Abstract
The global transcriptional regulator H-NS selectively silences bacterial genes associated with pathogenicity and responses to environmental insults. Although there is ample evidence that H-NS binds preferentially to DNA containing curved regions, we show here that a major basis for this selectivity is the presence of a conserved sequence motif in H-NS target transcriptons. We further show that there is a strong tendency for the H-NS binding sites to be clustered, both within operons and in genes contained in the pathogenicity-associated islands. In accordance with previously published findings, we show that these motifs occur in AT-rich regions of DNA. On the basis of these observations, we propose that H-NS silences extensive regions of the bacterial chromosome by binding first to nucleating high-affinity sites and then spreading along AT-rich DNA. This spreading would be reinforced by the frequent occurrence of the motif in such regions. Our findings suggest that such an organization enables the silencing of extensive regions of the genetic material, thereby providing a coherent framework that unifies studies on the H-NS protein and a concrete molecular basis for the genetic control of H-NS transcriptional silencing.
INTRODUCTION
H-NS is an abundant global regulator of transcription and a determinant of nucleoid structure in many bacteria (1,2). This protein is primarily a repressor acting both as a selective silencer of specific genes or regions of the chromosome and also more locally in concert with other transcription factors within a regulatory region (3). Genes under H-NS control in enteric bacteria are often responsive to abrupt environmental changes. Such genes include those activated by enhanced osmolarity of the growth medium (exemplified by proV) and also genes associated with pathogenicity, notably Shigella flexneri virF and the pathogenicity-associated islands in uropathogenic bacteria (4–9). H-NS binding to DNA in vitro is facilitated by structural characteristics of DNA (1) but by itself this mechanism may not provide sufficient selectivity for precise silencing. However, by combining recent findings we derive here an AT-rich sequence motif for directing sequence-specific DNA binding by H-NS. Depending on their context such motifs can effect local repression or act more globally altering the packaging of DNA thereby silencing the packaged genes. Importantly, the proximity of these sites within putative silenced regions indicates how H-NS silencing can be nucleated and propagated over a substantial length of the bacterial chromosome. We conclude from an analysis of genes containing H-NS binding sites that a major role of H-NS, and also possibly its paralog, is to regulate functional systems as a whole rather than selectively targeting individual regulatory or structural genes.
RESULTS
Identification of an H-NS DNA binding motif
In addition to structurally determined DNA binding (1), two recent findings suggest that H-NS binding can be highly sequence specific. First, two tight binding sites of identical sequence with a Kd of ∼15 nM were identified in the Escherichia coli proU operon (10). Importantly, this sequence also directed tight H-NS binding when placed in a completely different sequence context, thus excluding any more extensive structure-specific recognition. Second, the H-NS DNA binding domain, in isolation, selectively footprinted particular sequences of varying extents within the regulatory regions of several H-NS sensitive genes (11). Although both results are indicative of sequence-specific binding neither individually is sufficient to identify a ‘consensus’ sequence. However, inspection of the footprinted sequences revealed that they all contained at least one section with high similarity to the high-affinity sites in proV. From the identified sequences a template motif was derived (Table 1). Because the derived motif depended on the assumption that high-affinity binding sites are related in sequence we used the motif to predict three potential H-NS binding sites in the fis promoter from positions −150 to +1. These predicted sites, although imperfect fits to the derived motif, confirmed a footprinting analysis of this promoter (Supplementary Data) and thus provide a validation of the derived sequence template for H-NS binding in vitro (Table 1). Combining the sequences for all identified sites yields the 10 bp motif tCGATAAATT (Figure 1). Two features are of note. The base composition, 78% AT, is fully consistent with the documented preferred composition of H-NS targets in vivo (12). Second, a centrally positioned T-A base step is found in all identified sites. This step would confer thermal instability and both axial and torsional flexibility on the binding sites (13).
Table 1.
