrac clustering results as near-full-length sequences for three recent large-scale studies of microbial communities in the mouse and human gut, and the Guerrero Negro microbial mat. We then repeated this analysis for short sequences (100-, 150-, 200- and 250-base reads) resembling those produced by pyrosequencing. The results show that sequencing effort is best focused on gathering more short sequences rather than fewer longer ones, provided that the primers are chosen wisely, and that community comparison methods such as UniFrac are surprisingly robust to variation in the region sequenced.
INTRODUCTION
The vast majority of life on earth is microbial, and the vast majority of these microbial species have not been cultured in the laboratory (1). Consequently, our primary source of information about most microbial species consists of fragments of their DNA sequences. The DNA encoding the 16S rRNA gene has been widely used to specify bacterial taxa, since the region can be amplified using PCR primers that bind to conserved sites in most or all species, and large databases are available relating 16S rRNA sequences to bacterial phylogenies. As the cost of sequencing decreases, especially through techniques such as pyrosequencing [(2) and references therein], methods for comparing different communities based on the sequences they contain become increasingly important. In particular, techniques such as UniFrac (3) allow us to compare many microbial samples in terms of the phylogeny of the microbes that live in them. Such methods are particularly valuable as we begin to search for gradients that affect microbial distribution, and thus need to characterize many communities in an efficient and cost-effective fashion. Gradients of interest include different disease states in humans or animal models (4), or physical or chemical gradients in natural environments such as temperature or nutrient gradients in hot springs (5).
Our ability to apply phylogenetic diversity measures such as UniFrac to microbial community data relies on our ability to build phylogenetic trees from fragments of the 16S rRNA sequence. Because the accuracy of phylogenetic reconstruction depends sensitively on the number of informative sites, and tends to be much worse below a few hundred base pairs [see, for example, (6)], the short sequence reads produced from pyrosequencing, which are 100 nt on average for the GS 20 (Genome Sequencer 20 DNA Sequencing System, 454 Life Sciences, Inc., Bradford, CT, USA) and 200–300 nt for the newer GS FLX (Genome Sequencer FLX System, 454 Life Sciences, Inc., Bradford, CT,USA), may be unsuitable for performing phylogenetically based community analysis. However, this limitation can be at least partially overcome by using a reference tree based on full-length sequences, such as the tree from Phil Hugenholtz's 16S rRNA ARB Database (7), and then using an algorithm such as parsimony insertion (8) to add the short sequence reads to this reference tree. These procedures are necessarily approximate, and may lead to errors in phylogenetic reconstruction that could affect later conclusions about which communities are more similar or different. One substantial concern is that because different regions of the rRNA sequence differ in variability (9), conclusions drawn about the similarities between communities from different studies might be affected more by the region of the 16S rRNA that was chosen for sequencing than by the underlying biological reality.
Here we address these effects directly by asking how primer choice affects our ability to recover patterns of similarity between microbial communities obtained using full-length or near-full-length 16S rRNA sequences, using two complementary strategies. First, we ask whether microbial communities that come from a globally dispersed set of over 200 different physical locations (10), including soil, fresh water and marine sediment, form distinct clusters by habitat type or instead cluster by which region of the 16S rRNA was sequenced. Second, we test for the recovery of UniFrac clusters from near-full-length sequences for three different studies: a set of communities from lean and obese mice (4), a set of communities from the gastrointestinal tract of three healthy human individuals (11) and a set of sequences from the Guerrero Negro microbial mat [Harris, J.K., Walker, J.J. and Pace, N.R. unpublished data, and (12)]. In each case, we ask whether the same relationships would have been recovered if a smaller fragment of the sequence had been used. In particular, we are concerned with the trade-off that pyrosequencing offers: given finite resources, is it more efficient to collect a large number of 100-base 16S rRNA fragments, or to collect a smaller number of near-full-length rRNA sequences using traditional methods?
