Abstract
Targeted chromatin remodelling is essential for many nuclear processes, including the regulation of V(D)J recombination. ATP-dependent nucleosome remodelling complexes are important players in this process whose activity must be tightly regulated. We show here that histone acetylation regulates nucleosome remodelling complex activity to boost RAG cutting during the initiation of V(D)J recombination. RAG cutting requires nucleosome mobilization from recombination signal sequences. Histone acetylation does not stimulate nucleosome mobilization per se by CHRAC, ACF or their catalytic subunit, ISWI. Instead, we find the more open structure of acetylated chromatin regulates the ability of nucleosome remodelling complexes to access their nucleosome templates. We also find that bromodomain/acetylated histone tail interactions can contribute to this targeting at limited concentrations of remodelling complex. We therefore propose that the changes in higher order chromatin structure associated with histone acetylation contribute to the correct targeting of nucleosome remodelling complexes and this is a novel way in which histone acetylation can modulate remodelling complex activity.
INTRODUCTION
The highly condensed chromatin structure in the eukaryotic nucleus provides a formidable obstacle to prevent protein access to DNA. A fundamental question is how different regulatory elements co-operate to ensure that this chromatin packaging is disrupted only at specific loci in the appropriate cell type. One important group of players in this process is ATP-dependent nucleosome remodelling complexes. These complexes increase the accessibility of proteins to nucleosomal DNA by one of the several mechanisms: Sliding the histone octamer along the DNA template, localized disruption of histone/DNA contacts or trans-displacement of the histone octamer (1,2). Since these complexes function in an energy-dependent manner and play important roles in controlling the accessibility of DNA sequences, their activity needs to be tightly regulated.
During gene activation, remodelling complexes can be targeted to promoters via their association with transcriptional activators. For example, SWI/SNF is known to associate with acidic activators, such as Swi5, Gcn4 and Hap4 (3,4). A second potential means of targeting remodelling complexes is via interaction between specific domains, found in some subunits of remodelling complexes, and modified histone tails. For example, between bromodomains and acetylated histone tails as well as between PHD fingers and histone H3 trimethylated at lysine 4 (5). Indeed, the bromodomain in the Swi2/Snf2 subunit of SWI/SNF complex stabilizes the binding of this complex to an acetylated promoter nucleosome (6,7). Similarly, the interaction of the double bromodomain of Rsc4 with acetylated H3 Lys14 is essential for gene activation by the RSC complex (8) and the WSTF bromodomain helps to target the remodelling complex WINAC to vitamin D receptor-regulated promoters (9).
Although this suggests how remodelling complexes can be targeted to specific promoters, these are not necessarily the only mechanisms of targeting. For example, it has been well established that acetylation of histone tails results in a less-condensed higher order chromatin structure via sedimentation velocity and electron microscopy studies as well as by using DNase I as a probe for accessibility (10–12) and it was shown recently that acetylation at histone H4 K16 alone is sufficient to prevent chromatin folding into 30 nm fibres (13). Consequently, the more open structure of acetylated chromatin domains might be a further mechanism to target nucleosome remodelling complexes.
One situation where the more open structure of acetylated chromatin might be important in targeting nucleosome remodelling complexes is during V(D)J recombination: Here the generation of antigen receptor diversity relies on the stochastic use of different gene segments within the immunoglobulin or T-cell receptor loci (14) and thus chromatin needs to be remodelled at any one of the numerous points within the locus rather than at one specific promoter region. V(D)J recombination is initiated by only two proteins, RAG1 and RAG2, that introduce double-strand breaks at conserved recombination signal sequences (RSS) (15). Nucleosomes are known to inhibit the initiation of recombination by preventing RAG1 and RAG2 from binding to RSSs (16–18). Since nucleosomes are preferentially positioned over RSSs (19), they must be remodelled for initiation of recombination. The SWI/SNF remodelling complex has been shown to facilitate RAG cutting on mono-nucleosomes in vitro (20) as have BRG1 and SNF2h, the catalytic subunits of the hSWI/SNF and ISWI complexes, respectively (21). However, these remodelling complexes are present and active in most cell types and thus regulation of their targeting would seem important so that nucleosomes at RSSs are remodelled only in the correct cell- and locus-specific manner.
Increased histone acetylation has been correlated with loci undergoing recombination (22–25). Furthermore, although histone acetylation stimulates recombination in vivo (18), we, and others, found that it does not facilitate RAG cutting on mono-nucleosomes in vitro (16,18). Instead, we proposed that the more open structure associated with arrays of acetylated nucleosomes in vivo (10–12) might either directly increase accessibility to RAG proteins or might increase accessibility to elements that indirectly increase RAG cutting.
Here, we have investigated mechanistically how histone acetylation increases RAG cutting: Using arrays of acetylated and non-acetylated nucleosomes in vitro, we find that acetylation increases the accessibility to remodelling activities, which thus enhances their ability to mobilize nucleosomes. No effect of acetylation on remodelling activity was detected when mono-nucleosomes were used as the substrate. From these studies we suggest that one way in which histone acetylation could enhance the initiation of V(D)J recombination in vivo is by helping to control the accessibility of nucleosome remodelling complexes to their substrates. We further suggest that similar locus-wide increases in histone acetylation might play a more general role in directing remodelling complex activity during gene activation.
MATERIALS AND METHODS
DNA constructs
pMAB1 has a restriction site for Xho I in the VκL8 RSS spacer (19). A 3.15 kb Eco RI fragment that encompasses both the 12- and 23-RSSs (that lie 200 bp apart in an inverted orientation) was taken from pMAB1 and ligated to give pMAB2; this plasmid was used for the chromatin reconstitution. Fragments for mono-nucleosome reconstitution were prepared from pFM210 and pMAB5 (which carry 12 bp spacer from VκL8 with one modification) as described (19).
Proteins and nucleosome assembly
RAG1 (amino acids 384–1008) and RAG2 (amino acids 1–387) were purified as described (19). Full length RAG2 increases cutting on both free DNA and chromatin templates by the same amount compared to truncated RAG2 (21). This suggests that there are no chromatin-specific effects of the C-terminus in these in vitro assays and that the truncated form of RAG2 is a reasonable tool for these studies. The chromatin remodelling factors ACF, CHRAC and ISWI were purified according to (26). Recombinant HMGB1 (truncated form) was a generous gift from Dr Kevin Hiom. Histones were prepared from CV1 cells (11); chromatin assembly was performed as described (27) and was monitored by micrococcal nuclease digestion and supercoiling analysis (11). All experiments shown were confirmed using three different preparations of acetylated and non-acetylated histone preparations. In all cases, equivalent supercoiling and nucleosome spacing was confirmed prior to use in RAG cutting and accessibility assays. Mono-nucleosome assembly was by salt-urea dialysis (18); sliding assays were performed according to (28).
