Abstract
Translation requires the specific attachment of amino acids to tRNAs by aminoacyl-tRNA synthetases (aaRSs) and the subsequent delivery of aminoacyl-tRNAs to the ribosome by elongation factor 1 alpha (EF-1α). Interactions between EF-1α and various aaRSs have been described in eukaryotes, but the role of these complexes remains unclear. To investigate possible interactions between EF-1α and other cellular components, a yeast two-hybrid screen was performed for the archaeon Methanothermobacter thermautotrophicus. EF-1α was found to form a stable complex with leucyl-tRNA synthetase (LeuRS; KD = 0.7 μM). Complex formation had little effect on EF-1α activity, but increased the kcat for Leu-tRNALeu synthesis ∼8-fold. In addition, EF-1α co-purified with the archaeal multi-synthetase complex (MSC) comprised of LeuRS, LysRS and ProRS, suggesting the existence of a larger aaRS:EF-1α complex in archaea. These interactions between EF-1α and the archaeal MSC contribute to translational fidelity both by enhancing the aminoacylation efficiencies of the three aaRSs in the complex and by coupling two stages of translation: aminoacylation of cognate tRNAs and their subsequent channeling to the ribosome.
INTRODUCTION
The fidelity with which mRNA is decoded into proteins is essential for maintenance of the genetic code and is dependent on the specific coupling of amino acids to tRNAs by aminoacyl-tRNA synthetases (aaRSs). Once synthesized, the aminoacyl-tRNA (aa-tRNA) molecules are selectively bound and delivered to the ribosome by elongation factors (EF-1α in eukaryotes and archaea or EF-Tu in bacteria), providing the growing polypeptide chain with substrates for translation elongation. Based on active site architectures, the aaRSs are divided into two classes (class I and II) comprised of 10 members each (1,2). While the two classes of aaRSs perform the same essential function, class I aaRSs approach tRNAs from the minor groove of the tRNA acceptor stem and may be rate-limited by aa-tRNA release, and class II aaRSs approach the major groove of tRNAs and are rate-limited by a step prior to product release (3). These distinctions between class I and class II enzymes may compel an association of translation EFs with class I aaRSs for efficient aa-tRNA release. Conversely, class II aaRSs may not require stable interactions with EFs for efficient product release. Once synthesized, aa-tRNAs are selectively bound by EF-1α in a ternary complex with GTP (EF-1α·GTP·aa-tRNA) and delivered to the ribosome. Upon codon–anticodon recognition on the ribosome, GTP is hydrolyzed, releasing the aa-tRNA in the ribosomal A site. Finally, the inactive EF-1α·GDP complex dissociates from the ribosome, and is then re-activated with GTP for the next round of aa-tRNA selection.
An intricate network of protein–protein interactions is required for efficient translation in eukaryotes and archaea. In yeast, e.g. the accessory factor Arc1p stably interacts with the cytoplasmic methionyl- and glutamyl-tRNA synthetases (MetRS and GluRS, respectively) (4). In this ternary complex Arc1p facilitates the binding of the two tRNA substrates, thus increasing the catalytic efficiencies of both MetRS and GluRS. In mammalian cells, a multi-synthetase complex (MSC) exists, composed of nine aaRS activities (leucyl-, lysyl-, prolyl-, isoleucyl-, methionyl-, glutamyl-, glutaminyl-, arginyl- and aspartyl-tRNA synthetases) and three auxiliary proteins (5–8). Although the function of this complex in translation has not been fully elucidated, it has been proposed to play roles in both the exclusion of certain synthetases from the nucleus (9,10) and the control of non-canonical functions of aaRSs beyond translation (11,12). MSCs have also been reported in bacteria (13), and in archaea a smaller complex was described in Methanothermobacter thermautotrophicus that contained three of the same aaRS activities as the mammalian MSC, namely leucyl- (LeuRS), lysyl- (LysRS) and prolyl-tRNA synthetases [ProRS (14,15)]. Steady-state kinetic analyses showed that the catalytic efficiencies of both LysRS and ProRS were enhanced several-fold when in complex as compared to the free enzymes, while aminoacylation by LeuRS remained unchanged.
In addition to the larger MSC, higher eukaryotes contain a smaller complex composed of valyl-tRNA synthetase (ValRS) and EF-1α, which enhances the catalytic activity of ValRS almost 2-fold (16–20). This interaction has also been proposed to play a role in substrate channeling, whereby the newly synthesized aa-tRNA is directly transferred to the ribosome via EF-1α without diffusion to the cytoplasm (21). The stable complex between EF-1α and ValRS, a class I aaRS, correlates with the ability of EFs to form complexes with and enhance the rate of aminoacylation by class I aaRSs (3). EF-1α has also been shown to stimulate the dissociation of Asp-tRNAAsp from AspRS, providing further support for substrate channeling in translation (22). Additionally, mammalian tryptophanyl- and phenylalanyl-tRNA synthetases have been suggested to associate with EF-1α, although these interactions have not been characterized in vitro and the possible cellular roles of these associations remain unknown (23,24).
To investigate if aaRS:EF-1α complexes exist outside of the mammalian model systems, we undertook a systematic search for proteins interacting with EF-1α in archaea, which identified LeuRS as a stable partner. While the functional effects of complex formation on the activity of EF-1α were modest, the catalytic activity of LeuRS was significantly enhanced when compared to free enzyme. These data indicate the existence of a stable EF-1α·LeuRS complex in archaea and further suggest the association of EF-1α with the archaeal MSC. When considered in conjunction with earlier data, these associations enhance the overall rate of aminoacylation by all three aaRSs in complex, and facilitate transfer of newly synthesized aa-tRNAs to the ribosome via EF-1α.
