Abstract
Hepadnaviruses are DNA viruses that replicate by protein-primed reverse transcription, employing a specialized reverse transcriptase (RT), P protein. DNA synthesis from the pregenomic RNA is initiated by binding of P to the ε signal. Using ε as template and a Tyr-residue for initiation, the RT synthesizes a DNA oligo (priming) as primer for full-length DNA. Priming strictly requires prior RT activation by chaperones. Active P–ε complexes have been reconstituted in vitro, but whether in addition to the heat-shock protein 70 (Hsp70) system the Hsp90 system is essential has been controversial. Here we quantitatively compared Hsp70 versus Hsp70 plus Hsp90 RT activation, and corroborated that the Hsp70 system alone is sufficient; however, Hsp90 as well the Hsp70 nucleotide exchange factor Bag-1 markedly stimulated activation by increasing the steady-state concentration of the activated metastable RT form P*, though by different mechanisms. Hsp90 inhibition in intact cells by geldanamycin analogs blocked hepadnavirus replication, however not completely and only at severely cytotoxic inhibitor concentrations. While compatible with a basal level of Hsp90 independent in vivo replication, unambiguous statements are precluded by the simultaneous massive upregulation of Hsp70 and Hsp90.
INTRODUCTION
Hepatitis B virus (HBV), the causative agent of B-type hepatitis in humans, is the prototype of the hepadnaviruses, small DNA-containing viruses that replicate through reverse transcription (1,2); due to various experimental restrictions with HBV, duck hepatitis B virus (DHBV) provides an important model system (3). In all hepadnaviruses, a more than genome length transcript, the pregenomic RNA (pgRNA), acts as mRNA for the capsid protein and the viral reverse transcriptase (RT), termed P protein, and the same RNA is packaged into viral capsids and there it is reverse transcribed into progeny DNA genomes.
Hepadnavirus replication differs distinctively from that of retroviruses, reflected by the presence in P proteins of an extra terminal protein (TP) domain. Both pgRNA encapsidation and initiation of DNA synthesis are triggered by the binding of P to a 5′-proximal RNA stem-loop, ε, on the pgRNA. Once the RT is bound to ε, a Tyr-residue in the TP domain, rather than a tRNA, serves for the protein-primed synthesis of a 3 or 4 nt, ε-encoded, and covalently TP-linked DNA oligonucleotide (‘priming reaction’; Figure 1A). Subsequently, the complex translocates to a 3′ proximal RNA element, DR1*, and the oligo is extended into complete minus-strand DNA which remains covalently bound to the RT; plus-strand DNA synthesis eventually yields the relaxed circular (RC) DNA present in virions released from the cell.
Figure 1.
(A) Current model of replication initiation by hepadnaviral P proteins. DHBV P protein, with its terminal protein (TP) and reverse transcriptase/RNase H (RT/RH) domains linked through a dispensable spacer, is unable to bind Dε RNA without prior chaperone-mediated conversion into a metastable, active conformation (P*). In vitro activation strictly requires Hsc70, Hsp40 and ATP. The necessity for Hop and Hsp90 has been controversial (31,33). Dε RNA binding is accompanied by structural changes in the RNA and the RT, enabling the synthesis of a short DNA oligonucleotide templated by a bulged region within Dε (priming); its 5′-terminal nucleotide is covalently linked to a Tyr residue in the TP domain. (B) Different states of hepadnaviral RT activation. P represents the non-activated state. Chaperones and energy are required to produce, and maintain, activated metastable P* which is able to bind Dε RNA. The steady-state P* concentration depends on the rates of P* production and decay. Upon energy depletion P* decays within minutes whereas P–Dε RNA complexes are stable over hours (31).
Mechanistic studies have long been hampered by the lack of soluble recombinant P protein. Initial cell-free studies (4) performed in rabbit reticulocyte lysate (RL) showed that in vitro translated RT from DHBV though not from HBV (5) was capable of performing the authentic, ε-dependent priming reaction when provided with DHBV ε (Dε) RNA and dNTPs (Figure 1A); this system also revealed that formation of an active replication initiation complex is a multi-step process requiring specific interactions between P protein and Dε (6–10) that induce functionally crucial structural rearrangements in both the protein and the RNA (7,11). Most notably here, complex formation was found to be strictly dependent on cellular chaperones (12,13) which are abundantly present in RL; this very fact, however, also precluded clear-cut distinctions as to which chaperones are absolutely required for P activation, and which are not.