MATERIALS AND METHODS
Data sets
For the first part of the analysis, 16S rRNA data sets were obtained from 111 studies of physical environments, covering 202 samples, as previously described (10). This data set consisted of 21 173 unique sequences. The three 16S rRNA data sets for the second part of the analysis were the following: (i) sequences from the bacteria in the distal cecum of 19 mice, consisting of a mixture of obese individuals homozygous for a mutation in the leptin gene and heterozygous and wild-type lean individuals, for a total of 3732 sequences (3453 unique sequences) (4), (ii) 16S rRNA sequences from the bacteria in five sites along gut transects of three healthy human individuals as well as stool for a total of 11 738 sequences (7761 unique sequences) (11) and (iii) 16S rRNA sequences from the bacteria in a depth profile of the hypersaline Guerrero Negro microbial mat, in Baha Mexico, consisting of about 11 738 sequences (11 164 unique sequences), obtained from 10 depths ranging from 0 to 40 mm (Harris, J.K., Walker, J.J. and Pace, N.R., unpublished data). These samples are from the location described in ref. (12).
Alignment and phylogeny
Near-full-length 16S rRNA sequences were aligned using the NAST alignment tool at the greengenes web site (13) and added to the greengenes 1280 nt alignment. Sequences were clipped from positions 86 to 1326 (relative to the Escherichia coli 16S rRNA sequence), and only sequences spanning this range were retained. Duplicate sequences were merged during this step, and the number of times that each duplicate fragment appeared in each sequence was recorded as a count of the number of each type of sequence in each environment.
Clipped sequences were generated from this full-length alignment. To simulate pyrosequencing data, sequences were extended, starting from the primers, for either 100, 150, 200 or 250 bp in the forward or reverse direction depending on the orientation of the primer. Primers were chosen from the European Ribosomal RNA Database (14) and were as follows: F343 TACGGRAGGCAGCAG, R357 CTGCTGCCTYCC GTA, F517 GCCAGCAGCCGCGGTAA, R534 ATTA CCGCGGCTGCTGGC, F784 RGGATTAGATACCC C,R798 GGGGTATCTAATCCC, F917 GAATTGACG GGGRCCC, R926 CCGTCAATTYYTTTRAGTTT, F1099 GYAACGAGCGCAACCC, R1114 GGGTTGC GCTCGTTRC. All positions are given relative to the E. coli sequence, the position of which was mapped to the position within the alignment. These clipped sequences were then realigned with NAST, yielding a clipped alignment. Some sequences were excluded at this step because they failed to satisfy NAST's conditions for alignment (at least 75% identity to a template sequence and coverage of at least half the E. coli sequence in the alignment).
Each sequence was imported into ARB (8) and inserted using the LanemaskPH filter (7) into the greengenes CoreSet tree (15) using the parsimony insertion algorithm in ARB (8). Sequences from the data set being investigated in each case, i.e. the mouse or the human data set, were excluded from the tree prior to import (eliminating these sequences was necessary because otherwise each clipped sequence would have matched perfectly to the full-length version already in the tree). The resulting tree was exported for UniFrac analysis.
UniFrac analysis
Briefly, UniFrac measures the distance between two environments in terms of the fraction of evolutionary history that separates the organisms in the two environments. More specifically, for each pair of environments, UniFrac measures the fraction of the total branch length in a phylogenetic tree that leads to sequences from one community or the other but not both. UniFrac requires that each sequence be assigned to one or more environments. To compare sequences from many different environments, the UniFrac value is determined for all pairs of environments to produce a distance matrix. We use this distance matrix to cluster the environments using the hierarchical clustering algorithm called UPGMA (Unweighted Pair-Group Method with Arithmetic mean, a technique that merges the closest pair of environments or clusters of environments at each step), or to perform dimensionality reduction using PCoA (Principal Coordinates Analysis, a geometric technique that converts a matrix of distances between points in multivariate space into a projection that maximizes the amount of variation along a series of orthogonal axes) (3). For all of the analyses described here, we created environment files using the original authors’ annotations. Many sequences that were not identical as full-length sequences but were identical over the region of the sequence that remained after clipping were dropped from the analysis, reducing the effective sample size. We performed all analyses using the unweighted UniFrac algorithm, which is a qualitative metric of β-diversity and is unaffected by the presence of duplicate sequences. UniFrac analyses, including jackknifing, were performed as previously described (16).