Derivation of a DNA binding motif template for H-NS
| Gene | Site | Fit |
|---|---|---|
| (/10) | ||
| proV 1 | TCGATATATT | 9 |
| proV 2 | TCGATATATT | 9 |
| gadA | CGGATAAATC | 7 |
| gadX | TCGTTAAAAT | 9 |
| virF | TAGATTAAGT | 7.5 |
| cspA | CCGATTAATC | 7.5 |
| hns | TCCTTACATT | 8 |
| OriC | TGTATACTTT | 6 |
| Template | tCGtTAaATt | |
| a t | ||
| fis 1 | GCGATAATTA | 7 |
| fis 2 | GCATTTAAAA | 5.5 |
| fis 3 | GCGACTAAAA | 5.5 |
Figure 1.
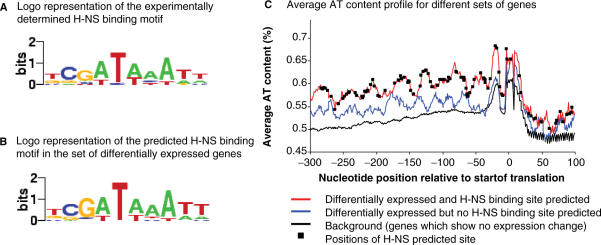
(A) Logo representation of the experimentally determined H-NS binding motif derived from sequences in Table 1. (B) Logo representation of the predicted H-NS binding motif derived from identified occurrences in the set of differentially expressed genes responding to H-NS removal under different growth conditions. (C) Average AT content profiles of sequences from −300 to +100 bp relative to the start of translation for sets of genes differentially expressed in hns cells but containing or lacking a predicted H-NS binding site. Logos were generated using enoLOGOS (http://biodev.hgen.pitt.edu/cgi-bin/enologos/enologos.cgi).
The binding motif was initially derived exclusively from in vitro footprinting data. However, in vivo binding sites have been identified by H-NS-DNA ChIP-on-chip experiments (12,14–16). Although such experiments do not provide the resolution of in vitro footprinting, they give valuable genome-scale information on locations where H-NS binds DNA. We can therefore ask whether sequences related to the binding motif occur at or close to the identified segments. We found that of the top 99 highest scoring sites identified by Grainger et al. (16) the motif occurred in 59 within 250 bp of the reported signal peak (Supplementary Data 1). As a control, we randomly picked 99 regions of 500 bp in length and asked what fraction contained a high-affinity H-NS binding site. This calculation was repeated 1000 times and revealed that the average number of expected binding sites was 39, with a standard deviation of ∼4.5 binding sites. Therefore, our observation that 59 of the 99 high-confidence regions have a high-affinity H-NS binding site is statistically significant (Z-score = 4.5; P-value <10−3). We conclude that the H-NS binding motif that we identified in vitro is also characteristic of many in vivo binding sites. This analysis also revealed that sites identified by the ChIP-on-chip protocol show a significant enrichment for AT. The average AT content of the high-confidence H-NS bound sites was 61.4%. Control experiments where we analyzed AT content for a thousand random 500 bp regions from the genome showed that the average expected value was 49.2%. Thus, the observed value of 61.4% represents a difference of >21 SDs from what is expected by chance. In another independent control experiment, we first identified 1000 random 500 bp segments that had a predicted binding site and analyzed their AT content. This investigation revealed that the average expected AT content for random 500 bp segments that contained a motif was 52%. We therefore conclude that the sites identified by ChIP-on-chip are, on average, substantially enriched in AT content but randomly selected sequences that may or may not contain a site are not.