RAG cleavage assay
Assembled nucleosome arrays were either untreated or incubated with 0.075% sarkosyl followed by purification through a G25 spin column (Pharmacia). Although this can result in loss of the regular nucleosome repeat, we find that incubation of these stripped templates with ACF regenerates the regular repeat structure identically on acetylated and non-acetylated chromatin (Supplementary Figure 1). Furthermore, we find that RAG cutting on sarkosyl-stripped chromatin is very similar (Figures 2, 4, 7B and Supplementary Figure 4) suggesting that the chromatin structure is not markedly different on acetylated and non-acetylated chromatin following sarkosyl stripping. Aliquots of 25 μl (equivalent to 100 ng DNA) were supplemented to a final concentration of 1 mM MnCl2, 1× McNAP (30 mM creatine phosphate, 3 mM ATP, 2.5 mM MgCl2, 1 mM DTT, 1 mM creatine phosphokinase) and incubated with RAG1 and RAG2 (∼50–200 ng of each protein) in the presence of 500 ng HMGB1 for 1 h at 30°C. Where indicated, ACF and ACF bromodomain mutant were added. The reaction was stopped with 50 μl stop mix (2.5% sarkosyl, 100 mM EDTA, pH 8.0) and incubated with 50 μg/ml RNase A for 30 min at 37°C followed by digestion with 0.02% SDS and 300 μg/ml proteinase K at 37°C overnight. DNA was extracted twice with phenol–chloroform and ethanol precipitated.
Figure 2.
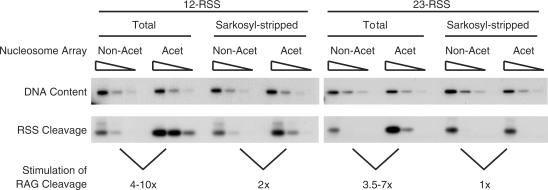
Increased RAG cutting on the acetylated nucleosome arrays. RAG cutting was performed on the acetylated (Acet) and non-acetylated (Non-Acet) nucleosome arrays either in the presence of Drosophila extract (Total) or following removal of proteins (Sarkosyl-stripped). Verification that the PCR reactions were in the semi-quantitative range (for each set of RAG cut templates) was achieved by confirming that the signal increased proportionately to the amount of input DNA (Supplementary Figure 2). The DNA content panel is a control PCR and shows that an equivalent amount of DNA was used per reaction. The stimulation in RAG cutting was normalized to the amount of DNA present. This stimulation of RAG cutting on the acetylated templates compares favourably with the acetylation-mediated increase in transcription (of ∼5-fold) reported previously with similarly reconstituted templates (27).
Figure 4.
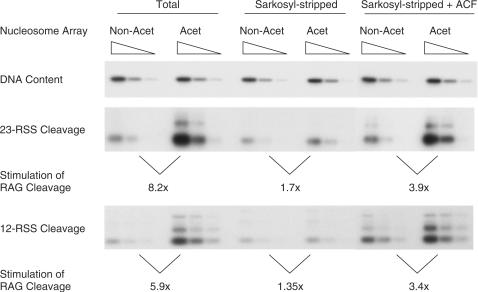
Addition of purified remodelling complexes stimulates RAG cutting on acetylated arrays. Acetylated (Acet) and non-acetylated (Non-Acet) arrays were used in RAG cutting assays either in the presence of Drosophila extract (Total), following sarkosyl stripping and purification on a G-25 spin column (Sarkosyl-stripped) or following addition of the purified remodelling complex, ACF, to sarkosyl-stripped material. The upper panel shows that an equivalent amount of DNA was used in each reaction. The lower panels show cutting at the 23- and 12-RSS, respectively. Similar results to those with ACF were obtained when CHRAC and ISWI were added to the stripped chromatin templates (data not shown and Supplementary Figure 4). The fold increase in RAG cutting was normalized to the amount of DNA present and compares cutting on acetylated and non-acetylated chromatin under the conditions indicated. The similar level of RAG cutting on stripped acetylated and non-acetylated templates in the absence of remodelling complexes (middle lanes) confirms that the RSSs are equivalently protected on the two templates. In this experiment, ACF does not enhance cutting on acetylated chromatin to the same level as in the total extract; this could be due to the presence of insufficient ACF since in other cases we could recapitulate the same fold increase as seen in the total extract (e.g. Figure 7B). The slower mobility PCR products detected for 12-RSS cutting are lost upon cleavage with a restriction enzyme that cuts within the non-conserved spacer of the 12-RSS (data not shown). Since the sizes of these slower mobility products correspond to those predicted for cleavage at the 23-RSS, we suggest that they are due to RAG cutting at the 23-RSS.
Figure 7.
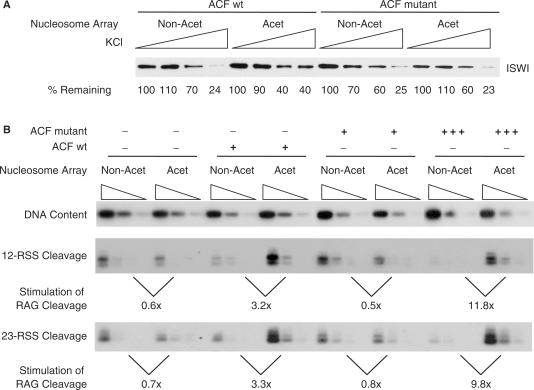
Binding and remodelling by wild-type ACF and ACF bromodomain mutant complexes on acetylated and non-acetylated chromatin (A) The ACF1 bromodomain causes only a slight increase in binding of ACF to acetylated chromatin. Chromatin templates were assembled with acetylated (Acet) or non-acetylated (Non-acet) histones, bound by ACF complexes containing wild-type ACF1 or ACF1 bromodomain mutant, followed by washes with increasing concentrations of salt. Western blotting shows the amount of the ISWI subunit that remained bound. As a control, the gel was Coomassie stained; this showed that the chromatin was equivalently assembled (data not shown). (B) RAG cutting is enhanced on acetylated chromatin by the ACF bromodomain mutant complex. Acetylated (Acet) and non-acetylated (Non-Acet) chromatin was assembled, sarkosyl-stripped and purified on a G-25 column prior to addition of ACF complexes containing wild-type ACF1 or ACF1 bromodomain mutant as indicated. Equivalent amounts of remodelling complex, determined by protein amounts, were added (+); addition of 2.5-fold ACF bromodomain mutant is indicated (+++). The upper panel shows that an equivalent amount of DNA was used in each reaction. The lower panels show cutting at the 23- and 12-RSS, respectively.