MATERIALS AND METHODS
Media, strains and plasmid construction
Media preparation and transformation of yeast host strain MaV203 with the bait vector pDBLeu and prey vector pDEST22 were performed according to the manual for ProQuest Two-Hybrid System (Invitrogen) and as described (15). All primers were from Integrated DNA Technologies. To construct the yeast two-hybrid bait vector containing the M. thermautotrophicus tuf gene (encoding EF-1α; MTH1058), the corresponding sequence was isolated by PCR using genomic M. thermautotrophicus DNA as template, the primers 5′-GTCGACCATGGCTAAAGA-3′ and 5′-GCTAGCTTATTTTGCTGG-3′ flanked by SalI and NheI sites, and Pfu DNA polymerase (Stratagene). The tuf PCR product was cloned into PCR-Blunt II-TOPO vector (Invitrogen), sequenced, and subsequently sub-cloned into the yeast ProQuest Two-Hybrid bait vector pDBLeu using the SalI and NheI restriction sites. Construction, amplification and screening of the M. thermautotrophicus cDNA-based yeast two-hybrid library were as previously described (15).
His6 fusion derivatives of LeuRS, LysRS and ProRS (MTH1508, MTH 1542 and MTH611, respectively) were prepared as previously described (15). C-terminal His6 tagged fusion derivatives of EF-1α and AlaRS (MTH1683) were prepared by inserting the corresponding PCR amplified genes into pET11a and pET33b vectors, respectively. For the His6-EF-1α construct, forward primer 5′-CATATGGCTAAAGAAAAAGAACACA TGA-3′ and reverse primer 5′-TGCTCTTCCGCATTTTGCTGGTACGAGGTCTATG-3′ were used. Cloning EF-1α into pET11a was done by isolating the respective NdeI and SapI fragment and ligating into NdeI and SapI digested pET11a. For the His6-AlaRS construct, forward primer 5′-GCTAGCATGATTACCATGTCCCATCAGCTTGAA-3′ and reverse primer 5′-GCG GCCGCCCTTCCTCACAGTAC TGAGTGCAGCT-3′ were used. Cloning AlaRS into pET33b was done by isolating the respective NheI and NotI fragment and ligating into NheI and NotI digested pET33b.
Protein production and purification
His6-LeuRS, His6-ProRS and His6-LysRS were produced and purified as previously described (15). His6-AlaRS was produced by transforming Escherichia coli BL21-RIL (Stratagene) with pET33b-alaS. The resulting transformants were grown and induced to produce protein using the Overnight Express Auto-induction System 1 (Novagen) following the manufacturer's protocol. Cell-free extract was produced by passing cells in buffer A [50 mM NaH2PO4 (pH 8.0), 300 mM NaCl and 10 mM imidazole] containing a protease inhibitor mixture tablet through a French pressure cell (Complete Mini, EDTA-free; Roche Applied Science) followed by centrifugation at 70 000 × g for 45 min. To reduce the amount of contaminating E. coli proteins, the supernatant was incubated at 60°C for 10 min followed by ultracentrifugation at 100 000 × g for 1 h. The supernatant from ultracentrifugation was loaded onto a Ni-NTA2+ column, washed extensively with buffer A, and eluted with an imidazole gradient (0–250 mM) in the same buffer. His6-EF-1α was produced by transforming E. coli BL21-RIL with pET11a-tuf and purified as previously described (15). Fractions containing His6-EF-1α and His6-AlaRS, as determined by SDS-PAGE Coomassie Brilliant Blue staining were pooled, concentrated by ultrafiltration (Amicon 30, Millipore) and stored at −80°C.
tRNA purification
Purification of in vitro transcribed tRNAPro and tRNALeu, and total tRNA from M. thermautotrophicus were as previously described (15). In vitro transcribed M. thermautotrophicus tRNALys and tRNAAla were inactive in aminoacylation (data not shown).
Aminoacylation assays
l-[U-14C] leucine (306 mCi/mmol), l-[U-14C] lysine (312 mCi/mmol), l-[U-14C] proline (241 mCi/mmol) and l-[U-14C] alanine (164 mCi/mmol) were all from Amersham Biosciences. A pre-reaction mixture containing 250 mM KCl, 100 mM Na-HEPES (pH 7.5), 10 mM dithiothreitol, 10 mM MgCl2, 50 μg/ml BSA, 6 mg/ml M. thermautotrophicus total tRNA or in vitro transcribed tRNA and enzymes at concentrations indicated for specific experiments was pre-incubated for 20 min at room temperature. The appropriate radiolabeled amino acid was then added to the mixture and the temperature increased to 50°C. After 1 min, 5 mM ATP was added to start the reaction. Aliquots were spotted onto 3MM paper pre-soaked in 5% trichloroacetic acid (TCA) (w/v), washed in TCA, and the radioactivity counted.
Fluorescence anisotropy
Fluorescent labeling and anisotropy measurements were based upon previously published procedures (25). LeuRS, AlaRS, LysRS, ProRS and EF-1α were labeled with an amine-reactive, extrinsic fluorophore, Alexa Fluor® (AF) 488 tetrafluorophenyl ester (Molecular Probes, Eugene, OR, USA) as previously described at a molar ratio of 1:10 enzyme:fluorophore. After labeling, protein was visualized on a 10% SDS-polyacrylamide gel and subjected to ultraviolet illumination, which confirmed that the final labeled product contained little or no free fluorophore. Prior to use in fluorescence anisotropy measurements, the activity of LeuRS-AF, AlaRS-AF, LysRS-AF and ProRS-AF were verified by aminoacylation assays and protein concentrations determined via active site titration (with the exception of LysRS). The concentrations of labeled LysRS-AF and EF-1α-AF were determined by dye binding (BioRad).
Equilibrium dissociation constants were determined by measuring the fluorescence anisotropy of LeuRS-AF, AlaRS-AF or EF-1α-AF (100 nM each) as a function of increasing concentrations of an unlabeled protein using a Fluorolog-3 spectrofluorimeter (Horiba Jobin Yvon) as previously described (15,25). The concentration ranges of unlabeled protein used in fluorescence anisotropy experiments with LeuRS-AF and AlaRS-AF were 50–6400 nM unlabeled EF-1α. Alternatively, labeled EF-1α-AF was incubated in the presence of unlabeled AlaRS (50–7000 nM). All measurements were carried out at least three times and the titration curves were fitted assuming a 1:1 binding stoichiometry.