In vivo function of an H-NS binding motif
In the absence of other data, the binding of H-NS to a site in vivo, or even the presence of a site, is not necessarily indicative of a biologically relevant role. To assess the functionality of the H-NS binding motif, we asked whether its occurrence correlated with genes known to be regulated by H-NS in vivo. To test whether such a correlation exists, we scanned two sets of genes previously identified by microarray analysis to be derepressed in an E. coli strain lacking H-NS (17). Since H-NS is only one component of a complex regulatory system which includes RpoS and LRP as well as other DNA binding proteins (1,18), derepression could be either a direct or an indirect consequence of H-NS removal and therefore not all genes identified by these screens are likely to be direct H-NS targets. The first set of scanned genes was initially compiled to include genes responding to H-NS removal under different growth conditions. We found that of the 57 genes scanned 33 (or 57.9%) contained one or more sequences related to the H-NS binding motif within a region spanning positions −300 to +100 bp relative to the first nucleotide of the translation start site (see Methods section). The positive hits included the genes ade, fimD, gadA, hlyE, ompF, osmC, proV (Supplementary Data 2), all of which have previously been independently shown to be regulated by H-NS. The second set of scanned genes were initially characterized as those which were differentially expressed in isogenic hns+ and hns− strains on either reduction or increase of the natural superhelical density (17). In this set, we found that of the 211 genes scanned 98 (or 46.4%) registered as positive hits (Supplementary Data 3). These included several additional genes, including the hde locus and many genes involved in flagella formation, previously known to be under H-NS control. Combining both sets 116 putative target genes were identified, 32 of which contained more than one copy of the motif.
Another question of interest was whether the genes containing the H-NS motif differ in base composition from those that do not. Since the preferred in vivo targets of H-NS are generally AT-rich (12), we compared the base composition of these two classes of genes for the region from −300 to +100 bp relative to the translation start site (see Methods section). We found that for the first set of scanned genes (expression change in the absence of H-NS) the AT content of the genes containing the H-NS motif was, on average, consistently ∼10% above the average AT content both of the genes apparently lacking the H-NS motif and also the average composition in this region for all E. coli genes (Figure 1; P-value <10−3; Wilcoxon signed-rank test). For the second set of scanned genes (initially assayed under conditions of hyper- and hypo-negative supercoiling), the same differential was observed between the genes containing the H-NS motif and the average composition in this region for all E. coli genes (P-value <10−3). However, for this set the AT content of the genes apparently lacking the H-NS motif was intermediate between those containing the H-NS motif and all E. coli genes (P-value <10−3). By including the additional putative H-NS motifs identified in our sequence screen, we refined the logo for H-NS binding (Figure 1) and observed little change from the input motif. Again, the most highly conserved feature of the individual identified motifs was a centrally placed T-A base step. In the very few cases where this was absent it was replaced by C-A, the second most flexible base step (13).
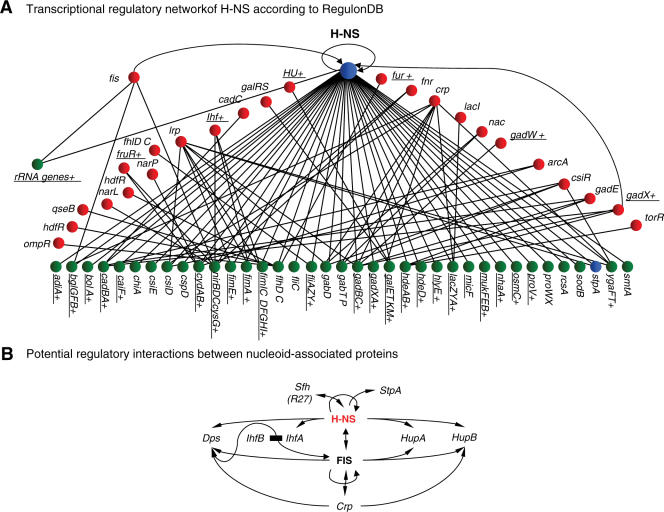
By their nature, microarray screens used to identify genes whose expression is hns sensitive would not necessarily reveal all genes under direct H-NS control. For example, one such well-characterized gene, bgl, remains uninduced under most laboratory conditions (19). We therefore scanned the genes compiled in the RegulonDB database (20) as well-characterized targets of H-NS. A total of 24 out of 39 (or 61.5%) contained sequences with a reasonable match to the binding site logo (Figure 2A). We noted that the bgl regulatory region, in addition to containing sequences closely related to the template, also contained several similar sequences that fitted the template less closely. Using the criterion that such ‘secondary’ sequences should contain a run of at least six consecutive base-pairs and a T-A base step we identified 10 motifs of this type associated with the bgl regulatory elements (Supplementary Data). A similar pattern of multiple primary and secondary sequences was also observed in a number of other genes known to be sensitive to H-NS. These included eltAB, flgA/flgB, hdeAB, htdA, micF, spvR, virF and yghJ (6,7,21–23).