Restriction enzyme accessibility
General accessibility to Pvu I: 150 μl aliquots (equivalent to 600 ng DNA) of chromatin assembly reactions were either left untreated, or incubated with 10 units of apyrase, or 15 mM AMP-PMP, and 15 mM MgCl2 for 30 min at 26°C. Prior to Pvu I cleavage all samples were supplemented with MgCl2 to 10 mM, and the control (untreated) samples were supplemented with ATP regenerating system (McNAP). The reactions were then digested with 75 units of Pvu I for 0, 5, 10, 15 and 30 min at 26°C, prior to addition of stop mix (2.5% sarkosyl, 100 mM EDTA). Recovered DNA was digested with Xho I and analysed on a 1% agarose gel by Southern blotting.
Ligation-mediated polymerase chain reaction (LMPCR)
This was performed as described (29). Primer 405-FM25 (5′-GCGGTGACTCGGGAGATCTGAAGTG-3′) was annealed to primer 406-FM11 (5′-CACTTCAGATC-3′) at a final concentration of 10 μM. A 10 pmol of the annealed linker FM25/11 was then ligated to the blunt-ended DNA fragments produced by RAG-cleavage with 200 units of T4 DNA ligase at 16°C overnight. For 12 bp-spacer RSS signal ends, ligation products were amplified by 16 cycles of PCR using linker-specific primer 405-FM25 plus the plasmid-specific primer 407-NEB1233 (5′-GCGGATAACAATTTCACACAGGA-3′). For 23 bp-spacer RSS signal ends, ligation products were amplified using linker-specific primer 405-FM25 plus the plasmid-specific primer 408-DR1 (5′-CAACGGTGGTATATCCAGTG-3′). PCR cycles were: 20 s at 95°C, 20 s at 60°C and 60 s at 72°C in a total volume of 25 μl. One microlitre of the LMPCR reaction was then used for a second PCR amplification (nested PCR) using primer 414-RSS/FM25-C (5′-ACTCGGGAGATCTGAAGTGCACAGT-3′) and primer 409-nested NEB1233 (5′-TTCACACAGGAAACAGCTATGACCATGATT-3′) for 12 bp-spacer RSS signal ends and primer 414-RSS/FM25-C and primer 410-nested DR1 (5′-CCAGTGATTTTTTTCTCCATTTTAGCTTCC-3′) for 23 bp-spacer RSS signal ends, respectively. Each cycle comprised 20 s at 95°C, 20 s at 63.5°C and 60 s at 72°C in a total volume of 25 μl; 20 cycles were performed. Samples were analysed on a 2% agarose gel by Southern blot analysis.
For a comparison of the DNA content, ligation products were amplified by 15 cycles of PCR using the plasmid-specific primer 421-F1 (5′-CATTTCCGTGTCGCCCTTATTCC-3′) plus the plasmid-specific primer 422-B15 (5′-CCATAGTTGCCTGACTCCCCGTC-3′). Each cycle comprised 20 s at 95°C, 20 s at 60°C and 60 s at 72°C in a total volume of 25 μl. Samples were analysed on a 2% agarose gel by Southern blot analysis. To verify that the PCR was in the quantitative range, the amount of DNA template was titrated and quantification using a phosphorimager verified that the amount of product increased proportionately to the input DNA.
Southern blotting and hybridization
PCR samples: Samples were run on a 2% agarose gel at 80 V for 1 h. Southern blotting was as described (19). The probes were primer 414-RSS/FM25-C for 12 bp-spacer and 23 bp-spacer RSS signal ends and primer 421-F1 for a non-RSS DNA fragments, respectively. Hybridization was at 45°C. Samples digested with Pvu I and Xho I were analysed as above except that 25 ng of each sample was run on a 1% agarose gel at 90 V for 2.5 h. The probes were pMAB2 for Pvu I/Xho I-digested samples, and a 359 bp Afl III-Hind III fragment from pMAB1 for Xho I/Alw NI-digested samples.
HMGB1 and remodelling complex binding assays
Acetylated or non-acetylated histones were reconstituted onto a 32P-labelled 284 bp MAB5 fragment (19) by salt dialysis under conditions that gave a mixture of mono-nucleosomal and free DNA templates. These were incubated with increasing amounts of HMGB1 (6.67–667 ng) in 50 mM NaCl, 10 mM Tris pH 8, 0.1 mM EDTA 5% glycerol and were electrophoresed at 80 V on a 0.8% agarose gel in 0.5 × TBE. Complexes were detected by autoradiography. Nucleosome arrays were assembled on magnetic bead bound DNA using Drosophila embryo extract and acetylated or non-acetylated histones. Following assembly, the chromatin was washed with a high salt buffer (Ex-750; 10 mM Tris pH 8, 1.5 mM MgCl2, 0.5 mM EGTA, 0.75 M KCl, 10% glycerol) to remove the bulk of the chromatin-associated proteins. HMGB1 was then incubated with this chromatin at approximately stoichiometric amounts (500 ng HMGB1/1.2 μg DNA template) in Ex-50 (as above with 50 mM KCl) for 30 min. The chromatin was then washed with increasing amounts of salt (Ex-50, 100, 200, 300, 400) prior to electrophoresis on a 15% acrylamide gel. Proteins were detected by silver staining.
Binding of remodelling complexes to chromatin templates was performed using chromatin assembled on beads in the same way. Assembled chromatin was sarkosyl washed as described for RAG cutting experiments, followed by washing in Ex-100 to remove the sarkosyl. The chromatin-bound beads were then resuspended in Ex-100 buffer and McNAP and incubated with 2 μg of ACF complex containing wild-type ACF1 or ACF1 bromodomain mutant for 30 min at 30°C. Bound proteins were released by washes with increasing amounts of salt (Ex buffer with 650, 750, 850 and 950 mM KCl). Chromatin was electrophoresed on a 10% gel; the binding of wild-type and bromodomain mutant ACF1 was detected by western blotting with anti-FLAG (M2) antibody and ISWI was detected with anti-ISWI antibody (26).