Co-immunoprecipitation of EF-1α with LeuRS
Total 100 µl agarose beads coated with Protein A were washed with 900 µl Na-HEPES (pH 7.5) and resuspended in 1 ml of the same buffer. Anti-EF-1α was then added to the beads, which were shaken at room temperature for 20 min, and washed three times [20 mM Na-HEPES (pH 7.5), 150 mM NaCl, 10% glycerol and 0.1% Triton X–100]. Cells were resuspended in a buffer containing Tris–HCl (pH 8.0), 150 mM NaCl, 1% Triton X-100, 1 mM phenylmethanesulfonyl fluoride; EF-1α (0.6 µM) was added, followed by shaking at 4°C for 1 h, and the beads then washed two times in the same buffer. Labeled LeuRS-AF (0.6 µM) was then added and the mixture was shaken at 4°C for 1 h. Following incubation, the beads were washed with 900 µl each of buffer 1 [50 mM Na-HEPES (pH 7.5), 500 mM NaCl, 0.2% Triton X-100 and 5 mM EDTA], buffer 2 [50 mM Na-HEPES (pH 7.5), 150 mM NaC1, 0.1% Triton X-100, 5 mM EDTA and 0.1% SDS] and buffer 3 [10 mM Tris–HC1 (pH 8.0) and 0.1% Triton X-100]. The beads were finally resuspended in 20 µl of SDS-PAGE loading buffer and incubated at 100°C for 5 min. Supernatants were resolved by electrophoresis on a 10% SDS-PAGE gel and then visualized by fluorescent scanning.
Nucleotide exchange and GTP hydrolysis by EF-1α
EF-1α was assayed for its ability to bind [3H]-GDP retained on nitrocellulose filters. Nucleotide exchange activity of 0.5 µM EF-1α was measured at 50°C in the presence of 100 µM [3H]-GDP (546 c.p.m./pmol) in a buffer containing 20 mM Tris–HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2 and 1 mM DTT. The exchange reaction was started by the addition of GDP or GTP as previously described (26). The solution was filtered onto nitrocellulose filters following the reaction, washed twice with 3 ml cold buffer, dried and radioactivity counted by liquid scintillation.
Measurements of the intrinsic GTPase activity of EF-1α [γ-32P]-GTP were performed in a buffer containing 20 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 3 M NaCl, 50 µM [γ-32P]-GTP (76 c.p.m./pmol), 1 mM DTT and 0.4 µM EF-1α in the presence or absence of LeuRS or BSA at concentrations indicated. Reaction mixtures were incubated at 50°C, and aliquots removed periodically, quenched by addition of ice-cold 1% charcoal (w/v) in 5.6% perchloric acid, and liberation of [γ-32P]-Pi measured using a charcoal adsorption assay on 3MM Whatman filters (27,28). The remaining [γ-32P]-GTP was then estimated by scintillation counting.
Elongation factor protection of aminoacyl-tRNA against spontaneous deacylation
M. thermautotrophicus EF-1α (15 μM) was activated in a buffer containing 50 mM Tris–HCl (pH 7.5), 1 mM GTP, 100 mM di-potassium glutarate, 10 mM MgCl2, 25 mM KCl, 5 mM DTT, 1.5 M NH4Cl, 3 mM PEP and 30 µg/ml pyruvate kinase (28). 15 µM Thermus thermophilus bacterial EF-Tu (15 μM) was activated in a buffer containing 50 mM Tris–HCl (pH 7.5), 70 mM KCl, 0.5 mM GTP, 7 mM MgCl2, 1 mM DTT, 2.5 mM PEP and 30 µg/ml pyruvate kinase (29). To test the protective effects of the archaeal EF-1α or bacterial EF-Tu against spontaneous hydrolysis of aa-tRNA, [14C]-Leu-tRNALeu (0.9 µM) was incubated at 50°C in the presence of 4 µM activated EF-1α or EF-Tu in activation buffer. At specific time intervals, aliquots were removed, precipitated with cold trichloracetic acid, filtered on nitrocellulose filters, dried and radioactivity counted.
Co-Purification of LeuRS, LysRS, ProRS and detection of EF-1α by immunoblotting
LeuRS, LysRS, and ProRS were co-purified from M. thermautotrophicus cell-free extract as previously described (15). Proteins were separated by SDS-PAGE and electroblotted onto nitrocellulose as previously described (30). Each fraction was then assayed for the presence of EF-1α by immunoblotting using antibody specifically directed against M. thermautotrophicus EF-1α.
RESULTS
Identification of proteins interacting with EF-1α
An M. thermautotrophicus cDNA library (cloned into the pDEST22 prey vector) was screened, searching for proteins that interact with the archaeal EF-1α (encoded in the bait vector pDBLeu). A total of 1.4 × 105 transformants were screened for the protein–protein interaction phenotype and approximately 100 clones were selected for plasmid isolation and sequencing. Proteins identified from the screen were divided into four categories based on functional annotation (Table 1). Two clones, each identified multiple times, contained genes involved in protein modification: a transglutaminase-like protein (MTH412) and a chaperonin (MTH218). Both clones have been isolated in previous screens and would not be expected to form specific interactions with EF-1α (14,15). Eight clones, each identified once in the screen, contained genes involved in methanogenesis (MTH965, MTH1130, MTH1135, MTH1161, MTH1162, MTH1163, MTH1164 and MTH1165). Methanogenesis protein subunits are known to associate as higher order complexes and would not be expected to associate with EF-1α specifically. Additionally, these and similar methanogenic proteins were identified in previous screens of this M. thermautotrophicus library, suggesting that the interactions are non-specific. The third category included an ATP synthase subunit (MTH957), signal peptidase (MTH1448), magnesium chelatase subunit (MTH673) and three proteins of unknown function (MTH674, MTH1843 and MTH1858). Similar to the second category, these proteins were found in previous screens and likely do not form specific interactions with EF-1α. The fourth category was composed of proteins involved in translation, LeuRS (MTH1508) and AlaRS (MTH1683), which were each identified once in the screen (Table 2). To gain insight into possible links between EFs and other cellular components involved in translation, associations between EF-1α and LeuRS and AlaRS were further investigated in vitro.