RESULTS
UniFrac clustering by environment is robust to variation in the length and location of the amplified region
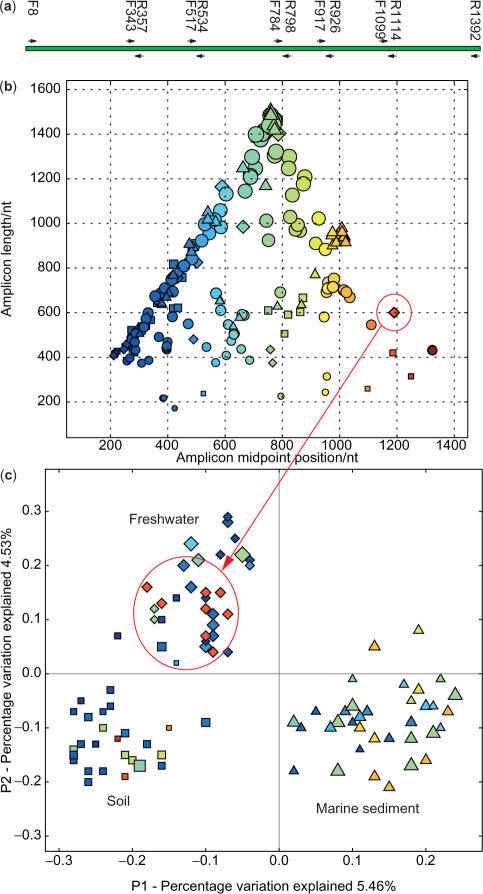
We tested for a sample of 202 samples from diverse physical environments (10) whether clustering was more robust by sample type or by the length or location of the amplicon. Figure 1 shows that samples from the same type of physical environment (same shape on Figure 1b and c) cluster together, and that samples that used an amplified fragment of a similar size (size of symbol in Figure 1) or location (color of symbol in Figure 1) did not. For clarity, Figure 1c shows only the 31 soil, 28 freshwater and 38 marine sediment samples, which fall into three discrete clusters. Therefore, at least for differences of the magnitude of differences between soil, water and sediment, there is no clear effect of the amplicon size and length on the observed clustering, and we can conclude in this sample that most of the variation in the observed sequences is accounted for by environment type rather than by methodological artifacts (Figure 1c).
Figure 1.
16S community samples from a broad range of physical environments cluster by environment type, not by primers. (a) Popular sequencing primers, as shown in the European rRNA database. The concern is that sequences amplified using the same primer pair might artifactually cluster together, even if the microbial communities differ, due to primer bias. (b) Distribution of community samples according to midpoint and length of amplicon. Symbol indicates environment type (squares = soil, triangles = marine sediment, diamonds = fresh water, circles = other environments), size indicates length of amplicon (larger symbols indicate longer amplicons) and color spectrum indicates position of midpoint in the sequence (blue → red = start → end of sequence). (c) Distribution of community samples in UniFrac principal coordinates anaylsis (PCoA), colors and symbols same as in (b) above. Samples clearly cluster by environment type, rather than by amplicon, as symbols of the same color and shape are found in each of the environment type clusters. Circles in (b) and (c) show a single point on the primer length graph split into several related but distinct samples on the environment graph (six from different rivers, two from different lakes).