Chaperones, initially termed heat-shock proteins (Hsps), are essential in all cells to prevent accumulation of misfolded proteins. The major eukaryotic cytosolic chaperone systems, besides the 60 kDa Hsp60 chaperonins (GroEL in bacteria), are Hsp70 including its constitutive form Hsc70 and Hsp90; in addition to ATP, their chaperoning activities depend on, and are modulated by, various co-chaperones (14–16). Hsp70 (DnaK in bacteria), in concert with Hsp40 (DnaJ in bacteria) and/or other J domain proteins (16), has usually more generalized roles such as folding of non-native, e.g. nascent or misfolded, proteins whereas few examples are known for a direct regulation of the activity of native substrates; in bacteria, these include the transcription factor σ32 (17,18), and proteins involved in phage and plasmid replication (19); in eukaryotes, a classic example is clathrin-coated vesicle uncoating via Hsc70 and the large J domain protein auxilin (20). Hsp90, not required for the biogenesis of most proteins (21), acts as a more specialized chaperone in activating various near-native client proteins (22,23), including many important regulatory molecules such as steroid hormone receptors and kinases (24,25). A comprehensive list of interaction partners may be found at http://www.picard.ch. Usually Hsp90 action requires cooperation with the Hsp70 system (26,27), mediated by the Hsp70-Hsp90 organizing protein (Hop) (25).
Based on data from the RL system and on the interference with viral replication in DHBV transfected cells by the Hsp90 inhibitor geldanamycin (GA) chaperone activation of the hepadnaviral RT was suggested to occur similar to that of steroid hormone receptors (12,13); there, ligand-binding competence is gained by sequential reactions with Hsp70 acting early, and Hsp90 late in the cycle (28,29) with additional factors being involved such as p23 and immunophilins (30). Importantly, Hsp90 appears absolutely required for receptor maturation.
Recent successes in producing recombinant DHBV P protein from Escherichia coli as fusions with solubility enhancing domains such as NusA or GrpE (8,31), or GST (32,33) enabled the in vitro reconstitution of priming-active RT complexes entirely from purified components, allowing to dissect the roles of individual chaperones. For the GST-fused P protein, a strict dependence on both the Hsp70 and Hsp90 systems was reported (33) except when the entire RNase H domain was removed (34); our own data (31), mainly using the NusA-fused P protein (NusDP), indicated instead that Hsc70 and Hsp40 plus ATP are necessary and sufficient to generate the activated, metastable state P* (Figure 1B).
To resolve these seemingly discordant results and given the limited knowledge on the molecular mechanisms of client protein activation by chaperones, in particular by Hsp90 (23), we compared in detail how the Hsp70 and the Hsp90 systems, alone or in combination, influence in vitro hepadnavirus RT activation (Figure 1B). In addition, we specifically modulated the Hsp70 and Hsp90 chaperoning activities. For Hsp90, we used mutants defective for ATPase activity or interaction with Hop. For Hsc70, we affected the ADP–ATP exchange rate by the Hsp70 nucleotide exchange factor Bag-1 (35). Bag-1 belongs to a family of proteins (36) which all contain at least one copy of a conserved, roughly 50-amino-acid-long Bag-domain that mediates their interaction with Hsp70 (36–38). A hallmark of Hsp70 activity is the cyclic binding and release of substrates, driven by ATP hydrolysis and ADP–ATP exchange (16). Bag-1 accelerates this cycle by increasing the ADP–ATP exchange rate (39) which, depending on the Bag-1 isoform (40) and the Bag-1 to Hsp70 ratio (39), may increase or decrease overall chaperone activity (35,39–42)
Corroborating our previous conclusion we found that the Hsp70 system plus ATP was sufficient to activate P protein, yet activation was significantly stimulated both by Bag-1 and by Hsp90 plus Hop, though by different mechanisms. Hence the hepadnaviral RT represents an unusual chaperone client that can be fully matured, in vitro, into an active state by the Hsp70 system yet the level of activation remains tunable by the Hsp90 system. We also re-examined the effects of Hsp90 inhibition on virus replication in intact cells. Only under special conditions did we observe a significant though incomplete reduction of viral DNA synthesis. Notably, this inhibition required inhibitor concentrations that exerted substantial cytotoxicity and induced a strong heat-shock response, including the upregulation of Hsp70 and Hsp90 itself. Hence whether or not Hsp90 is essential for hepadnavirus replication in vivo remains to be determined.
MATERIALS AND METHODS
Recombinant DHBV RT
DHBV P proteins with an internal deletion of the dispensable spacer region and an additional deletion of 25 amino acids from the C terminus were expressed in E. coli as fusion proteins with GrpE (termed GrpDP) and NusA (termed NusDP) and purified as previously described (31). Notably, this procedure removes most of the bacterial chaperones DnaK and GroEL, which have been reported to be present in up to stoichiometric amounts in recombinant GST-P preparations (32). From sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) analysis and scanning the Coomassie Blue stained bands, we estimate that the bacterial chaperones accounted for between 10, and 20%, of the full-length RT protein in both the GrpDP and NusDP preparations used (Supplementary Figure S1).