Table 1.
Proteins identified as interacting with M. thermautotrophicus EF-1α
| Function | ORFa | Descriptionb |
|---|---|---|
| Translation | ||
| MTH1508 | Leucyl-tRNA synthetase | |
| MTH1683 | Alanyl-tRNA synthetase | |
| Protein modification | ||
| MTH412 | Transglutaminase | |
| MTH218 | Chaperonin | |
| Methanogenesis | ||
| MTH965 | Tungsten formyl methanofuran dehydrogenase, subunit F | |
| MTH1130 | Methyl coenzyme M reductase II, gamma subunit | |
| MTH1135 | Methyl viologen-reducing hydrogenase, gamma subunit | |
| MTH1161 | N5-methyltetrahydromethanopterin: CoM methyltransferase, C-subunit | |
| MTH1162 | N5-methyltetrahydromethanopterin: CoM methyltransferase, D-subunit | |
| MTH1163 | N5-methyltetrahydromethanopterin: CoM methyltransferase, E-subunit | |
| MTH1164 | Methyl Coenzyme M reductase, alpha subunit | |
| MTH1165 | Methyl Coenzyme M reductase, gamma subunit | |
| Other | ||
| MTH957 | ATP synthase subunit C | |
| MTH1448 | Signal peptidase | |
| MTH673 | Magnesium chelatase subunit | |
| MTH674 | Unknown function | |
| MTH1843 | Unknown function | |
| MTH1858 | Unknown function | |
aORFs are numbered according to the annotated genome sequence of M. thermautotrophicus.
bPredicted functions of the corresponding proteins are taken from the TIGR Comprehensive Microbial Resource (http://cmr.tigr.org/tigr-scripts/CMR/shared/AnnotationSearch.cgi).
Table 2.
Yeast two-hybrid interactions with EF-1α
| Plasmids | - His +25 mM 3-AT | - Ura + 0.2% 5-FOA | Description |
|---|---|---|---|
| Control A | − | ++ | No interaction standard |
| Control B | + | + | Weak interaction standard |
| Control C | ++ | +/− | Moderate interaction standard |
| Control D | +++ | − | Strong interaction standard |
| pDBLeu- EF-1α/pDEST-LeuRS | +++ | +/− | LeuRS insert in prey vector |
| pDBLeu- EF-1α/pDEST-AlaRS | ++ | +/− | AlaRS insert in prey vector |
| pDBLeu- EF-1α/pPC86 | +/− | + | No insert in prey vector |
Growth phenotypes on selective media for interactions between reference proteins (A–D controls, ProQuest Two-Hybrid System, Invitrogen), and between EF-1α (encoded in the bait vector pDBLeu) and LeuRS or AlaRS (library inserts in the prey vector pDEST). Growth on selective media SC -Leu, -Trp, -His, +25 mM 3-AT or on SC -Leu, -Trp, -Ura, +0.02% 5-FOA where − indicates no growth, + weak growth, ++ moderate growth and +++ strong growth.
Association of EF-1α with LeuRS and AlaRS
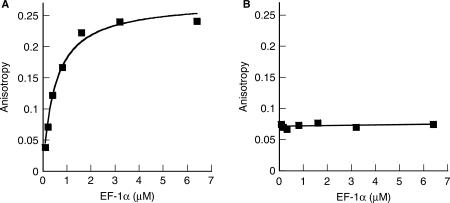
To investigate possible interactions between EF-1α and the two aaRSs, fluorescence anisotropy experiments were employed to determine equilibrium dissociation constants (KDs) as previously described (25). The affinity between LeuRS-AF and EF-1α·GDP was investigated in the presence of increasing concentrations of unlabeled EF-1α, which resulted in a significant increase in anisotropy and a KD of 730 ± 130 nM (Figure 1A). Although determination of the binding affinities between each labeled aaRS and EF-1α·GTP would also be of interest, it is technically impractical to maintain EF-1α in the GTP-bound state under the conditions required for fluorescence measurements. The binding of AlaRS-AF to EF-1α could not be detected by fluorescence anisotropy (Figure 1B), however, regardless of which of the two proteins was labeled with the fluorescent dye (data not shown). This suggests that EF-1α and AlaRS do not form a specific complex, associate only transiently in the cell, or that experimental conditions were not conducive to complex formation.
Figure 1.
Fluorescence anisotropy experiments. The binding affinities of EF-1α to A, Alexa Fluor-labeled LeuRS-AF and (B), labeled AlaRS-AF were measured using 100 nM of each labeled aaRS as a function of increasing concentrations of unlabeled partner protein. Representative data sets are shown, with values representing the means of three independent experiments.
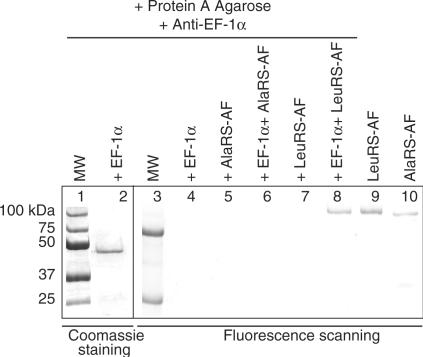
The interaction between EF-1α and LeuRS was further investigated using antibodies against EF-1α in co-immunoprecipitation experiments (Figure 2). Unlabeled EF-1α, bound by antibody specifically directed against EF-1α (Figure 2, lane 2), was incubated with fluorescently labeled LeuRS-AF and the complex was extensively washed. LeuRS-AF co-immunoprecipitated specifically with EF-1α, but not in its absence (Figure 2, lanes 7–8). Consistent with the lack of detectable affinity between AlaRS-AF and EF-1α in the fluorescence anisotropy experiments, labeled AlaRS-AF failed to co-immunoprecipitate with EF-1α (Figure 2, lane 6).
Figure 2.