Recovery of UniFrac clusters by specific primer pairs is influenced by both length and location
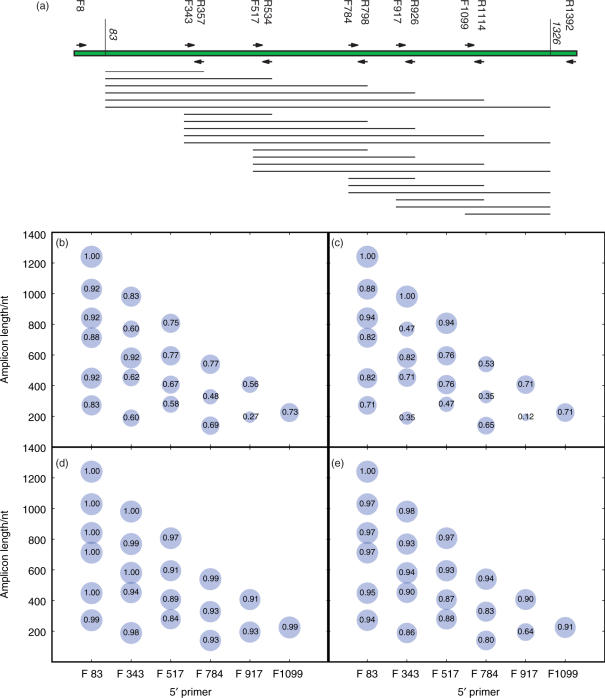
For the remainder of the analyses, we used a combined data set of well-characterized near-full-length 16S rRNA sequences from the human (11), mouse (4) and Guerrero Negro (Harris, J.K., Walker, J.J. and Pace, N.R., unpublished data) sequences described above. For each combination of the following popular primers in the European Ribosomal RNA Database, F343, R357, F517, R534, F784, R798, F917, R926, F1099 and R1114, and the clipped ends of the near-full-length-sequences at position 83 and 1326, we clipped out the corresponding sequence from each of the reads, simulating the effects of using different primer pairs (all numbering is relative to the E. coli sequence, following the usual convention) (Figure 2a). We measured the extent to which the amplicons inferred from each pair of primers recaptured the topology of the clusters obtained using the full-length sequences. Cluster recoveries varied from 12% (mouse, F917 to R1114) to 100% (several of the near-full-length sequences) (Figure 2c). We repeated this analysis using all sequences (Figure 2b) or only the mouse sequences (Figure 2c) to ensure that the inclusion of extremely disparate sequences did not unduly influence the result. Repeating these analyses using the correlation between the UniFrac distances between each pair of samples (Figure 2d and e) indicated that, although the correlations between the distances for corresponding pairs in the two analyses was typically very good (83–100%, except that F917-R1114 is an outlier at 64% recovery when only the mouse sequences were used), there was still substantial variation depending on amplicon. We did not see any systematic biases in clustering due to primer choice. The primary effect of using different amplicons was to degrade the quality of the clustering rather than to cluster specific types of amplicon together in a manner unrelated to the original community (data not shown). Although longer amplicons generally gave better clustering and better correspondence with the distances, using longer amplicons could in some cases lead to worse cluster recovery. For example, F784-R926 provided markedly better cluster recovery for the mouse samples than did F784-R1114 (Figure 2c). Consequently, expending effort on collecting longer sequences, e.g. by assembling overlapping reads, may not always lead to better characterization of the microbial community, and short amplicons are often sufficient if the amplicons are chosen carefully.
Figure 2.
UniFrac clustering with artificially shortened amplicons tends to recapture the same patterns as the full-length sequences. (a) Primer sequences as in Figure 1a, showing the artificial amplicons that were obtained by clipping the sequences using each primer pair. Sequences were truncated at positions 83 and 1326 (relative to the E. coli sequence) because this was the limit of the amplified region of the near-full-length sequences in the three samples (human, mouse, Guerrero Negro). Each line shows one of the sequences that represents a bubble in the other panels. (b) and (c) Cluster recovery rate for the clipped sequences using all three data sets, or only the mouse data set, respectively. The size of each bubble is proportional to the recovery rate, and the number inside each bubble shows the recovery rate (i.e. the fraction of nodes in the cluster that were recovered using the clipped sequences). The x-axis shows the starting primer, and the y-axis shows the length of each amplicon. Surprisingly, although longer amplicons generally gave better cluster recoveries, some long amplicons gave very poor cluster recovery (e.g. F343-R1114 recovered only 47% of the nodes in the cluster diagram for the mouse data set). (d) and (e) Pearson correlation coefficients between the pairwise UniFrac distance scores using the full-length sequences and each set of clipped sequences from all three data sets, or from only the mouse data set, respectively. In general, the correlation between the UniFrac distances was very high even when the cluster recovery was low, suggesting that UniFrac distances are robust to primer choices (although the details of the clustering in the tree can be relatively sensitive, especially in nodes that were not jackknife-supported). Results for the Guerrero Negro data set and the human data set alone were essentially identical (data not shown).