Chaperones
Genes for rat Hsc70 (43), human Hsp40 (Hdj1) (44), human Hop (45), murine Bag-1S (42) and the mutant Bag-1S-R182A (Bag-1S Arg to Ala substitution at amino acid position 182) were cloned into a modified pET30a vector (Novagen) downstream of a hexa-histidine tag and a TEV protease cleavage site (cloning details are available upon request). The proteins were expressed in E. coli BL21 Star cells (Invitrogen) supplemented with the Codon Plus RIL plasmid (Stratagene) and purified by immobilized metal affinity chromatography (IMAC) using HisTrap columns and FPLC equipment (GE Healthcare). Subsequently the His-tags were cleaved off with tobacco etch virus (TEV) protease and removed by immobilization to IMAC resin. The purified proteins were dialyzed against storage buffer [20 mM Tris pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, 1 mM DTT, 0.1% (v/v) complete EDTA-free protease inhibitor cocktail (Roche)] and stored at –80°C. The Bag-1C expression construct was obtained by insertion of a PCR-amplified DNA fragment encoding amino acids 96–219 of the murine Bag-1S protein (corresponding to amino acids 232–355 of murine Bag-1L, GenBank Accession Number AF022223) into the vector pET30a. The resulting plasmid encodes a non-cleavable hexa-histidine tag at the carboxy-terminus of Bag-1C. Expression and purification were performed as described above. Human Hsp90β was purified to homogeneity from baculovirus-infected insect cells essentially as previously described (46). Native dimeric state and ATPase activity were confirmed by gel filtration and ATPase assay, respectively. Wild-type yeast Hsp90 (Hsp82) and its deletion variants yHsp90 ΔMEEVD (lacking 5 C terminal amino acids) and yHsp 90Δ16 [lacking 16 N terminal amino acids (47)] were cloned into the vector pET28b (Novagen) downstream of a hexa-histidine tag (48). Proteins were expressed in E. coli BL21 Star cells and purified by IMAC, anion exchange chromatography and gel filtration essentially as previously described (48), and stored at –80°C. An SDS–PAGE analysis of all recombinant proteins used is shown in Supplementary Figure S1.
Reconstitution of P protein–Dε RNA complexes
Standard reconstitution reactions contained 150 nM (150 ng) GrpDP, 4.2 µM (3 µg) Hsc70, 5.3 µM (2 µg) Hsp40, 0.6 µM (0.5 µg) Hsp90, 0.9 µM (0.5 µg) Hop, 2 µl TKD buffer (100 mM Tris–HCl, pH 7.5; 250 mM KCl; 10 mM DTT), 1 µl of an ATP regenerating system (50 mM ATP, 25 mM MgCl2, 250 mM creatine phosphate, 100 U/ml phosphocreatine kinase), 10 U of RNasin RNase inhibitor (Promega), 1 µM in vitro transcribed Dε RNA (31) and H2O up to a final volume of 10 µl. In some experiments, lower GrpDP concentrations were used as indicated. Where appropriate, Bag-1S or mutant Bag1 proteins were included at the indicated concentrations. To allow for P–Dε complex formation samples were incubated at 30°C for 3.5 h (reconstitution reaction). Prior analyses had shown that the Hsc70/Hsp40 only, as well as the Hsp90/Hop, supplemented reactions remained in a linear phase for at least 5 h. Enzymatic activities of reconstituted P–Dε complexes were subsequently examined by adjusting the reactions to [α32P]-dATP priming conditions as previously described (31,49). Due to the distinct buffer conditions, including the presence of 0.2% (v/v) NP-40 detergent, no further RT activation by the chaperones occurs whereas already formed P–Dε complexes remain largely stable (31). The priming signal intensity is therefore an approximate measure of the amount of P–Dε complexes formed during the reconstitution reaction. Samples were separated by SDS–PAGE and 32P-labeled RT proteins were detected by autoradiography or phosphorimaging (BAS-1500, Fuji) and quantified using MacBas software (Fuji).