RESULTS
Acetylation stimulates RAG cutting
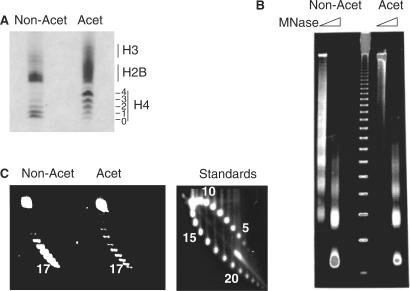
We reported previously that histone acetylation stimulates V(D)J recombination in vivo but has no effect on RAG cutting on isolated mono-nucleosomes in vitro (18). Therefore, we firstly asked if acetylation enhances RAG cutting in vitro when arrays of nucleosomes are used as the chromatin template. We assembled plasmid DNA (pMAB2) carrying 12- and 23-spacer RSSs into highly acetylated or non-acetylated chromatin using Drosophila chromatin assembly extracts. These extracts were depleted of endogenous histones and subsequently ‘reprogrammed’ with non-acetylated or highly acetylated histones prepared from CV1 cells that had been either untreated or treated with the histone deacetylase inhibitor, trichostatin A (Figure 1A). Assembly results in highly chromatinized templates that contain physiologically spaced nucleosomes and a full complement of chromatin-associated proteins (27). Accurate comparison of RAG cutting on the acetylated and non-acetylated chromatin templates requires that both types of chromatin template have equivalent nucleosome density and spacing: Micrococcal nuclease assays confirmed that the nucleosome spacing was equivalent (average of 180 bp; Figure 1B). Furthermore, our previous studies using psoralen-crosslinking and electron microscopy found that plasmids assembled with acetylated and non-acetylated histones that have the same superhelical density also contain the same absolute number of nucleosomes (27). Supercoiling analysis demonstrated that the plasmids used here were equivalently chromatinized with an average of 17 nucleosomes per plasmid (Figure 1C). Since the plasmid used is 3150 bp, this confirmed that the templates were fully assembled into chromatin. Before each experiment, we verified that the plasmids were equivalently assembled into acetylated and non-acetylated chromatin via these assays.
Figure 1.
Assembly of acetylated and non-acetylated chromatin. (A) Triton acid urea gel showing acetylated (Acet) and non-acetylated (Non-Acet) histones purified from CV1 cells. Numbers indicate the lysines modified. (B) Micrococcal nuclease ladders of the assembled non-acetylated (Non-Acet) and acetylated (Acet) arrays. The marker is a 123 bp ladder. (C) Supercoiling analysis of acetylated (Acet) and non-acetylated (Non-Acet) templates. The standards show supercoiling analysis of templates with known nucleosome densities.
To investigate if histone acetylation stimulates RAG cutting on these chromatin templates, we initially performed RAG cutting reactions in the presence of total Drosophila extract. After purification of the DNA, RAG cutting was detected by ligation of a linker onto the blunt double-strand break followed by semi-quantitative LMPCR. As can be seen in Figure 2, RAG cutting is enhanced on the acetylated template by between 3.5- and 10-fold compared to the non-acetylated chromatin; control experiments showed that in all cases a similar amount of total DNA was recovered (upper panel) and that the PCR was in the semi-quantitative range (Supplementary Figure 2).
If acetylation directly facilitates accessibility of RAGs to RSSs, we would predict a similar increase in RAG cutting after sarkosyl stripping the chromatin templates, an established method to remove most chromatin-associated proteins, including chromatin remodellers (30). If, however, acetylation increases RAG cutting via components in the extract, we would expect RAG cutting on sarkosyl-stripped acetylated and non-acetylated arrays to be equivalent. Using purified templates we find acetylation causes only a minimal increase in cutting, indicating that a component in the Drosophila extract is required to enhance cutting on the acetylated arrays (Figure 2). Moreover, the inability of acetylation alone to enhance RAG cutting is fully consistent with earlier data that nucleosomes, which are known to inhibit RAG cutting (16,17), are preferentially positioned at RSSs (19).
Remodelling complexes preferentially increase RAG cutting on acetylated chromatin
We next investigated what might be the component in Drosophila extract that cooperates with acetylation to stimulate RAG cutting. Since Drosophila embryo extracts have an abundance of nucleosome remodelling complexes, and since nucleosomes at RSSs must be remodelled to initiate recombination (16–18), good candidates to enhance RAG cutting are nucleosome remodelling complexes. Previously, we found that the nucleosome remodelling complex, NURF, is unable to mobilize nucleosomes off the favoured nucleosome position conferred by RSSs (19). However, other studies have shown that another remodelling complex found in Drosophila extracts, ACF, does not always mobilize nucleosomes to their most thermally stable position (31). We therefore tested if this, and other, nucleosome remodelling complexes are capable of mobilizing nucleosomes off the energetically favoured RSSs. As can be seen in Figure 3, ACF, as well as its catalytic subunit, ISWI, can indeed cause nucleosome mobilization off RSSs. The direction of this mobilization depends on the activity of the specific remodelling complex (28).
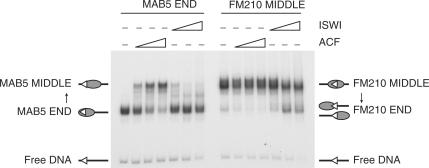
Figure 3.
Remodelling complexes mobilize nucleosomes off RSSs. A sliding assay on mono-nucleosomes reconstituted onto fragments carrying a 12-RSS using recombinant ACF and ISWI is shown. FM210 has a nucleosome positioned over the RSS in the middle of the fragment whereas MAB5 has a nucleosome positioned over the RSS at the left end of the fragment. Preferential sliding of nucleosomes by ACF from the end of the DNA fragment to the middle and vice versa for the purified catalytic subunit, ISWI, has been demonstrated previously (54). Similar mobilization from the end position to the middle position was also observed using CHRAC, a second ISWI-containing remodelling complex isolated from Drosophila extracts (data not shown).