Figure 2.
(A) Analysis of known transcriptional regulatory network of H-NS from RegulonDB for occurrence of the binding motif template. Genes containing a predicted strong H-NS binding site are underlined. (B) Potential regulatory interactions between nucleoid-associated proteins. The arrows indicate that one protein may directly affect the expression of another but do not indicate whether such regulation would be positive or negative.
Although the genes silenced by H-NS appear to contain multiple putative sites for H-NS binding, there are other genes for which H-NS appears to bind to more isolated sites. One such gene is nir (3) where H-NS binds to the upstream region in concert with FIS and IHF to repress promoter activity. This region contains two potential H-NS binding sites (GGGTATATTG and TTGTATAAAT) correlating with the H-NS footprint. Both contain the ‘core’ of the consensus and the presence of two T-A steps indicates that the sequences are unlikely to confer coherent intrinsic curvature (24).
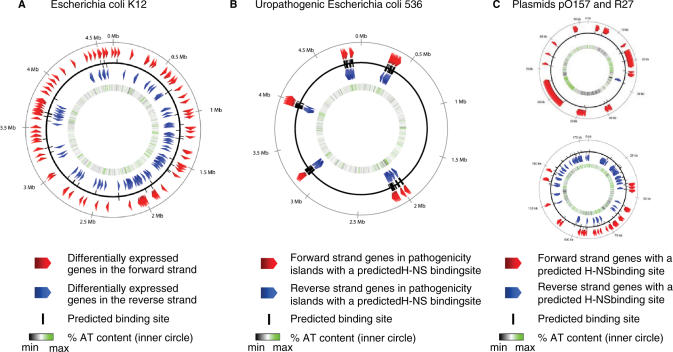
Since it is well established that H-NS regulates many genes correlated with enteric pathogenicity of E. coli and Salmonella typhimurium ssp., we asked whether putative H-NS binding sites could be identified within plasmids such as pO157 and R27 that confer pathogenicity and also within the pathogenicity-associated islands in the uropathogenic E. coli strain 536 (9). In all cases, we observed a substantial number of positive hits, respectively 19, 80 and 211 genes (Figure 3). In many cases, these hits identify genes of unknown function. We note that particularly in the case of the pathogenicity-associated islands of strain 536 there is a strong tendency for the identified genes to be clustered. On average within the islands there is a strong predicted site spaced approximately every 2.5 genes (Table 2). Although certain predicted sites can be associated with more than one gene (e.g. when transcription is divergent), the analysis also reveals that there are regions within all of the pathogenicity islands where the spacing between strong sites is ≥8 genes. These regions could conceivably represent gaps between potential silenced domains.
Figure 3.
Maps of occurrence of sequences with close matches to the binding motif template in selected regions of (A) E. coli K12, (B) uropathogenic E. coli 536 (C) plasmids pO157 (top) and R27 (bottom). The predicted sites are oriented with respect to the DNA strands.
Table 2.
Average number of genes between identified H-NS binding motifs in the pathogenicity-associated islands in E. coli U536
| Island | Average number of genes between motifs |
|---|---|
| PAI1 | 2.40 |
| PAI2 | 2.40 |
| PAI3 | 2.38 |
| PAI4 | 2.71 |
| PAI5 | 2.95 |
| AVG | 2.57 |
DISCUSSION
By combining DNA footprinting data from different sources, we have identified a DNA sequence that correlates with H-NS binding at specific locations in the E. coli genome. This sequence, or sequences closely related to it, is found in the regulatory regions of genes that are silenced by H-NS (e.g. bgl and proU) and also those (e.g. nir) in which H-NS acts in cooperation with other abundant DNA-binding proteins. In the former class, but not in the latter, abundant AT-rich sequences which fit less well to the consensus are interdigitated with the putative high-affinity binding sites. We suggest that this organization facilitates the ‘spreading’ of bound H-NS around nucleating high-affinity sites and the consequent formation of a repressive higher-order structure that is responsible for silencing.