Relative affinity of in vitro activated RT for Dε RNA
P–Dε complexes were reconstituted at standard conditions from 150 nM GrpDP and increasing amounts of Dε RNA (ranging from 2.5 nM to 2 µM) in the presence of standard concentrations (see before) of either Hsc70 and Hsp40, or in the additional presence of Hsp90 and Hop. Functional P–Dε complexes were detected by 32P-dATP priming assays. Priming signal intensities (I[Dε]) for each Dε concentration [Dε] were quantified by phosphorimaging. To determine the parameters Kd (a nominal apparent dissociation constant) and Imax (theoretical maximum priming intensity at saturating concentration of Dε) for each of the two data sets hyperbolic regression analyses according to following equation were performed:
Detection of P–Dε complexes by RNA gel shift assay
For direct detection of P–Dε complexes by RNA gel shift assays (50), reconstitution reactions were assembled under standard conditions using 150 ng of NusDP protein, except that TKD buffer was replaced by TMNK buffer (100 mM Tris–HCl, pH 7.5; 10 mM MgCl2; 75 mM NaCl; 100 mM KCl, 20 mM DTT) supplemented with EDTA-free protease inhibitor cocktail, and the Dε RNA was 32P labeled (31). Samples were analyzed by native gel electrophoresis in 0.5× TBE buffer using 5% polyacrylamide gels (acrylamide:bis-acrylamide ratio 80:1) containing 2.5% (v/v) glycerol. Signals were detected by autoradiography.
Hsp90 inhibition by GA analogs 17-DMAG and 17-AAG
17-(Dimethylaminoethylamino)-17-demethoxygeldanamycin (17-DMAG) and 17-allylamino-17-demethoxygeldanamycin (17-AAG) were obtained from Invivogen, and 1 mM stock solutions in H2O (17-DMAG) and DMSO (17-AAG) were prepared. The impact of the drugs on the chicken hepatoma cell line LMH was assessed by incubating cells, cultured as previously described (51), in the presence of the indicated drug concentrations for 24 or 48 h; for 17-AAG, DMSO at the concentration corresponding to that at the highest drug dose was used as negative control. For transfections with the DHBV expression vector pCD16 (52) TransIT-LT1 (Mirus) reagent was used as recommended by the manufacturer. In brief, cells in 6-well plates were incubated at 50–60% confluency with the transfection reagent/DNA mixture for 14–16 h; thereafter, medium was changed and the cells were maintained for the indicated time periods. Drug was added immediately after the transfection phase, or added 1 or 6 h prior to transfection, and maintained at the same concentration throughout the experiments. Cells were harvested and cytoplasmic lysates were prepared as described (51). Total protein concentration in the lysates was determined using the bicinchoninic acid (BCA) assay as recommended by the manufacturer (Pierce). Viral DNA from cytoplasmic nucleocapsids was isolated and detected by Southern blotting as described (53). Primary antibodies for western blotting were T9026 (anti-tubulin; Sigma); AC88 (anti-Hsp90α/β; Stressgen); and SPA-757 (anti-Hsc70/Hsp70; Stressgen). For detection, the blots were incubated with appropriate peroxidase or alkaline phosphatase conjugated secondary antibodies, followed by ECL+ (GE Healthcare) or CDP* reagent (Roche), respectively. Chemiluminescent signals were recorded on X-ray film or on a Fuji LAS 3000 instrument, and quantified using AIDA software. In vitro inhibitory activity of 17-DMAG was addressed by performing P–Dε complex reconstitutions, as described above, in the presence of the indicated concentrations of the drug and subsequent priming assays.
RESULTS
Stimulatory but non-essential role of Hsp90 plus Hop in RT activation in vitro
Initiation of DNA synthesis by P protein requires first that the Dε template RNA be bound, for which chaperone assistance is obligatory. Successful initiation results in the covalent attachment of the Dε templated DNA oligo to the RT. Hence, in the presence of α-32P labeled dNTPs, P protein in priming-competent complexes becomes radioactively labeled and can sensitively and quantitatively be detected by autoradiography (‘priming assay’; see Figure 1A). In our previous study, most experiments had been performed with a NusA-fused DHBV P protein (NusDP). Because the GrpE-fused protein (GrpDP) appeared to have similar properties (31) yet a smaller heterologous part (23 kDa for GrpE versus 55 kDa for NusA), GrpDP was used in the current study. In addition, instead of His-tagged Hsp40 and commercial bovine Hsc70 we here employed E. coli-derived preparations of tag-free human Hsp40 (Hdj1) and rat Hsc70; this excluded any contamination of the Hsc70/Hsp40 only reactions with eukaryotic Hsp90.
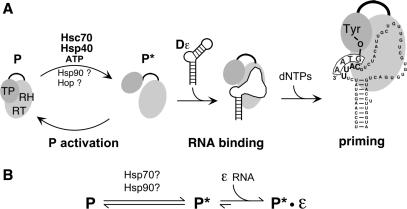
To establish that all these components were functional GrpDP (150 nM) was incubated under standard conditions (see Materials and Methods section for details) with various combinations of Dε RNA, Hsc70, Hsp40, Hsp90 and Hop, always in the presence of an ATP-regenerating system. Potential priming products were analyzed by SDS–PAGE and autoradiography. As shown in Figure 2A, GrpDP could be activated by Hsc70 plus Hsp40 (lane 3) but not by either of the two chaperones alone (lanes 1 and 2). Hsp90, in the absence of Hsc70, or Hsp40, or Dε RNA (lanes 4, 5 and 6), was unable to promote any P activity. However, when all basic Hsp70 components and, in addition, Hop were present, Hsp90 caused an ∼5-fold, Hop-dependent increase in signal strength (lane 7).