Next, we tested if addition of these same nucleosome remodelling complexes can recapitulate the increased RAG cutting observed on acetylated templates. To this end, chromatin templates were prepared as for Figure 2 and remodelling complexes and other chromatin-bound proteins were removed via sarkosyl stripping. Following purification of the chromatin on a spin column, the RAG cutting assay was performed either with or without addition of purified remodelling complexes. Similar to our observation in Figure 2, control experiments show an increase in RAG cutting on the acetylated compared to the non-acetylated templates in the presence of Drosophila extract. This increased cutting is lost following sarkosyl stripping, confirming that acetylation does not notably enhance RAG cutting alone. However, addition of the purified ISWI containing remodelling complex, ACF, to the sarkosyl-stripped templates, markedly enhanced RAG cutting on the acetylated arrays (Figure 4). Similar increased RAG cutting on the acetylated chromatin was observed upon addition of the remodelling complex CHRAC or its catalytic subunit ISWI (data not shown and Supplementary Figure 4). These data are therefore consistent with the idea that acetylation enhances the ability of nucleosome remodelling complexes to remodel nucleosomes at RSSs, which then allows increased RAG cutting.
Acetylation enhances accessibility to nucleosome remodelling complexes
We next wanted to investigate how remodelling activity is specifically increased on the acetylated chromatin. Three main possibilities exist: Firstly, acetylation might increase the accessibility to the remodelling complexes; the increase in RAG cutting would then be due to remodelling complexes gaining access to, and remodelling, more templates. Alternatively, (or additionally) acetylation might increase the activity of remodelling complexes once they have gained access to the chromatin template. Thirdly, the ACF1 subunit of ACF and CHRAC has a bromodomain (26) which might facilitate binding and/or retention of the remodelling complex to acetylated chromatin once it has gained access, thereby increasing its local concentration and the probability of nucleosome remodelling.
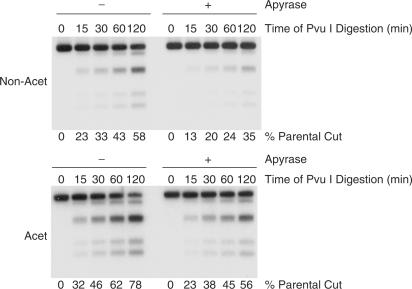
To begin to differentiate these possibilities, we performed a time course of digestion with the restriction enzyme, Pvu I. This enzyme has a number of sites throughout the plasmid and thus can be used as a probe to determine how many of these sites are accessible. As can be seen in Figure 5, cutting by Pvu I is substantially increased on the acetylated compared to the non-acetylated arrays (left panel). Cutting by restriction enzymes has previously been used as an assay for nucleosome remodelling activity [reviewed in (2)] and the observed cutting by Pvu I reflects both the number of its sites that were initially not covered by a nucleosome plus those that become available due to nucleosome mobilization during the reaction. To determine the number of these Pvu I sites that are accessible on the chromatin templates without remodelling, we inhibited ATP-dependent nucleosome remodelling activity by addition of apyrase. Now, although total cutting is reduced, digestion of the acetylated chromatin remains markedly higher than the non-acetylated template (Figure 5, right panel). Similarly, inhibition of remodelling activity by addition of a 5-fold excess of non-hydrolysable AMP-PNP resulted in higher cutting on the acetylated compared to the non-acetylated templates (data not shown). Together, these data indicate that acetylation increases the accessibility of these chromatin templates; previous studies using DNase I as the probe for accessibility showed similar increased accessibility for acetylated chromatin (11).
Figure 5.
Increased accessibility of acetylated arrays. Time course of Pvu I digestion on acetylated (Acet) and non-acetylated (Non-Acet) arrays. The percent parental band cut was calculated following quantification with a phosphorimager. The increased cutting on acetylated arrays was observed in seven independent experiments and with three different preparations of (normalized) acetylated and non-acetylated histones. The right panels show Pvu I cutting when most ATP-dependent nucleosome remodelling complex activity was inhibited by removal of ATP with apyrase. Similar experiments in which remodelling activity is inhibited by addition of a large excess of AMP-PNP also showed increased Pvu I digestion on the acetylated chromatin (data not shown). We suggest that the difference in the level of accessibility between acetylated and non-acetylated chromatin measured here is less than the observed difference in the level of RAG cutting because Pvu I (28 kDa) is much smaller than ACF (600 kDa). Therefore, Pvu I would be expected to access compacted non-acetylated chromatin more readily thereby resulting in a smaller difference in cutting compared to acetylated chromatin.
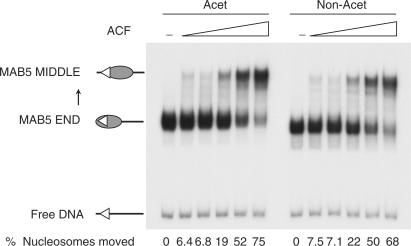
We next wished to determine whether acetylation additionally increases the ability of remodelling complexes to mobilize nucleosomes once they have gained access to the templates. Previous studies showed that acetylation of histone H4 peptide substrates at K12 and K16 actually decreases ATPase activity (32,33). To investigate if the hyper-acetylated histones used here cause a direct effect on remodelling activity, we titrated increasing amounts of remodelling factor and compared the degree of mobilization of acetylated and non-acetylated mono-nucleosomes. This showed that acetylation had no significant effect on the ability of ACF to mobilize mono-nucleosomes (Figure 6). Similar results were obtained with the catalytic subunit, ISWI (Supplementary Figure 3). Furthermore, we find that once the nucleosome has been mobilized off the RSS, RAG cutting is increased similarly on acetylated and non-acetylated nucleosomes (Supplementary Figure 3).
Figure 6.
Acetylation does not increase sliding on mono-nucleosomes by ACF or ISWI. Sliding assay using mono-nucleosomes reconstituted with acetylated and non-acetylated histones onto a 12-RSS fragment from MAB5. Increasing amounts of ACF are indicated above the gel. ISWI also caused equivalent mobilization of acetylated and non-acetylated nucleosomes (Supplementary Figure 3).