Reads of 100–200 nucleotides, such as those produced by pyrosequencing, can yield the same clustering as full-length sequences if the correct regions are chosen for sequencing
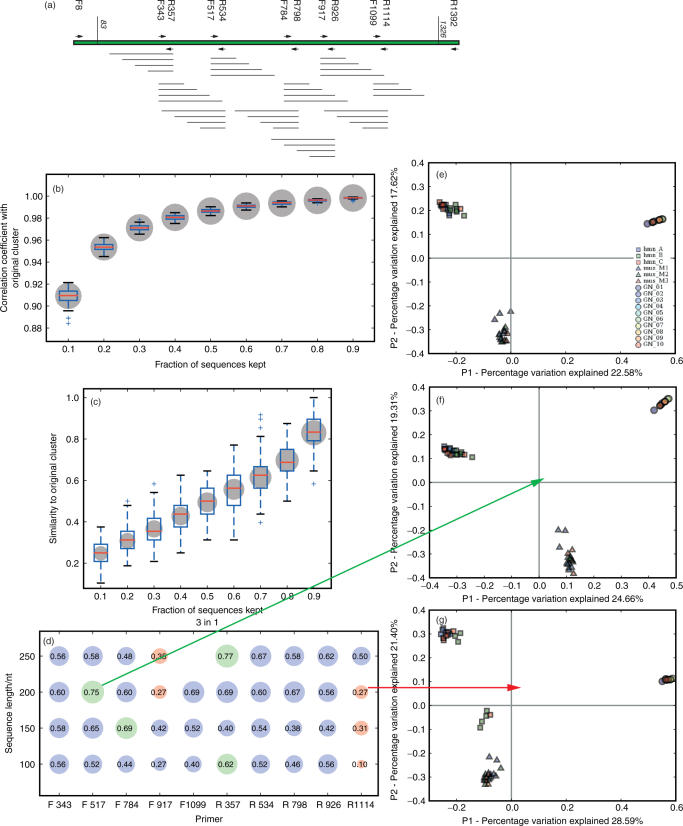
We next tested whether read lengths of 100, 150, 200 and 250 bases, starting at each of the forward and reverse primers described above (Figure 3a), could recapture the same conclusions that would be drawn from the full-length sequences. We note that the short sequences generated by pyrosequencing are likely to be problematic for inferring phylogeny by themselves due to the small number of characters, but that the workflow described in the Materials and Methods section for incorporating them into an existing phylogenetic tree built with full-length sequences appears to work well in practice. We compared the effects of collecting fewer sequences from each sample (through jackknifing) with the effects of collecting very short sequences from each sample (by clipping the sequences). Jackknifing had a much less severe effect on the overall pattern of UniFrac distances (Figure 3b) than on cluster recovery (Figure 3c): even when only 10% of the sequences in each sample were used, the correlations between the pairwise distances inferred from the small sample and the pairwise distances inferred using all available sequences averaged 91%. When half of the sequences were used, the pairwise distances were almost 99% accurate relative to the full sample. Consequently, relatively small samples of sequences are sufficient for accurate estimation of the difference in evolutionary history between two samples. In contrast, the cluster recovery (i.e. the fraction of the nodes in the dendrogram produced by hierarchical clustering of the full data set that were also found in the dendrogram produced by hierarchical clustering of the jackknifed data set) was much more sensitive to sample size and increased roughly proportionally to sample size, with only 82% cluster recovery on average when 90% of the sequences were kept, and 37% cluster recovery when 30% of the sequences were kept. Thus, interpretation of the details of the hierarchical clustering produced by UniFrac should be performed with caution. However, we note that some of the samples in our data set may not have been significantly different to begin with, which would lead to unstable clustering due to random placement of effectively identical communities into a strictly bifurcating tree structure. In contrast, the clipped sequences, which for 100 bases represent only about 8% of the amplicon, performed relatively well (Figure 3d): the best primer, R357, recovered 62% of the nodes, and was thus equivalent to jackknifing with 60–70% of the sequences. Figure 3f shows an example of good PCoA clustering with the 200-base fragments from F517 and Figure 3g an example of poor clustering with the 200-base fragments from R1114: the former recaptures the same pattern as the near-full-length sequence (Figure 3e), whereas the latter artifactually produces two separate clusters of the human samples (shown as squares), and fails to recapture the between-species separation to the same extent. We note that both good and poor choices of primers are available at any length: even with 250-base sequences, F917 provides very poor cluster recovery, whereas R357 generally provides relatively good cluster recovery. It is important to note that hypervariable regions, which have been used in a previous study using the GS 20 pyrosequencer (17), are not the best choice for community characterization. However, they are excellent for measuring the diversity of OTUs (Operational Taxonomic Units, i.e. groups of sequences defined by similarity because species definitions are typically not available) because of the difficulty of integrating these hypervariable regions into a phylogenetic tree.