Figure 2.
Chaperone dependence of priming-active P–Dε complex formation. (A) Stimulatory role of Hsp90. P protein–Dε complexes were reconstituted from GrpDP (150 nM) in the presence of the indicated chaperones and subsequently subjected to priming assays. Samples were separated by SDS–PAGE and the 32P-labeled P protein was visualized by autoradiography. (B) Effect of Hsp90 concentration on complex reconstitution. P protein–Dε complexes were reconstituted from GrpDP (10 nM), and fixed concentrations of Hsc70, Hsp40, Hop and Dε RNA plus varying concentrations of Hsp90 as indicated. Active complexes were detected by priming assays. 32P-labeled P protein was visualized by phosphorimaging (inset), and the quantified values were plotted against the Hsp90 concentration. (C) Effect of P protein concentration on complex reconstitution. P–Dε complexes were reconstituted with increasing concentrations of GrpDP in the presence of fixed concentrations of Hsc70 and Hsp40 only (upper panel), or in the additional presence of Hsp90 and Hop (lower panel). Complex formation was analyzed by priming assays as in (A).
Hence these data corroborated that for RT activation Hsc70 and Hsp40 plus energy are sufficient and necessary, whereas Hsp90 plus Hop have a stimulatory but non-essential role. Because these results remained in contrast to the strict requirement for Hsp90/Hop reported by others (33), we suspected that the low amounts of recombinant RT used there (10 nM) could have obscured detectable priming activity in the absence of Hsp90/Hop. We therefore performed reconstitution assays in which decreasing concentrations of GrpDP (from 160 to 5 nM) were incubated with fixed amounts of the various chaperones. Beforehand, we titrated the Hsp90 concentration required for maximal stimulation at 10 nM GrpDP concentration. Stimulation showed an initial linear increase which leveled off at ∼0.6 µM Hsp90 (Figure 2B); this Hsp90 concentration was used in the subsequent experiments. Decreasing the amount of GrpDP reduced, expectedly, the strength of the priming signals (Figure 2C). Without Hsp90/Hop, signals became barely detectable, except upon long-term exposure, around 10 nM P protein. Inclusion of Hsp90/Hop increased the signals, in the range of 4- to 5-fold at the higher, and up to ∼8-fold at the lowest P concentrations, resulting in an easily detectable signal. Hence the seemingly strict Hsp90/Hop dependence of P activation may be due to quantitative rather than qualitative differences between the assay systems used.
Stimulatory activity of Hsp90 depends on its ability to interact with Hop but can occur independent from Hsp90 ATPase activity
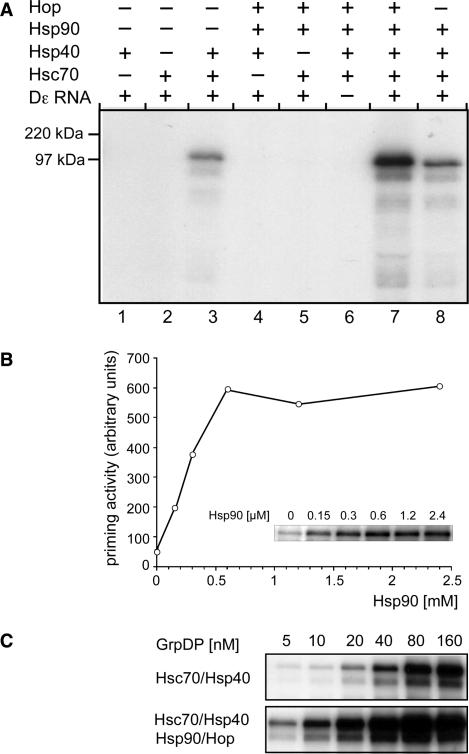
Complete passage through the Hsp90 chaperoning cycle requires its intrinsic ATPase activity (23,54,55) but Hsp90 can also display nucleotide-independent ‘passive’ chaperoning activity (56,57). To dissect whether Hsp90 ATPase activity is necessary for the stimulatory activity on the RT, ATP depletion is unsuitable because it would simultaneously block the Hsp70 system, which is itself required for Hsp90 stimulation (Figure 2A). Therefore we employed a variant Hsp90, yHsp90Δ16, which is essentially deficient in ATP hydrolysis (47). Second, the dependence on Hop of Hsp90 stimulation had suggested that an interaction between the two proteins was necessary. To address this point, we used the mutant yHsp90ΔMEEVD, which lacks the primary C terminal Hop interaction motif (58). Both proteins were available as the yeast Hsp90 homologs. We therefore first established that wild-type yeast Hsp90 can functionally substitute for the human Hsp90 used in the previous experiments; this was indeed the case, with comparable (3.9-fold) stimulation of the Hsc70/Hsp40-mediated activation (Figure 3). Only a slight increase (1.3- to 1.4-fold) was seen with yHsp90ΔMEEVD but yHsp90Δ16 stimulated activation significantly (2.9-fold), yet only in the presence of Hop. These results were reproduced in three independent experiments. These data suggest that Hsp90 stimulation of RT activation does not require ATP hydrolysis, yet it is still dependent on Hop as an adaptor to Hsc70/Hsp40.