Co-immunoprecipitation of EF-1α with AlaRS and LeuRS. SDS-PAGE visualized by Coomassie brilliant blue staining and fluorescence scanning of unlabeled EF-1α with fluorescently labeled AlaRS-AF and LeuRS-AF. Lanes 1 and 3, protein molecular weight markers visualized by Coomassie staining and fluorescence scanning, respectively. Lane 2, immunoprecipitation of unlabeled EF-1α. Lanes 4, 5 and 7 show the EF-1α antibody bound by Protein A in the presence of either EF-1α, AlaRS-AF or LeuRS-AF alone, respectively. Lane 6 shows the co-immunoprecipitation experiment in the presence of EF-1α and AlaRS-AF. Lane 8 shows the co-immunoprecipitation of both LeuRS-AF and EF-1α. Lanes 9 and 10 indicate stock solutions of labeled LeuRS-AF and AlaRS-AF, respectively.
Effects of the EF-1α·LeuRS complex on GTP hydrolysis by EF-1α
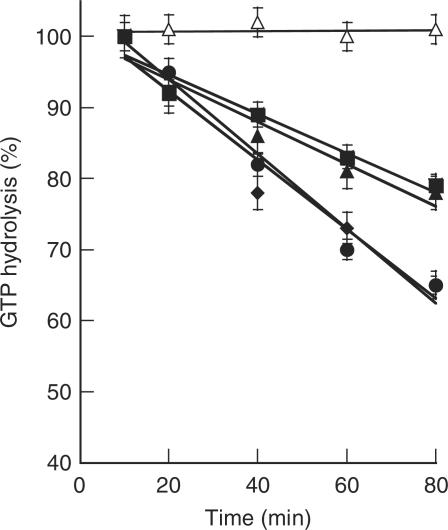
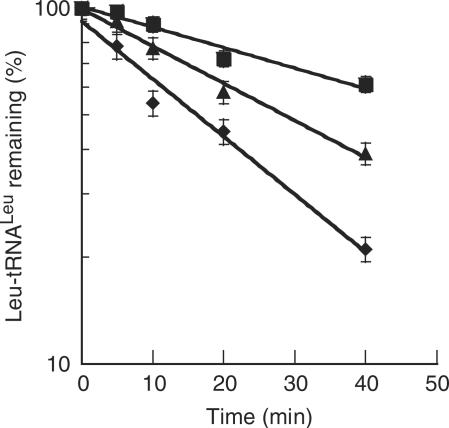
Efficient protein synthesis requires EF-1α·GTP to bind and deliver aa-tRNAs to the ribosomal A site during translation elongation. Once codon–anticodon interactions have been established on the ribosome, EF-1α·GTP is hydrolyzed, resulting in the EF-1α·GDP form. The nucleotide exchange factor then exchanges GDP for GTP, activating EF-1α for another round of aa-tRNA selection. The archaeal EF-1α could be successfully activated with GTP (Figure 3) and the protective effects of EF-1α on the labile M. thermautotrophicus Leu-tRNALeu ester bond were moderate under experimental conditions, providing an ∼2-fold increase in the half life of the aa-tRNA while the bacterial T. thermophilus EF-Tu increased the aa-tRNA half life by almost 3-fold (Figure 4). The relatively low level of aa-tRNA protection by EF-1α made it impractical to quantify the effects of complex formation on this particular activity. As an alternative, the effects of LeuRS on the GTP hydrolysis activity of EF-1α were investigated. Upon complex formation between LeuRS and EF-1α, the rate of GTP hydrolysis by EF-1α was enhanced by ∼1.5-fold as compared to EF-1α alone (Figure 3).
Figure 3.
Effects of complex formation on GTP hydrolysis by EF-1α. Measurements of GTP hydrolysis by 0.4 µM EF-1α·[γ-32P]GTP alone (filled square) were performed in the presence of BSA (filled triangle), or in the presence of a 5-fold (filled diamond) or 10-fold (filled circle) excess of LeuRS. Open triangle no enzyme control.
Figure 4.
Protection of Leu-tRNALeu. Protection of 0.3 µM M. thermautotrophicus [14C]Leu-tRNALeu by 3.0 µM T. thermophilus EF-Tu (filled square), 3.0 µM M. thermautotrophicus EF-1α (filled triangle), or no enzyme (filled diamond).
Effects on aminoacylation of complex formation between EF-1α and LeuRS
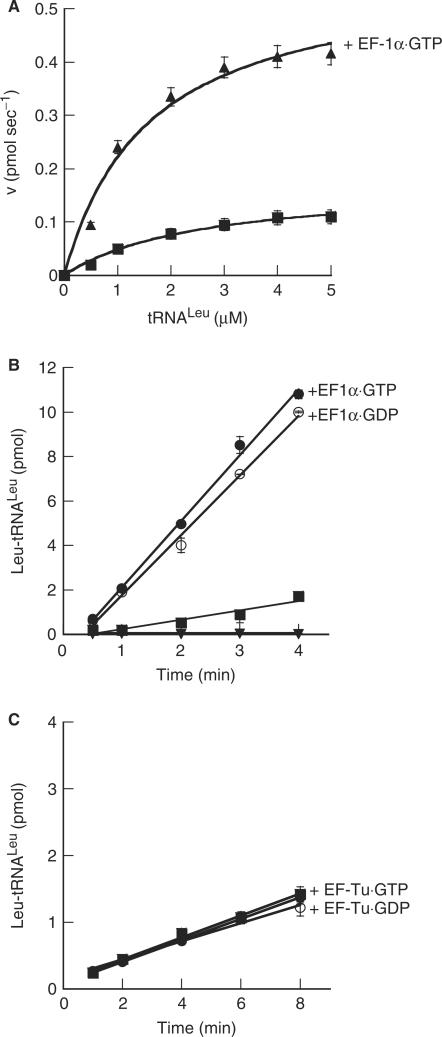
The potential impact of EF-1α on the activity of LeuRS was monitored with respect to Leu-tRNALeu synthesis. Steady-state aminoacylation kinetics of LeuRS in the presence of archaeal EF-1α·GTP or EF-1α·GDP both indicated that complex formation specifically enhances the kinetics of tRNALeu aminoacylation, leading to an 8-fold increase in the kcat for Leu-tRNALeu synthesis and an overall enhancement in the rate of aminoacylation by LeuRS (Table 3; Figure 5A and B). The enhancement seen in leucylation by EF-1α occurred irregardless of the guanine nucleotide bound by EF-1α (Figure 5B). Addition of T. thermophilus EF-Tu, which protects archaeal Leu-tRNALeu, did not result in a significant increase in aminoacylation by LeuRS as compared to the free enzyme (Figure 5C). This indicated that aminoacylation by LeuRS is specifically enhanced by the archaeal EF-1α, regardless of the bound guanine nucleotide, and is not an artifact of aa-tRNA protection (Figure 4) or a GTPase activity (Figure 5C).