Figure 3.
UniFrac analysis of short clipped sequences simulating 454 reads, using data from all three sequence sets (human, mouse, Guerrero Negro). (a) Diagram showing clipped reads of 100, 150, 200 and 250 bases starting with each of the forward and reverse primers. Note that F1099 + 250 is not available because it exceeds the end of the near-full-length sequences we used for the analysis. (b) Correlation in UniFrac distances between jackknifed data sets and full data sets (ranging from 0, no correlation, to 1, perfect correlation). Size of bubble reflects average strength of correlation. Note that the y-axis on this plot ranges from 0.88 to 1, so all the correlations are very strong. The x-axis shows fraction of sequences retained in the jackknifing. Box plots show quartiles, medians, 95% quantiles and outliers for n = 100 jackknife replicates. (c) Cluster recovery using the same jackknifed data as (b). Note that cluster recovery is always much lower and more variable than distance recovery, indicating that many of the details of the clustering are not supported by jackknifing. (d) Cluster recovery from each primer for each read length. Best primer at each read length is shown in green; worst is shown in red. Number inside each bubble indicates the cluster recovery (size of each bubble is also proportional to cluster recovery, same scale as (b) above. (e) UniFrac PCoA clustering of the full-length sequences (legend key: hmn = human, A, B and C are three separate individuals (12); mus = mouse, M1, M2 and M3 are the three different mothers and their offspring; GN = Guerrero Negro, 10 samples are 10 different sediment layers from shallowest to deepest). (f) UniFrac PCoA clustering of an example of good cluster recovery, F517 with 200-base reads. Note that the clustering is almost identical to that of the full-length sequences, with a slight rotation of the coordinate axes, and the relative ordering of points within each cluster is preserved. (g) UniFrac PCoA clustering of an example of poor cluster recovery, R1114 with 200-base reads. The human samples are apparently split into two separate groups, suggesting the wrong biological conclusion.