Figure 3.
Efficient Hsp90 stimulation requires Hop but not the ATPase activity of Hsp90. P–Dε RNA complexes were reconstituted from GrpDP and Hsc70/Hsp40 in the absence of Hsp90 and Hop, or in the additional presence of Hop and wild-type yeast Hsp90 (lane wt), or the yeast Hsp90 variants ΔMEEVD or yHsp90Δ16 (lanes Δ16). The stimulatory activity of yHsp90Δ16 was abolished when Hop was omitted (lane Δ16–Hop). Complex formation was monitored by priming assays as in Figure 2. Relative signal intensities are given below each lane, with the signal from the unsupplemented reaction set at 1.0.
The Hsc70 nucleotide exchange factor Bag-1 stimulates Hsc70/Hsp40-mediated RT activation
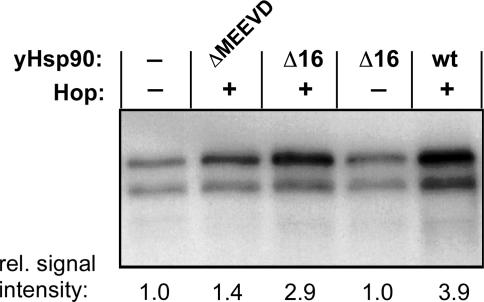
A typical feature of authentic chaperoning activity is the transient, iterative and ATP-consuming interaction with substrates. Stable substrate binding by Hsp70 requires ATP hydrolysis; Hsp40 serves to stimulate the Hsc70 ATPase activity, leading to increased levels of Hsc70ADP. At the concentrations of Hsc70 and Hsp40 used here, Hsp40 is not limiting (31). Initiation of a new chaperoning cycle requires that the Hsc70-bound ADP (Hsc70ADP) can be exchanged for ATP; this naturally slow reaction might therefore be rate limiting. To test this conjecture, we exploited the nucleotide exchange factor Bag-1 and supplemented the minimal reconstitution system consisting of GrpDP, Dε RNA, Hsc70 and Hsp40 plus ATP, with various concentrations of Bag-1, in the form of Bag-1S (Figure 4A). Bag-1S significantly stimulated Hsc70/Hsp40-mediated P protein activation in a concentration-dependent manner (Figure 4B). The activating effect was optimal at concentrations around 5 µM, corresponding to an equimolar ratio of Bag-1S to Hsc70. In this range, we observed a reproducible 4- to 5-fold increase in priming competent P–Dε complexes. Further rising the Bag-1S concentration lowered the stimulatory activity. Stimulation was not due to non-specific stabilization of the RT because bovine serum albumin had no effect (Figure 4B). These data indicate that in the minimal Hsc70/Hsp40 reconstitution system the exchange of ADP for ATP on Hsc70 is rate limiting for P protein activation.
Figure 4.
Effect of Bag-1 on GrpDP activation. (A) Schematic representation of the examined Bag-1 variants. The Bag-domain and the ubiquitin-like domain are shaded in dark and light gray, respectively. Numbers refer to amino acid positions. The Arg to Ala mutation at amino acid 182 in Bag-1S-R182A is indicated. (B) Bag-1S enhances Hsc70/Hsp40-mediated P protein activation. P protein–Dε complexes were reconstituted from GrpDP, Hsc70 and Hsp40 in the presence of varying amounts of Bag-1S (open circles) or BSA (filled triangles), and complex formation was monitored by priming assays. Priming signals were quantified and plotted against the concentration of the respective protein added, with the value obtained without addition of Bag-1S and BSA set to 100%. The relative priming activities are given as means ± SD from between two and five independent experiments, each determined in duplicate. (C) The Bag domain is sufficient to stimulate P protein activation and acts by affecting nucleotide exchange on Hsc70. Standard Hsc70/Hsp40 reconstitution reactions were supplemented with the indicated amounts of Bag-1C (open circles), or the non-functional mutant Bag-1S-R182A (filled triangles). Relative priming activities were determined as in (A).