We have validated the identified binding site by three procedures—by showing that it occurs with high frequency in vivo at H-NS binding sites identified by a ChIP-on-chip protocol, by a search for its occurrence in genes shown to be differentially regulated in hns mutant strains and by direct comparison to reported characterized footprints. We have further shown that sequences closely related to the consensus are associated with genes implicated in pathogenicity. Such genes have previously been shown to be under H-NS control. Our search was restricted to a region spanning −300 to +100 bp relative to the initiating ATG of translated genes and would miss sites located outside this region. Indeed, one of the high-affinity binding sites identified by Bouffartigues et al. (10) is located at +130 bp downstream of the translation start site. The genes identified are thus a minimal estimate of those containing the motif. Nevertheless, its identification within all the characterized regulatory regions analyzed (e.g. bgl, proV) implies that a high-affinity binding site or sites are characteristic of such regions. We infer that these sites are a basis for selective silencing. Our screen also identified putative binding sites in adjacent genes (e.g. csgD and csgE, fimA and fimI), and genes separated by one other gene within the same operon (e.g. flgC, flgE, flgG). The pattern of sites associated with adjacent genes was particularly apparent both in the R27 plasmid and the pathogenicity-associated islands of E. coli U536.
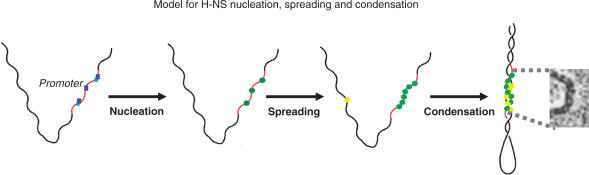
Our results are consistent with a silencing mechanism in which H-NS binds first to nucleating high-affinity sites (10,25), corresponding in principle to the logo identified here, and then polymerizes resulting eventually in the formation of a supercoiled interwound filament containing two DNA duplexes joined by protein bridges, concomitantly constraining a DNA loop (21,26–29) (Figure 4). This mode of binding also explains previous observations showing that H-NS binds preferentially to DNA containing intrinsically curved regions since the DNA in plectonemes is bent both within the interwindings and at the apical loops. The frequent location of nucleating sites associated with adjacent genes and also their repetition within operons would allow the filament to silence an extensive region. Such a mechanism would be consistent with the silencing of pathogenicity-associated islands in enteropathogenic Proteobacteria (9). Similarly, the AT-rich composition of regions adjacent to the sites we have identified could facilitate spreading of H-NS by interspersing high-affinity nucleating sites with lower affinity AT-rich secondary sites. In support of this notion, in the pO157 plasmid the higher AT content of regions containing H-NS binding sites relative to those that do not, be a simple consequence of the composition of the site itself since there are extensive regions enriched in AT close to the initiation codon that lack identified motifs (Supplementary Data 5). In addition, we have shown that the presence of a motif per se is not necessarily correlated with substantial AT enrichment. AT-enriched regions are thus extensive and may contain zero to several examples of the motif. We suggest that nucleation of DNA binding of a structural component of a protein–DNA array, such as an H-NS-DNA fiber, followed by spreading, may constitute a general mechanism for organizing chromatin both in bacteria and eukaryotes.
Figure 4.
Proposed mechanism of silencing over extended regions. Initial nucleation of H-NS binding at high-affinity sites is following by spreading and condensation of H-NS on supercoiled DNA to generate a silenced fiber. Spreading and condensation could be either consecutive or concomitant. Blue squares represent high-affinity H-NS binding sequences (nucleating sites) and green and yellow circles represent bound H-NS on nominal distal and proximal sides, respectively, of a promoter. The inset of an H-NS-DNA complex is taken from ref. 26 with permission.