Table 3.
Steady-state aminoacylation kinetics of M. thermautotrophicus LeuRS
| Enzymea | Additionsb | KM tRNALeu (µM) | kcat (s−1) |
|---|---|---|---|
| LeuRS | None | 1.4 ± 0.03 | 0.22 ± 0.01 |
| LeuRS | EF-1α·GTP | 3.8 ± 0.6 | 1.7 ± 0.02 |
| LeuRS | BSA | 1.1 ± 0.4 | 0.34 ± 0.09 |
aEnzymes were added at a final concentration of 10 nM.
bAddition of other components at 3.5 µM each.
Figure 5.
Aminoacylation by LeuRS in the presence of archaeal EF-1α and bacterial EF-Tu. (A), Aminoacylation by 10 nM LeuRS alone (filled square) or in the presence of 3.5 µM M. thermautotrophicus EF-1α·GTP (filled triangle). (B), aminoacylation by 10 nM LeuRS alone (filled square) in the presence of 3.5 µM EF-1α·GTP (filled circle), EF-1α·GDP (open circle) or (C) 3.5 µM T. thermophilus EF-Tu·GTP (filled circle) or EF-Tu·GDP (open circle). Inverted filled triangle, Mt EF-1α alone.
Association of EF-1α with the archaeal MSC
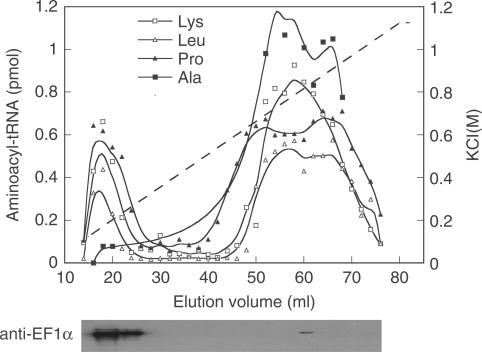
Formation of a stable complex between EF-1α and LeuRS raised the question of whether this association is part of the archaeal MSC. To purify the archaeal MSC, M. thermautotrophicus cell-free extracts were applied to a gel filtration and then to an anion exchange column and the fractions were assayed for aminoacylation activities of AlaRS, LeuRS, LysRS and ProRS as previously described (15). After extensive washing, the MSC eluted at low salt while AlaRS eluted with the free forms of the synthetases at high salt. Immunoblotting using polyclonal antibodies specifically directed against M. thermautotrophicus EF-1α indicated that the elongation factor co-purified with the MSC, but not AlaRS, in fractions from both gel filtration (data not shown) and anion exchange chromatography (Figure 6). Further attempts to co-immunoprecipitate EF-1α with components of the archaeal MSC other than LeuRS were unsuccessful (data not shown).
Figure 6.
Co-purification of LysRS, LeuRS, ProRS and EF-1α. M. thermautotrophicus cell-free extracts, initially applied to a Sephacryl S300 gel filtration column, were applied to a Q-Sepharose column and extensively washed prior to development with a KCl gradient. Aminoacyl-tRNA synthesis activities were monitored in the eluted fractions; Lys-tRNALys (open square), Leu-tRNALeu (open triangle), Pro-tRNAPro (filled triangle) and Ala-tRNAAla (filled square). The presence of EF-1α was assayed in each eluted fraction by immunoblotting with antibodies against EF-1α.
Effects of complex formation on the aminoacylation activities of the archaeal MSC
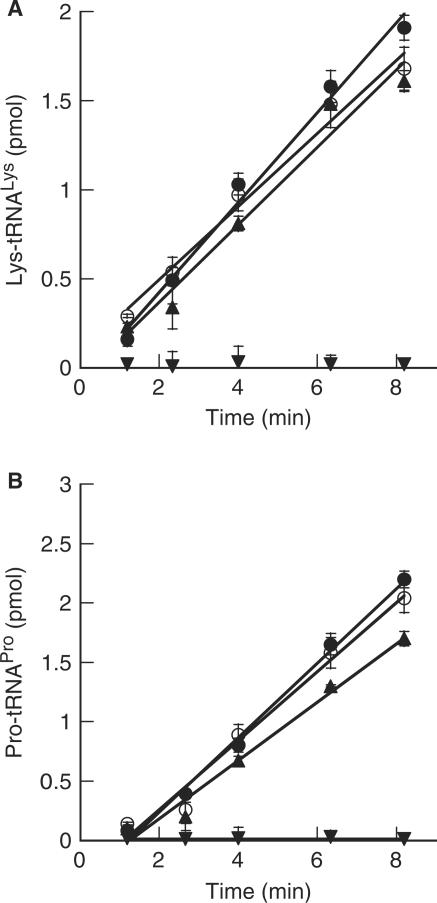
Previously, we have shown that the association between LeuRS, LysRS and ProRS led to an enhancement in the steady-state kinetics of tRNALys and tRNAPro aminoacylation by 3- and 5-fold, respectively (14,15). No enhancement of the catalytic efficiency by LeuRS, however, was observed when in the presence of LysRS or ProRS. Since EF-1α associates with the archaeal MSC, we explored the possible effects of EF-1α on the aminoacylation activities of ProRS and LysRS. Neither the activities of ProRS nor LysRS were affected by the presence of excess EF-1α in either the GDP- or GTP-bound forms (Figure 7A and B), while the addition of excess EF-1α resulted in enhanced aminoacylation by LeuRS (Figure 5B). Taken together with previous data, it is clear that LeuRS enhances the catalytic activities of both LysRS and ProRS, while aminoacylation by LeuRS is enhanced by EF-1α.