Since we are using GrpE–P protein fusion proteins, it is important to note that although GrpE is a functional E. coli homolog of Bag-1, it does not affect the nucleotide exchange rate of eukaryotic Hsc70 (35). This was confirmed in that activation of the NusDP fusion protein was similarly stimulated by Bag-1S (data not shown). Conversely, Bag-1 cannot stimulate bacterial Hsp70 (35). These data also indicate that residual bacterial DnaK and GroEL, amounting to at most 10–20% of the amount of full-length RT protein present in the recombinant preparations (see Materials and Methods section, and Supplementary Figure S1), cannot account for the observed RT activation. Furthermore, replacement of eukaryotic Hsc70 and Hsp40 by bacterial DnaK and DnaJ plus ATP under otherwise identical conditions did not lead to any RT activation (data not shown).
A functional Bag domain is necessary and sufficient for stimulation of Hsc70/Hsp40-mediated RT activation
In order to define the minimal domain of Bag-1S required for stimulation of replication initiation complex formation, we used a deletion mutant, Bag-1C, that comprises only the Bag domain. Specifically, it is the murine homolog of a fragment of human Bag-1M (amino acids 151–274) that has been shown to functionally interact with Hsc70 via enhancing its nucleotide exchange rate (39). Bag-1C stimulated the formation of priming-competent initiation complexes to a similar extent (∼4-fold), and with similar concentration dependence, as Bag-1S (Figure 4C). Stimulation was maximal at ∼5 µM Bag-1C and declined at higher concentrations, as with Bag-1S; at the highest concentration tested (20 µM) Hsc70/Hsp40 activation was inhibited.
To formally prove that the stimulation was caused by accelerated nucleotide exchange on Hsc70 we also tested a point mutant of Bag-1S, Bag-1S-R182A, in which the single Arg to Ala substitution in the Bag domain renders the protein deficient for promoting nucleotide exchange (59). As shown in Figure 4C, Bag-1S-R182A did neither stimulate nor inhibit P protein activation at any concentration tested. Together, these data demonstrated that the Bag domain is sufficient for enhancement of Hsc70/Hsp40-mediated P protein activation, provided its ability to accelerate nucleotide exchange on Hsp70 is intact. Hence, most likely Bag-1 stimulates RT activation by direct interaction of the Bag domain with the ATPase domain of Hsc70.
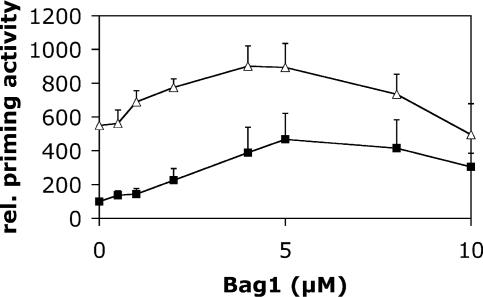
Combined Bag-1S plus Hsp90/Hop stimulation of RT activation
The above described data had shown that both Hsp90/Hop and Bag-1S stimulated Hsc70/Hsp40-mediated RT activation. We next analyzed which effects a combination of Bag-1S plus Hsp90/Hop had on the basal Hsc70/Hsp40 activation of P protein. Standard reconstitution assays were performed with Hsc70, Hsp40, Hop and Hsp90, plus increasing amounts of Bag-1S as described earlier, and compared with reactions lacking Hsp90 and Hop (Figure 5); to ensure that, in the simultaneous presence of Bag-1S stimulation, Hsp90 would not become limiting these experiments were performed at low GrpDP concentration (10 nM). As before, the Hsc70/Hsp40-only-mediated activation was maximally stimulated, by 4- to 5-fold, at an optimal Bag-1S concentration of around 5 µM. The Hsp90/Hop supplemented reactions, starting off at ∼5-fold higher levels, were similarly affected by Bag-1S, with essentially the same concentration dependence. In fact, up to the optimal Bag-1S concentration, the two curves were almost superimposable, with a nearly constant difference in relative priming efficiencies. Hence the extent of Hsp90/Hop stimulation was limited by the available levels of Hsc70/Hsp40-pre-activated P protein. The mutant Hsp90 protein defective for Hop interaction had essentially no effect (data not shown).
Figure 5.
Stimulation of P activation by Bag-1S plus Hsp90/Hop. P protein–Dε complexes were reconstituted from GrpDP (10 nM) with only Hsc70 and Hsp40 (filled squares), or in the additional presence of Hsp90 and Hop (open triangles), and supplemented with the indicated amounts of Bag-1S. Complex formation was monitored by priming assays. Signals were quantified by phosphorimaging, normalized to the signal obtained in the presence of Hsc70 and Hsp40 only, and plotted against the concentration of Bag-1S. Relative priming activities are given as means ± SD from at least four independent experiments.