The ability of H-NS both to bind locally and to silence longer regions of DNA is similar to the DNA-binding properties of another abundant global regulator, FIS. Both can bind with high specificity to individual sites and can also bind cooperatively to more extensive regions. These properties allow them to act both as transcription factors and as determinants of nucleoid structure. Another feature of the occurrence of H-NS binding sites is that many genes within a particular regulatory system appear to be targeted. This pattern is particularly evident within the genes responsible for flagellar biogenesis. The H-NS binding motif is associated with not only the genes encoding the structural genes of flagella but also with the specific sigma factor (fliA) and anti-sigma factor (flgM) genes for regulating flagella biosynthesis. In addition the H-NS binding motif is associated with papX, a gene encoded by the pO157 plasmid that is a functional homolog of Proteus mirabilis mrpJ (30) and represses the flagellar regulon. In this example, the pattern is indicative of a primary global regulator that controls both positive and negative secondary regulators as well as their targets. To assess the biological relevance of this pattern, we compared the occurrence of the motif with the in vivo binding of H-NS to flagellar genes. Peaks of binding detected by ChIP-on-chip are located near flgN, fliA, fliC (two separate ones), fliD, fliF and fliZ (14). Nearly all of these, with the exception of the second fliC peak and fliD, contain the H-NS binding motif. We conclude that there is a good correlation between the presence of the motif and in vivo binding for this set of genes and that this correlation is indicative of functionality. However, the effects of hns on the expression of the flagellar regulon are complex. In hns mutants, production of FliC protein in response to anoxia is enhanced (31). Nevertheless, the flhDC master operon is repressed in vitro by H-NS and expression from its promoter is increased in an hns mutant (32). Similarly an activator, HdfR, of the master operon, is also negatively regulated by H-NS in vivo (33) and its gene is associated with a putative H-NS binding site. These observations show that H-NS can repress certain individual transcriptons within the flagellar regulon, but the regulatory complexity is such that the outcome of H-NS removal is likely only predictable in the context of the complete control network. We suggest that H-NS acts as a system regulator that is involved in coordinating the expression of the flagellar regulon.
Another possible example of this mode of control by H-NS is the mxi-spa type III secretion regulon (34). In this system, the activator VirB displaces H-NS from repressed promoter sites but does not itself directly affect RNA polymerase function (35). Other such regulons would include the pili and fimbriae biosynthetic modules. An additional aspect of regulation by H-NS is the interdependent regulation of H-NS and its paralogs. One of the genes identified in our screen was sfh, a paralog of H-NS encoded by the R27 virulence plasmid (21,36). The paralog has the same DNA binding domain as H-NS and so is assumed to bind to the same DNA sites. Notably the regulatory regions of both the hns gene itself and that of its paralog StpA contain multiple sites with similarities to the H-NS binding site, although in neither case are there sites which correspond to the canonical sequence (Supplementary Data 12). We suggest that interactions between these different, but functionally related proteins serve to act homeostatically to maintain an appropriate overall total concentration of repressors in the cell. Concomitantly, the interactions of H-NS paralog and FIS with the regulatory regions of other genes encoding nucleoid-associated proteins (crp, hupA, hupB) would similarly act homeostatically to maintain nucleoid structure (Figure 2B).
MATERIALS AND METHODS
The complete genome, predicted proteome and coding sequence locations for the E. coli strain K12 and the plasmids were obtained from NCBI GenBank. The regions from −300 to +100 bp relative to the translation start site were scanned for the presence of the binding motif using several publicly available programs such as MAST, MotifScanner and ScanACE. The program WebLogo was used to automatically generate logos of the predicted binding sites. For the AT content analysis, the genes were categorized into three classes: (i) those that were differentially expressed and predicted to have an H-NS binding site (direct targets), (ii) those that were differentially expressed but did not contain a predicted H-NS binding site (indirect targets) and (iii) those that were not differentially expressed (control set). In order to calculate the mean AT content for each position within the sequences of each set, we used a window size of 11 bp (i.e. ±5 bp). We acquired ChIP-on-chip signal locations from Grainger et al. (16). A window of −250 to +250 bp relative to the signal center was analyzed for presence of the binding site motif. Other methods were as described above.