Figure 7.
Effects of the association of EF-1α and the archaeal MSC on the aminoacylation activities of LysRS and ProRS. Aminoacylation by A, 100 nM LysRS (filled triangle) and (B), 80 nM ProRS (filled diamond) alone or in the presence of 3.5 µM EF-1α·GTP (filled circle) or EF-1α·GDP (open circle). Inverted filled triangle EF-1α alone.
DISCUSSION
Complex formation between aaRSs and EF-1α
In the present study, a yeast two-hybrid screen identified AlaRS and LeuRS as proteins that potentially interact with archaeal EF-1α. Stable complex formation between EF-1α and LeuRS was confirmed in vitro while the association with AlaRS could not yet be further supported experimentally. This is in agreement with the recent suggestion that translation EFs may be predisposed to form complexes with certain class I aaRSs such as LeuRS, which are suggested to be rate-limited by release of aa-tRNA (3). In contrast, class II aaRSs such as AlaRS are rate-limited at a step prior to product release and may not require stable complex formation with EFs for efficient aminoacylation. The interaction with LeuRS in archaea adds to a growing list of associations formed between EF-1α and various aaRSs, suggesting that a significant proportion of these proteins may be sequestered within the cell (6,19,22–24). Our inability to detect interactions with other aaRSs in the yeast two-hybrid screen implies that either the heterologous yeast system hinders their detection, or that complexes with EF-1α are rarer in archaea than in eukaryotes.
Functional consequences of the association between LeuRS and EF-1α
Although complex formation resulted in only a modest enhancement in EF-1α·GTP hydrolysis, the effects on aminoacylation by LeuRS were more pronounced. The kcat for Leu-tRNALeu synthesis was enhanced by ∼8-fold in the presence of EF-1α, as compared to free enzyme. This is reminiscent of the complex identified between human EF-1α·GTP and ValRS, which led to an ∼2-fold enhanced aminoacylation (19). Distinct from the previously identified complexes, the enhancement in leucylation by EF-1α occurred irrespective of the bound guanine nucleotide (GTP or GDP). This suggests that the surfaces mediating the interactions between LeuRS with EF-1α allow the two proteins to remain in contact, regardless of whether aa-tRNA is bound by EF-1α or LeuRS. This would allow EF-1α to freely alternate between the GDP-bound enzyme and the GTP-bound form (capable of binding aa-tRNAs), while maintaining interactions with LeuRS. This would also explain the apparent stability of the complex, and its potential role in substrate channeling, since delivery of aa-tRNA to the ribosomal A site would not necessitate dissociation of LeuRS from EF-1α.
The archaeal MSC
Whether the LeuRS:EF-1α complex exists as a discrete binary complex, or instead is part of a larger macromolecular machine, is unclear. Preliminary findings suggest that EF-1α is linked to the archaeal MSC through its interactions with LeuRS (Figure 6). The association of EF-1α with the archaeal MSC may contribute to the overall efficiency of translation by enhancing aminoacylation by all three aaRSs, as the catalytic efficiencies of LysRS and ProRS were enhanced when in the presence of LeuRS, while EF-1α has been shown to enhance aminoacylation by LeuRS. The efficiency of translation may also be enhanced through substrate channeling of aa-tRNA directly from the aaRSs to EF-1α. As EF-1α has been shown to be linked to LeuRS, LysRS and ProRS through its interactions with LeuRS, these associations may allow substrate channeling of all three aa-tRNAs synthesized by the aaRSs associated in the archaeal MSC. In conjunction with earlier data, this suggests that complex formation between LeuRS, LysRS, ProRS and EF-1α may play a role in translation both by enhancing the catalytic efficiencies of all three aaRSs, and by providing a mechanism whereby aa-tRNAs synthesized by the archaeal MSC are channeled directly to the ribosome without diffusion into the cytoplasm.
ACKNOWLEDGEMENTS
We would like to thank C. Gruber and M. Smith for making the cDNA library, Z. Kelman for assistance with analysis of two-hybrid assay data and V. Godinic for advice on MaV203. We thank S. Ataide, J. Ling and H. Roy for critical reading of the manuscript. This work was supported by Grant GM 65183 from the National Institutes of Health. Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 2.Ribas De Pouplana L, Schimmel P. Two classes of tRNA synthetases suggested by sterically compatible dockings on tRNA acceptor stem. Cell. 2001;104:191–193. doi: 10.1016/s0092-8674(01)00204-5. [DOI] [PubMed] [Google Scholar]
- 3.Zhang CM, Perona JJ, Ryu K, Francklyn C, Hou YM. Distinct kinetic mechanisms of the two classes of aminoacyl-tRNA synthetases. J. Mol. Biol. 2006;361:300–311. doi: 10.1016/j.jmb.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt EC. The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- 5.Kerjan P, Cerini C, Semeriva M, Mirande M. The multienzyme complex containing nine aminoacyl-tRNA synthetases is ubiquitous from Drosophila to mammals. Biochim. Biophys. Acta. 1994;1199:293–297. doi: 10.1016/0304-4165(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 6.Quevillon S, Robinson JC, Berthonneau E, Siatecka M, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein-protein interactions and characterization of a core protein. J. Mol. Biol. 1999;285:183–195. doi: 10.1006/jmbi.1998.2316. [DOI] [PubMed] [Google Scholar]
- 7.Robinson JC, Kerjan P, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: quantitative analysis of protein-protein interactions and mechanism of complex assembly. J. Mol. Biol. 2000;304:983–994. doi: 10.1006/jmbi.2000.4242. [DOI] [PubMed] [Google Scholar]