Next, to determine whether acetylation influences the off-rate/retention of remodelling complexes once they have gained access to their nucleosome templates, we bound purified ACF complex to acetylated and non-acetylated chromatin and then challenged this binding with salt washes of increasing stringency. Western analysis showed binding of ACF to acetylated chromatin is marginally increased compared to non-acetylated chromatin (Figure 7A, left side). This could be due to the increased accessibility of the acetylated chromatin; however, the ACF1 subunit has a bromodomain (26) and it seemed possible that this might affect the retention of the ACF complex via acetylated histone tails. We therefore performed the same experiment but used ACF1 where the bromodomain had been mutated (26). This time, no difference in off-rate is observed (Figure 7A, right side). To investigate if the marginal difference in binding of ACF to acetylated chromatin contributes to the increased remodelling, we performed a RAG cutting assay. As observed previously, addition of wild-type ACF complex stimulated RAG cutting on acetylated chromatin by more than 3-fold (Figure 7B). Mutation of the bromodomain eliminated this increase. However, upon addition of 2.5-fold higher amounts of ACF1 mutant-containing complex, enhanced RAG cutting on acetylated chromatin was again observed (Figure 7B, far right panel). Likewise, addition of the ISWI catalytic subunit, that lacks a bromodomain, also resulted in increased RAG cutting on acetylated chromatin (Supplementary Figure 4). Together, these data suggest that the more open structure of acetylated chromatin alone is sufficient to target remodelling complexes. However, when the concentration of the remodelling complex is limiting, bromodomain/acetylated histone tail interactions can promote some retention of remodelling complexes and thus contribute to increased nucleosome remodelling.
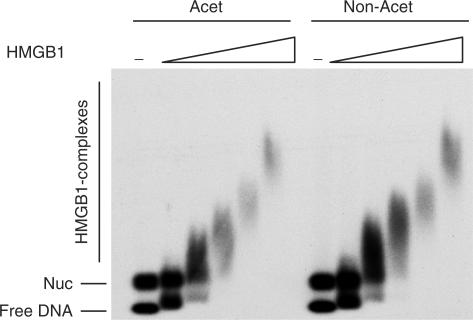
Finally, it is possible that acetylation enhances remodelling activity in yet another way: C-terminally truncated HMGB1 was included in all RAG cutting reactions since it causes a marked increase in RAG cutting in vitro, primarily at 23-RSSs (34). Recently, this truncated form of HMGB1 was also found to inhibit nucleosome mobilization by ACF by binding tightly to nucleosomal DNA (35). It therefore seemed possible that acetylation might alter the binding of truncated HMGB1 to nucleosomes. Indeed, if acetylation inhibited binding of truncated HMGB1 then this might lead to increased remodelling and thus RAG cutting specifically on the acetylated templates. However, as can be seen in Figure 8, acetylation makes no difference whatsoever to the binding of HMGB1 to mono-nucleosomes nor to its binding to nucleosome arrays (Supplementary Figure 5). Together, these data therefore support the hypothesis that a prime mechanism by which acetylation increases remodelling complex activity is by increasing their accessibility to chromatin templates.
Figure 8.
Binding of HMGB1 to purified acetylated and non-acetylated nucleosomes. Nucleosomes were assembled on the MAB5 fragment. One specific HMGB1/nucleosome complex is detected at the lowest concentration of added HMGB1. Addition of more HMGB1 results in non-specific binding. The dash indicates nucleosome without any added HMGB1.
DISCUSSION
V(D)J recombination involves the introduction of double-strand breaks into genomic DNA; such a process needs to be stringently regulated. Consistent with this, a number of levels of chromatin packaging contribute to the regulation. The mono-nucleosome imparts one level of protection. However, for the nucleosome-mediated repression to influence the initiation of recombination, the agent that mediates nucleosome remodelling needs to be regulated in the correct cell- and stage-specific manner. We describe here one way in which histone acetylation could contribute to this regulation, namely by modulating the accessibility of nucleosome remodelling complexes. Once the remodellers have been targeted they can, in turn, cause mobilization of nucleosomes off the RSSs to allow RAG cutting. We further suggest that similar regulation remodelling complex accessibility could contribute to their recruitment to acetylated, transcriptionally active domains.
Our data suggest that the ACF1 bromodomain/histone tail interaction does not make a major contribution to the binding and increased nucleosome remodelling by the ACF complex on acetylated chromatin. In contrast to tandem bromodomains, single bromodomain/acetylated histone tail interactions are weak [300–900 μM for GCN5 (36)]. Indeed, interaction between the WSTF bromodomain and acetylated histone tails alone was not sufficient to tether the remodelling complex WINAC at promoters (9). This suggests that it is unlikely that the single bromodomain in ACF1 can independently target the ACF remodelling complex in the nucleus. Consistent with this, we find that addition of just 2.5-fold higher amounts of ACF bromodomain mutant overcomes the requirement for the bromodomain (Figure 7B). Likewise, addition of the catalytic subunit, ISWI, results in increased cutting on acetylated chromatin. Therefore, it seems that the more open structure of acetylated chromatin alone is sufficient to target remodelling complexes and the extent to which the bromodomain/acetylated histone tail interaction contributes to this targeting in vivo will depend on the physiological concentration of remodelling complexes.
A number of properties of acetylated chromatin could modulate the accessibility to remodelling complexes. Firstly, acetylation is known to reduce compaction of the 30 nm fibre. Trypsinization studies showed that the histone tails play a key role in this compaction [reviewed in (37)]; acetylation of histone tails reduces but does not abolish this compaction as determined by altered sedimentation velocity and electron microscopy (10,12). Secondly, acetylated chromatin displays increased thermal untwisting. Indeed, acetylated nucleosomes constrain only about half the amount of DNA compared to non-acetylated nucleosomes (11). Since DNA at the entry/exit points loses contact with the histone octamer first in thermal denaturation experiments (38), and since the ends of acetylated nucleosomes are more accessible to DNase I (39), it has been proposed that DNA at the nucleosome entry/exit points will be more accessible in acetylated chromatin (11). ISWI binds DNA in the linker region close to nucleosomes (40). Therefore, it seems likely that the increased accessibility of these regions contributes to enhanced ISWI-dependent nucleosome remodelling.
A third means to increase the accessibility of acetylated chromatin is if proteins differentially associate with acetylated and non-acetylated chromatin. In vitro, these effects seem to be small: Removal of chromatin-associated proteins by sarkosyl-stripping demonstrated that the acetylated chromatin remains significantly more accessible than the non-acetylated (M.B., unpublished data). In vivo, however, the binding of chromatin-associated proteins is likely to enhance the difference in accessibility between the acetylated and non-acetylated arrays. Indeed, since histone H3 lysine 9 can be modified by either acetylation or methylation and since methylated H3 lysine 9 is a high affinity binding site for heterochromatin protein 1 (41,42), methylation will lead to the formation of a repressed chromatin structure; acetylation will reverse this effect.