SUPPLEMENTARY DATA
Available from NAR website and from URL http://www.mrc-lmb.cam.ac.uk/genomes/madanm/blang/hns/
ACKNOWLEDGEMENTS
This work was supported in part by Italian MIUR grants (PRIN 2005) to C.L.P. and C.O.G. E.B. was supported by La Fondation pour la Recherche Médicale. S.R. acknowledges the ANR for funding for the MASTRIT project. Funding to pay the Open Access publication charges for this article was provided by Medical Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nat. Rev. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 2.Rimsky S. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 2004;7:109–114. doi: 10.1016/j.mib.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Browning DF, Cole JA, Busby SJ. Suppression of FNR-dependent transcription activation at the Escherichia coli nir promoter by Fis, IHF and H-NS: modulation of transcription initiation by a complex nucleo-protein assembly. Mol. Microbiol. 2000;37:1258–1269. doi: 10.1046/j.1365-2958.2000.02087.x. [DOI] [PubMed] [Google Scholar]
- 4.Lucht JM, Dersch P, Kempf B, Bremer E. Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J. Biol. Chem. 1994;269:6578–6586. [PubMed] [Google Scholar]
- 5.Fletcher SA, Csonka LN. Fine-structure deletion analysis of the transcriptional silencer of the proU operon of Salmonella typhimurium. J. Bacteriol. 1995;177:4508–4513. doi: 10.1128/jb.177.15.4508-4513.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gowrishankar J, Manna D. How is osmotic regulation of transcription of the Escherichia coli proU operon achieved? A review and a model. Genetica. 1996;97:363–378. doi: 10.1007/BF00055322. [DOI] [PubMed] [Google Scholar]
- 7.Falconi M, Colonna B, Prosseda G, Micheli G, Gualerzi CO. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 1998;17:7033–7043. doi: 10.1093/emboj/17.23.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prosseda G, Falconi M, Giangrossi M, Gualerzi CO, Micheli G, Colonna B. The virF promoter in Shigella: more than just a curved DNA stretch. Mol. Microbiol. 2004;51:523–537. doi: 10.1046/j.1365-2958.2003.03848.x. [DOI] [PubMed] [Google Scholar]
- 9.Brzuszkiewicz E, Bruggemann H, Liesegang H, Emmerth M, Olschlager T, Nagy G, Albermann K, Wagner C, Buchrieser C, et al. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl Acad. Sci. USA. 2006;103:12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 2007;14:441–448. doi: 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]
- 11.Stella S. Ph.D. Thesis. University of Camerino; 2007. [Google Scholar]
- 12.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 13.Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oshima T, Ishikawa S, Kurokawa K, Aiba H, Ogasawara N. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 2006;13:141–153. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- 15.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grainger DC, Hurd D, Goldberg MD, Busby SJ. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 2006;34:4642–4652. doi: 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blot N, Mavathur R, Geertz M, Travers A, Muskhelishvili G. Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep. 2006;7:710–715. doi: 10.1038/sj.embor.7400729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouvier J, Gordia S, Kampmann G, Lange R, Hengge-Aronis R, Gutierrez C. Interplay between global regulators of Escherichia coli: effect of RpoS, Lrp and H-NS on transcription of the gene osmC. Mol. Microbiol. 1998;28:971–980. doi: 10.1046/j.1365-2958.1998.00855.x. [DOI] [PubMed] [Google Scholar]
- 19.Dole S, Nagarajavel V, Schnetz K. The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon downstream of the promoter. Mol. Microbiol. 2004;52:589–600. doi: 10.1111/j.1365-2958.2004.04001.x. [DOI] [PubMed] [Google Scholar]