100-base reads using primer R357 recapture a surprising amount of the biological signal in the data
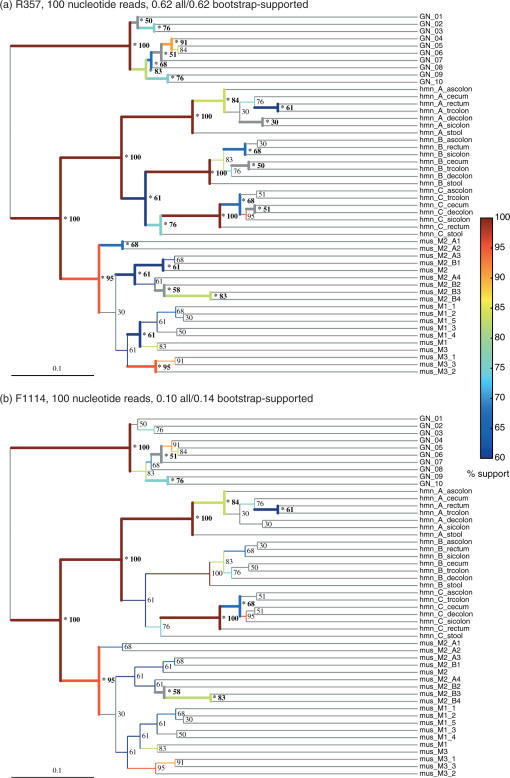
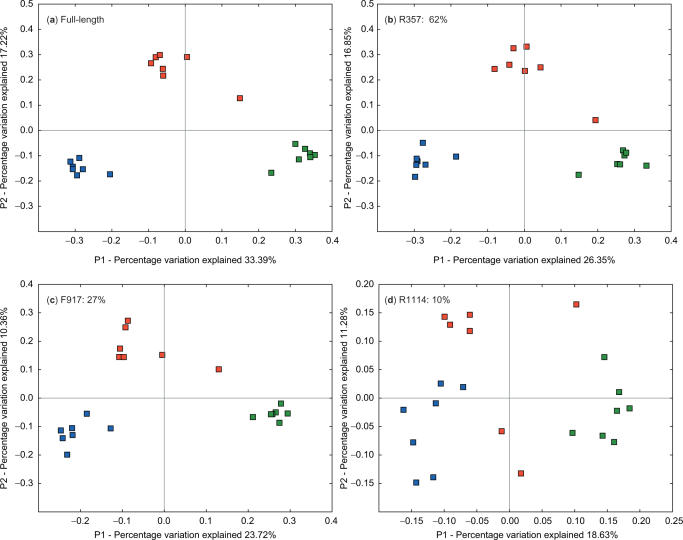
We tested whether different primers were able to recapture the jackknife-supported nodes by building the hierarchical clustering for the full-length sequences, performing jackknifing, and testing whether the jackknife-supported nodes were preferentially recovered (on the assumption that the jackknife-supported nodes were more likely to reflect significant biological differences). Interestingly, we found that jackknife-supported nodes were not significantly more likely to be recovered than other nodes (Figure 4), suggesting that either jackknifing is not effective for uncovering meaningful clusters or that the errors introduced by clipping the sequences are orthogonal to the errors introduced by undersampling. Because the jackknife-supported nodes tended to have clear biological interpretations in these samples (e.g. each of human, mouse and Guerrero Negro formed discrete clusters, each of the humans formed a discrete cluster, each of the mothers of the mouse litter and her offspring formed a discrete cluster), we favor the latter explanation. Thus, although some primer choices such as R357 appear to recapture the same results as full-length sequences very effectively, they are not guaranteed to recover all the clusters that would be jackknife-supported with full-length sequences. Because the human, mouse and Guerrero Negro samples are very different from one another, we also tested whether the same results applied within just the human sample. Figure 5 shows the PCoA clustering for the full-length sequences (Figure 5a) and for 100-base clipped sequences starting at R357 (Figure 5b, 62% cluster recovery), F917 (Figure 5c, 27% cluster recovery) and R1114 (Figure 5d, 10% cluster recovery). The three individuals, colored in red, green and blue, form well-defined samples in all except the R1114 sequences. Interestingly, F917 has good correlations in the UniFrac distances even though the cluster recovery in the hierarchical clustering is low, suggesting that PCoA may be a more useful guide than cluster recovery to detecting similarities in the data. Notably, R1114 fails to separate the red points from the others along PC2 (the second principal component axis), and does not separate the red points from the blue and green points along PC1 (the first principal component axis). It thus fails completely to recapture the essential patterns in the data, although there are still significant differences in location between the three clusters. Thus, the results hold both for comparing very diverse samples (human and mouse gut, and microbial mat) and for comparing relatively similar samples (different locations within the gut in different humans).
Figure 4.