There is a strong correlation between increased histone acetylation and the initiation of V(D)J recombination (22,23,43). Since nucleosomes need to be remodelled in a stochastic, locus-wide manner for the initiation of recombination, a plausible hypothesis is that the more accessible chromatin structure generated by histone acetylation facilitates the targeting of nucleosome remodelling complexes in vivo during V(D)J recombination. Consistent with this idea, subunits of remodelling complexes have been found to coincide with increased histone H3 acetylation throughout recombining loci. Indeed, the presence of the SWI/SNF ATPase, BRG1, correlated with increased histone H3 acetylation at two independent loci (the murine immunoglobulin heavy chain and the TCR-β locus), in cells where these loci are undergoing recombination (44). Recent studies have suggested yet a further signal that might contribute to the targeting of remodelling complexes: The PHD finger in the remodelling complex, NURF, as well as in the repressor ING2, interacts with histone H3 that is trimethylated at lysine 4 (H3K4me3) (5). Since acetylation and H3K4me3 occur on the same molecule of histone H3 (45), it is possible that these two modifications reinforce each other in targeting remodelling complexes although, it should be pointed out that to date, H3K4me3 has only been reported at the boundaries of domains undergoing V(D)J recombination (44).
As well as targeting remodelling complexes, these modifications also potentially target RAG proteins: The RAG2 C-terminal domain was found to interact with free histones that are modified in various ways, including acetylation and H3K4me2 (46). Whilst this might help to stimulate RAG cutting in vivo on acetylated chromatin, it is unlikely to have any effect in our in vitro assay since we used truncated RAG2 that lacks the C-terminal domain. Moreover, it should be pointed out that since RAG proteins are incapable of forming double-strand breaks at the RSS when it is constrained by a nucleosome (16–18), correct targeting of remodelling complexes is also needed for recombination to occur.
Increased locus-wide acetylation is not only associated with recombining loci but also with transcriptionally active domains (47). A reasonable possibility, therefore, is that the more open structure associated with domain-wide acetylation in vivo (10–12) directs the remodelling complex to the appropriate chromatin region. More precise recruitment of the remodelling complex to promoter elements might then be mediated either by transcription factors associated with the remodelling complex (3) or by a specific histone code of promoter nucleosome (5–7). Interestingly, using integrated mini-gene V(D)J recombination templates, it was found that a promoter proximal to the recombining RSS is required to obtain efficient RAG cutting (48). Since the orientation of the promoter was irrelevant to the enhanced recombination, a plausible scenario is that promoters help direct remodelling complex activity once accessibility has been gained to the acetylated locus.
The ability of histone acetylation to target remodelling complex activity requires that histone hyper-acetylation precedes remodeller recruitment. During gene activation, temporal studies have shown that at some promoters this is indeed the case (49,50). At the yeast HO promoter, however, SWI/SNF is present prior to recruitment of Gcn5 acetyltransferase (51). In this case, it seems likely that association of SWI/SNF with the activator, Swi5, provides an alternative means of directing the remodelling complex to the promoter (4).
One previous report suggested that histone acetylation synergizes with SWI/SNF to enhance RAG cutting on mono-nucleosomes (20). However, two other studies found no effect of acetylation on RAG cutting on mono-nucleosomes (16,18). Furthermore, acetylation of histone tail peptides was found to have either no effect on remodelling activity (52) or to actually inhibit ATPase activity (32,33). A possible reason for the differences with the study by Kwon et al. (20) might stem from mechanistic differences in remodelling by SWI/SNF and ISWI-containing complexes: Indeed, the histone tails appear to have distinct roles in remodelling by these two classes of complexes (53). More recent studies from the Oettinger group now report that acetylation does not affect remodelling complex activity even on nucleosome arrays (21). We suggest these latest differences might be due to the degree of chromatin assembly: In the Oettinger study, chromatin was assembled using 5S rDNA nucleosome positioning sequences and assembly was checked using a restriction enzyme that examines nucleosome assembly at only one site in the array of 12 nucleosomes. Therefore, it seems possible that other regions of the array might not be uniformly assembled. In our case, Drosophila extracts were used to assemble physiologically spaced chromatin and in all cases we checked that assembly of acetylated and non-acetylated chromatin was identical (Figure 1). We therefore suggest that our data are consistent with the idea that acetylation enhances remodelling complex activity by increasing their accessibility to nucleosome arrays and that this is a new way in which acetylation and remodelling complexes synergize to enhance locus activity.
Taken together, these data begin to build a picture of how the different levels of chromatin packaging cooperate to generate an active chromatin structure. Understanding how the acetylation is targeted to specific regions of recombining loci is a key question for future studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
ACKNOWLEDGEMENTS
We thank Kevin Hiom for the gift of HMGB1, Fraser McBlane for the RAG1 and RAG2 expression vectors and for helpful advice with the LMPCR, Cristina Chioda for cloning the ACF1deltaBrd-FLAG bacmid and Peter Verrijzer for the anti-ISWI antiserum. This work was supported by the Medical Research Council (UK). M.B. was the recipient of an EMBO long-term fellowship. K.P.N. was supported by a Wellcome Trust Career Development Fellowship. J.B. gratefully acknowledges support from the Lister Institute of Preventive Medicine and a University Research Fellowship from the University of Leeds. Funding to pay the Open Access publication charges for this article was provided by the Medical Research Council (UK).
Conflict of interest statement. None declared.