- 20.Salgado H, Gama-Castro S, Peralta-Gil M, Diaz-Peredo E, Sanchez-Solano F, Santos-Zavaleta A, Martinez-Flores I, Jimenez-Jacinto V, Bonavides-Martinez C, et al. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34:D394–D397. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Tauschek M, Strugnell R, Robins-Browne RM. The H-NS protein represses transcription of the eltAB operon, which encodes heat-labile enterotoxin in enterotoxigenic Escherichia coli, by binding to regions downstream of the promoter. Microbiology. 2005;151:1199–1208. doi: 10.1099/mic.0.27734-0. [DOI] [PubMed] [Google Scholar]
- 22.Doyle M, Fookes M, Ivens A, Mangan MW, Wain J, Dorman CJ. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science. 2007;315:251–252. doi: 10.1126/science.1137550. [DOI] [PubMed] [Google Scholar]
- 23.Yang J, Baldi DL, Tauschek M, Strugnell RA, Robins-Browne RM. Transcriptional regulation of the yghJ-pppA-yghG-gspCDEFGHIJKLM cluster, encoding the type II secretion pathway in enterotoxigenic Escherichia coli. J. Bacteriol. 2007;189:142–150. doi: 10.1128/JB.01115-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koo HS, Crothers DM. Chemical determinants of DNA bending at adenine-thymine tracts. Biochemistry. 1987;26:3745–3748. doi: 10.1021/bi00386a070. [DOI] [PubMed] [Google Scholar]
- 25.Rimsky S, Zuber F, Buckle M, Buc H. A molecular mechanism for the repression of transcription by the H-NS protein. Mol. Microbiol. 2001;42:1311–1323. doi: 10.1046/j.1365-2958.2001.02706.x. [DOI] [PubMed] [Google Scholar]
- 26.Schneider R, Lurz R, Luder G, Tolksdorf C, Travers A, Muskhelishvili G. An architectural role of the Escherichia coli chromatin protein FIS in organising DNA. Nucleic Acids Res. 2001;29:5107–5114. doi: 10.1093/nar/29.24.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dame RT, Wyman C, Goosen N. Structural basis for preferential binding of H-NS to curved DNA. Biochimie. 2001;83:231–234. doi: 10.1016/s0300-9084(00)01213-x. [DOI] [PubMed] [Google Scholar]
- 28.Dame RT, Noom MC, Wuite GJ. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- 29.Travers A, Muskhelishvili G. A common topology for bacterial and eukaryotic transcription initiation? EMBO Rep. 2007;8:147–151. doi: 10.1038/sj.embor.7400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Rasko DA, Lockatell CV, Johnson DE, Mobley HL. Repression of bacterial motility by a novel fimbrial gene product. EMBO J. 2001;20:4854–4862. doi: 10.1093/emboj/20.17.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landini P, Zehnder AJ. The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating expression of flagellar genes and lipopolysaccharide production. J. Bacteriol. 2002;184:1522–1529. doi: 10.1128/JB.184.6.1522-1529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 1999;181:7500–7508. doi: 10.1128/jb.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ko M, Park C. H-NS-Dependent regulation of flagellar synthesis is mediated by a LysR family protein. J. Bacteriol. 2000;182:4670–4672. doi: 10.1128/jb.182.16.4670-4672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beloin C, Dorman CJ. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol. Microbiol. 2003;47:825–838. doi: 10.1046/j.1365-2958.2003.03347.x. [DOI] [PubMed] [Google Scholar]
- 35.Turner EC, Dorman CJ. H-NS antagonism in Shigella flexneri by VirB, a virulence gene transcription regulator that is closely related to plasmid partition factors. J. Bacteriol. 2007;189:3403–3413. doi: 10.1128/JB.01813-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doyle M, Dorman CJ. Reciprocal transcriptional and posttranscriptional growth-phase-dependent expression of sfh, a gene that encodes a paralog of the nucleoid-associated protein H-NS. J. Bacteriol. 2006;188:7581–7591. doi: 10.1128/JB.00685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.