UniFrac hierarchical clustering recoveries from a good and a bad primer. Data shown are from 100-base reads starting at R357 and F1114 respectively, using the cluster diagram obtained from full-length sequences as a reference. Jackknife values are shown for each node (100 replicates), and edges are shown colored by jackknife values (gray for < 60%; color bar shows scale for values above 60%). Recovered nodes are marked with an asterisk and their edges are indicated with heavy lines. R357 (a) recovers essentially all the biological signal, including the grouping of the samples from the three human individuals A, B and C, the layer structure of the Guerrero Negro microbial mat, and the clustering of mice by mother. In contrast, F1114 (b) is able only to differentiate the three general environment types from each other, and fails to recapture many nodes that are jackknife supported at the 90% level and above.
Figure 5.
UniFrac PCoA analysis of full-length human sequences, and three 100-base clipped sequence sets simulating 454 reads. The three different individuals are colored separately, with the same coloring applied to all three graphs. R357 and F917 recapture the overall pattern (discrete clusters for each individual) extremely well. In contrast, R1114 distorts the pattern substantially, and suggests separation of the three samples only along PC1. The percentage next to each primer number indicates the percentage of nodes in the hierarchical clustering on the full-length sequences that was recovered in the clipped sequences.
DISCUSSION
We have shown that short sequence fragments from the 16S rRNA, including 100-base reads similar to those used in pyrosequencing, allow substantial resolution of biologically meaningful similarities and differences between microbial samples. We have demonstrated that it is possible to recapture the conclusions obtained from full-length sequences. However, accurate use of these short sequence reads for community comparisons requires judicious choice of primers, and also requires that the sequences be placed in the context of a larger phylogenetic tree based on full-length 16S rRNA sequences. The trade-off between obtaining more sequences and obtaining longer sequences is clear: 100-base sequences from R357 covering only 8% of the length of the near-full-length 16S rRNA sequences we used for this study provided comparable resolution to 70% jackknifes with the full-length sequences. In other words, the 454 GS 20 or GS FLX fragments are nearly ten times as efficient, base pair for base pair, at uncovering the structure of the set of microbial communities being studied (although the present results hold only for 16S rRNA sequences, and the performance of these techniques for metagenomic sequences remains to be characterized). Combined with the 10-fold cost advantage per base pair of pyrosequencing over Sanger sequencing, the elimination of the time-consuming and laborious requirement for cloning, and the ability to use primer barcoding techniques (18, 19) to characterize many environmental samples in parallel on a single sequencing run, we expect that pyrosequencing using the best of the primers we identified in this study, R357, will rapidly replace Sanger sequencing for microbial community analyses.
ACKNOWLEDGEMENTS
The work was support by a grant from the W.M. Keck Foundation to F.D.B. and R.K., and by a CCFAR developmental award to R.K. M.H. was supported by the NIH/CU Molecular Biophysics Training Program T32GM065103. Analyses were performed using the W.M. Keck RNA Bioinformatics Facility. We thank J.K. Harris, J.J. Walker and N.R. Pace, R.E. Ley and J.I. Gordon for helpful discussions, and J.K. Harris, J.J. Walker and N.R. Pace for sharing unpublished data. Funding to pay the Open Access publication charges for this article was provided by the W.M. Keck Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 2.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc. Natl Acad. Sci. USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 7.Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3:1–8. doi: 10.1186/gb-2002-3-2-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius J, Gutell R, et al. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980;8:2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc. Natl Acad. Sci. USA. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ley RE, Harris JK, Wilcox J, Spear JR, Miller SR, Bebout BM, Maresca JA, Bryant DA, Sogin ML, et al. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl. Environ. Microbiol. 2006;72:3685–3695. doi: 10.1128/AEM.72.5.3685-3695.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:394–399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuyts J, Perriere G, Van De Peer Y. The European ribosomal RNA database. Nucleic Acids Res. 2004;32:D101–D103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lozupone C, Hamady M, Knight R. UniFrac–an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371–384. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl Acad. Sci. USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Binladen J, Gilbert MT, Bollback JP, Panitz F, Bendixen C, Nielsen R, Willerslev E. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE. 2007;2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman C, Minkah N, Arens MQ, Tebas P, Bushman FD. Nucleic Acids Res. 2007. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. 18 June 2007 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]