REFERENCES
- 1.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 2.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 3.Neely KE, Hassan AH, Wallberg AE, Steger DJ, Cairns BR, Wright AP, Workman JL. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- 4.Yudkovsky N, Logie C, Hahn S, Peterson CL. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes. Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 6.Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 7.Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 8.Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujiki R, Kim MS, Sasaki Y, Yoshimura K, Kitagawa H, Kato S. Ligand-induced transrepression by VDR through association of WSTF with acetylated histones. EMBO J. 2005;24:3881–3894. doi: 10.1038/sj.emboj.7600853. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Garcia-Ramirez M, Rocchini C, Ausio J. Modulation of chromatin folding by histone acetylation. J. Biol. Chem. 1995;270:17923–17928. doi: 10.1074/jbc.270.30.17923. [DOI] [PubMed] [Google Scholar]
- 11.Krajewski WA, Becker PB. Reconstitution of hyperacetylated, DNase I-sensitive chromatin characterized by high conformational flexibility of nucleosomal DNA. Proc. Natl Acad. Sci. USA. 1998;95:1540–1545. doi: 10.1073/pnas.95.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 14.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal v(d)j recombination. Cell. 2002;109(Suppl):S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 15.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 16.Golding A, Chandler S, Ballestar E, Wolffe AP, Schlissel MS. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO J. 1999;18:3712–3723. doi: 10.1093/emboj/18.13.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon J, Imbalzano AN, Matthews A, Oettinger MA. Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol. Cell. 1998;2:829–839. doi: 10.1016/s1097-2765(00)80297-x. [DOI] [PubMed] [Google Scholar]
- 18.McBlane F, Boyes J. Stimulation of V(D)J recombination by histone acetylation. Curr. Biol. 2000;10:483–486. doi: 10.1016/s0960-9822(00)00449-8. [DOI] [PubMed] [Google Scholar]
- 19.Baumann M, Mamais A, McBlane F, Xiao H, Boyes J. Regulation of V(D)J recombination by nucleosome positioning at recombination signal sequences. EMBO J. 2003;22:5197–5207. doi: 10.1093/emboj/cdg487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon J, Morshead KB, Guyon JR, Kingston RE, Oettinger MA. Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Mol. Cell. 2000;6:1037–1048. doi: 10.1016/s1097-2765(00)00102-7. [DOI] [PubMed] [Google Scholar]
- 21.Patenge N, Elkin SK, Oettinger MA. ATP-dependent remodeling by SWI/SNF and ISWI proteins stimulates V(D)J cleavage of 5 S arrays. J. Biol. Chem. 2004;279:35360–35367. doi: 10.1074/jbc.M405790200. [DOI] [PubMed] [Google Scholar]
- 22.Huang J, Durum SK, Muegge K. Cutting edge: histone acetylation and recombination at the TCR gamma locus follows IL-7 induction. J. Immunol. 2001;167:6073–6077. doi: 10.4049/jimmunol.167.11.6073. [DOI] [PubMed] [Google Scholar]
- 23.Ye SK, Agata Y, Lee HC, Kurooka H, Kitamura T, Shimizu A, Honjo T, Ikuta K. The IL-7 receptor controls the accessibility of the TCRgamma locus by Stat5 and histone acetylation. Immunity. 2001;15:813–823. doi: 10.1016/s1074-7613(01)00230-8. [DOI] [PubMed] [Google Scholar]
- 24.McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- 25.Roth DB, Roth SY. Unequal access: regulating V(D)J recombination through chromatin remodeling. Cell. 2000;103:699–702. doi: 10.1016/s0092-8674(00)00173-2. [DOI] [PubMed] [Google Scholar]
- 26.Eberharter A, Vetter I, Ferreira R, Becker PB. ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts. EMBO J. 2004;23:4029–4039. doi: 10.1038/sj.emboj.7600382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nightingale KP, Wellinger RE, Sogo JM, Becker PB. Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26 gene in chromatin. EMBO J. 1998;17:2865–2876. doi: 10.1093/emboj/17.10.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eberharter A, Ferrari S, Langst G, Straub T, Imhof A, Varga-Weisz P, Wilm M, Becker PB. Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J. 2001;20:3781–3788. doi: 10.1093/emboj/20.14.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Gent DC, McBlane JF, Ramsden DA, Sadofsky MJ, Hesse JE, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 30.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 31.Kang JG, Hamiche A, Wu C. GAL4 directs nucleosome sliding induced by NURF. EMBO J. 2002;21:1406–1413. doi: 10.1093/emboj/21.6.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clapier CR, Nightingale KP, Becker PB. A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 2002;30:649–655. doi: 10.1093/nar/30.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corona DF, Clapier CR, Becker PB, Tamkun JW. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 2002;3:242–247. doi: 10.1093/embo-reports/kvf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Gent DC, Hiom K, Paull TT, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonaldi T, Langst G, Strohner R, Becker PB, Bianchi ME. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J. 2002;21:6865–6873. doi: 10.1093/emboj/cdf692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J. Mol. Biol. 2000;304:355–370. doi: 10.1006/jmbi.2000.4207. [DOI] [PubMed] [Google Scholar]
- 37.Hayes JJ, Hansen JC. Nucleosomes and the chromatin fiber. Curr. Opin. Genet. Dev. 2001;11:124–129. doi: 10.1016/s0959-437x(00)00168-4. [DOI] [PubMed] [Google Scholar]
- 38.Simpson RT. Mechanism of a reversible, thermally induced conformational change in chromatin core particles. J. Biol. Chem. 1979;254:10123–10127. [PubMed] [Google Scholar]
- 39.Simpson RT. Structure of chromatin containing extensively acetylated H3 and H4. Cell. 1978;13:691–699. doi: 10.1016/0092-8674(78)90219-2. [DOI] [PubMed] [Google Scholar]
- 40.Langst G, Becker PB. ISWI induces nucleosome sliding on nicked DNA. Mol. Cell. 2001;8:1085–1092. doi: 10.1016/s1097-2765(01)00397-5. [DOI] [PubMed] [Google Scholar]
- 41.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 42.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 43.McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination [see comments] Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- 44.Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc. Natl Acad. Sci. USA. 2003;100:11577–11582. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nightingale KP, Gendreizig S, White DA, Bradbury C, Hollfelder F, Turner BM. Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J. Biol. Chem. 2007;282:4408–4416. doi: 10.1074/jbc.M606773200. [DOI] [PubMed] [Google Scholar]
- 46.West KL, Singha NC, De Ioannes P, Lacomis L, Erdjument-Bromage H, Tempst P, Cortes P. A direct interaction between the RAG2 C terminus and the core histones is required for efficient V(D)J recombination. Immunity. 2005;23:203–212. doi: 10.1016/j.immuni.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 47.Hebbes TR, Clayton AL, Thorne AW, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikes ML, Meade A, Tripathi R, Krangel MS, Oltz EM. Regulation of V(D)J recombination: a dominant role for promoter positioning in gene segment accessibility. Proc. Natl Acad. Sci. USA. 2002;99:12309–12314. doi: 10.1073/pnas.182166699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 50.Reinke H, Gregory PD, Horz W. A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol. Cell. 2001;7:529–538. doi: 10.1016/s1097-2765(01)00200-3. [DOI] [PubMed] [Google Scholar]
- 51.Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 52.Georgel PT, Tsukiyama T, Wu C. Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J. 1997;16:4717–4726. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Logie C, Tse C, Hansen JC, Peterson CL. The core histone N-terminal domains are required for multiple rounds of catalytic chromatin remodeling by the SWI/SNF and RSC complexes. Biochemistry. 1999;38:2514–2522. doi: 10.1021/bi982109d. [DOI] [PubMed] [Google Scholar]
- 54.Langst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.