- 8.Wolfe CL, Warrington JA, Treadwell L, Norcum MT. A three-dimensional working model of the multienzyme complex of aminoacyl-tRNA synthetases based on electron microscopic placements of tRNA and proteins. J. Biol. Chem. 2005;280:38870–38878. doi: 10.1074/jbc.M502759200. [DOI] [PubMed] [Google Scholar]
- 9.Galani K, Grosshans H, Deinert K, Hurt EC, Simos G. The intracellular location of two aminoacyl-tRNA synthetases depends on complex formation with Arc1p. EMBO J. 2001;20:6889–6898. doi: 10.1093/emboj/20.23.6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golinelli-Cohen MP, Mirande M. Arc1p is required for cytoplasmic confinement of synthetases and tRNA. Mol. Cell. Biochem. 2007;300:47–59. doi: 10.1007/s11010-006-9367-4. [DOI] [PubMed] [Google Scholar]
- 11.Sampath P, Mazumder B, Seshadri V, Gerber CA, Chavatte L, Kinter M, Ting SM, Dignam JD, Kim S, et al. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell. 2004;119:195–208. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 12.Park SG, Ewalt KL, Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem. Sci. 2005;30:569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Harris CL. An aminoacyl-tRNA synthetase complex in Escherichia coli. J. Bacteriol. 1987;169:2718–2723. doi: 10.1128/jb.169.6.2718-2723.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Praetorius-Ibba M, Rogers TE, Samson R, Kelman Z, Ibba M. Association between archaeal prolyl-and leucyl-tRNA synthetases enhances tRNAPro aminoacylation. J. Biol. Chem. 2005;280:26099–26104. doi: 10.1074/jbc.M503539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Praetorius-Ibba M, Hausmann CD, Paras M, Rogers TE, Ibba M. Functional association between three archaeal aminoacyl-tRNA synthetases. J. Biol. Chem. 2007;282:3680–3687. doi: 10.1074/jbc.M609988200. [DOI] [PubMed] [Google Scholar]
- 16.Motorin Y, Wolfson AD, Orlovsky AF, Gladilin KL. Mammalian valyl-tRNA synthetase forms a complex with the first elongation factor. FEBS Lett. 1988;238:262–264. doi: 10.1016/0014-5793(88)80492-7. [DOI] [PubMed] [Google Scholar]
- 17.Bec G, Kerjan P, Zha XD, Waller JP. Valyl-tRNA synthetase from rabbit liver. I. Purification as a heterotypic complex in association with elongation factor 1. J. Biol. Chem. 1989;264:21131–21137. [PubMed] [Google Scholar]
- 18.Negrutskii BS, Budkevich TV, Shalak VF, Turkovskaya GV, El'skaya AV. Rabbit translation elongation factor 1 alpha stimulates the activity of homologous aminoacyl-tRNA synthetase. FEBS Lett. 1996;382:18–20. doi: 10.1016/0014-5793(96)00128-7. [DOI] [PubMed] [Google Scholar]
- 19.Negrutskii BS, Shalak VF, Kerjan P, El'skaya AV, Mirande M. Functional interaction of mammalian valyl-tRNA synthetase with elongation factor EF-1alpha in the complex with EF-1H. J. Biol. Chem. 1999;274:4545–4550. doi: 10.1074/jbc.274.8.4545. [DOI] [PubMed] [Google Scholar]
- 20.Jiang S, Wolfe CL, Warrington JA, Norcum MT. Three-dimensional reconstruction of the valyl-tRNA synthetase/elongation factor-1H complex and localization of the delta subunit. FEBS Lett. 2005;579:6049–6054. doi: 10.1016/j.febslet.2005.09.062. [DOI] [PubMed] [Google Scholar]
- 21.Negrutskii BS, Deutscher MP. Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc. Natl Acad. Sci. USA. 1991;88:4991–4995. doi: 10.1073/pnas.88.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed VS, Wastney ME, Yang DC. Mechanisms of the transfer of aminoacyl-tRNA from aminoacyl-tRNA synthetase to the elongation factor 1 alpha. J. Biol. Chem. 1994;269:32932–32936. [PubMed] [Google Scholar]
- 23.Sang LJ, Gyu PS, Park H, Seol W, Lee S, Kim S. Interaction network of human aminoacyl-tRNA synthetases and subunits of elongation factor 1 complex. Biochem. Biophys. Res. Commun. 2002;291:158–164. doi: 10.1006/bbrc.2002.6398. [DOI] [PubMed] [Google Scholar]
- 24.Yang XL, Otero FJ, Ewalt KL, Liu J, Swairjo MA, Kohrer C, RajBhandary UL, Skene RJ, McRee DE, et al. Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis. EMBO J. 2006;25:2919–2929. doi: 10.1038/sj.emboj.7601154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An S, Musier-Forsyth K. Trans-editing of Cys-tRNAPro by Haemophilus influenzae YbaK protein. J. Biol. Chem. 2004;279:42359–42362. doi: 10.1074/jbc.C400304200. [DOI] [PubMed] [Google Scholar]
- 26.Raimo G, Masullo M, Savino G, Scarano G, Ianniciello G, Parente A, Bocchini V. Archaeal elongation factor 1 beta is a dimer. Primary structure, molecular and biochemical properties. Biochim. Biophys. Acta. 1996;1293:106–112. doi: 10.1016/0167-4838(95)00233-2. [DOI] [PubMed] [Google Scholar]
- 27.Masullo M, De Vendittis E, Bocchini V. Archaebacterial elongation factor 1 alpha carries the catalytic site for GTP hydrolysis. J. Biol. Chem. 1994;269:20376–20379. [PubMed] [Google Scholar]
- 28.Raimo G, Masullo M, Lombardo B, Bocchini V. The archaeal elongation factor 1alpha bound to GTP forms a ternary complex with eubacterial and eukaryal aminoacyl-tRNA. Eur. J. Biochem. 2000;267:6012–6018. doi: 10.1046/j.1432-1327.2000.01678.x. [DOI] [PubMed] [Google Scholar]
- 29.Asahara H, Uhlenbeck OC. The tRNA specificity of Thermus thermophilus EF-Tu. Proc. Natl Acad. Sci. USA. 2002;99:3499–3504. doi: 10.1073/pnas.052028599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ibba M, Hennecke H. Relaxing the substrate specificity of an aminoacyl-tRNA synthetase allows in vitro and in vivo synthesis of proteins containing unnatural amino acids. FEBS Lett. 1995;364:272–275. doi: 10.1016/0014-5793(95)00408-2. [DOI] [PubMed] [Google Scholar]