Hsp90/Hop affect the quantity, not the quality, of Hsc70/Hsp40-activated RT molecules
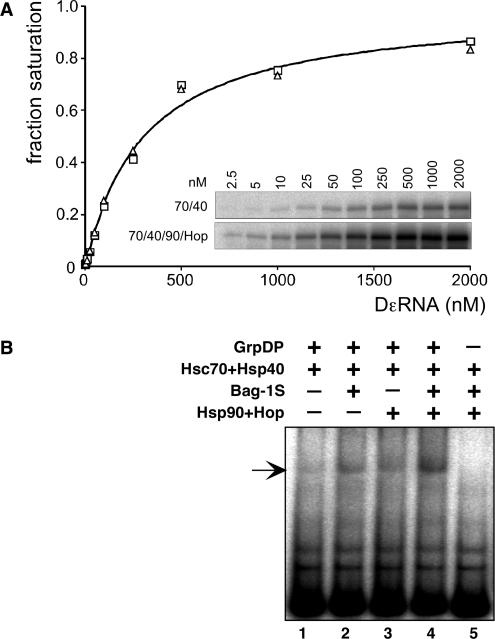
Because Dε must be bound to P to act as priming template and because P–Dε complexes are relatively stable (31), the priming signal intensities are generally proportional to the amount of these complexes present in the reaction. However, such proportionality is not necessarily direct. For instance, Hsp90 acting on the Hsc70/Hsp40 pre-activated P* molecules might create a distinct activated form, P**, that has an increased affinity for Dε RNA; or it could enhance the ability of the RT to utilize the bound RNA as template; in this scenario, the same amounts of P–Dε complexes would lead to stronger priming signals. To address the first point, we compared the relative affinities for Dε RNA of Hsp70 system versus Hsp70 plus Hsp90 system activated RT. Reconstitution reactions containing fixed concentrations of all protein components were incubated with increasing amounts of Dε RNA, then priming assays were performed (Figure 6A). The absolute priming signal intensities were, expectedly, higher in the Hsp90/Hop supplemented reactions, yet the Dε concentration dependence was virtually unchanged, with half-maximal signal intensities at ∼0.3 µM RNA, close to the value measured for in vitro translated P protein in RL (60). Hence Hsp90/Hop did not increase the apparent affinity of P* for its template RNA.
Figure 6.
(A) Hsp90 does not affect the relative affinity of P* for Dε RNA. P protein–Dε complexes were reconstituted with only Hsc70 and Hsp40 (open squares), or in addition with Hsp90 and Hop (open triangles), and varying concentrations of Dε RNA. Complex formation was monitored by priming assays (inset), and signal intensities were quantified by phosphorimaging. Values were normalized against the calculated maximum intensity at saturating concentration of Dε (I[Dε]/Imax; ‘fraction saturation’), and plotted against the Dε RNA concentration. A hyperbolic saturation curve for a nominal apparent Kd = 0.3 µM, as indicated, fits equally well to both data sets. (B) Hsp90 plus Hop, like Bag-1S, increase steady-state levels of P–Dε complexes. Complex formation upon RT activation with the indicated chaperones and co-chaperones was monitored via RNA gel shift assay. The increases in signal strengths via stimulation by Hsp90/Hop, Bag-1S, or a combination of both, paralleled those observed in the corresponding priming assays.
To address the second point, we used RNA gel shift assays to directly monitor the relative amounts of P–Dε complexes produced by the Hsp70 system, and in the additional presence of Bag-1S, Hsp90/Hop, or both (Figure 6B). Weak but specific signals, as indicated by the absence of a corresponding signal in the control lacking RT (lane 5), were obtained with Hsc70/Hsp40 alone (lane 1). Both Bag-1S (lane 2) and Hsp90/Hop (lane 3) increased the signal intensities, and clearly the strongest signals were obtained from the doubly supplemented reaction (lane 4). Although an exact quantitation is difficult, these data paralleled those in the previous priming experiments. Hence Hsp90/Hop did not substantially enhance the efficiency of template utilization by P*. While the increase in P–Dε complexes seen in the gel shift assay for the Hsp90/Hop and Bag-1S supplemented reactions would per se be compatible with an enhanced affinity of P for Dε RNA, this is highly unlikely in view of the RNA titration data reported above. Hence in combination, the two data sets strongly suggest that the Hsp90 system increases the number, not the quality of Hsp70 system activated RT molecules.
Pharmacological Hsp90 inhibition does not unambiguously prove an essential role for Hsp90 in hepadnavirus RT activation in intact cells
The ansamycin antibiotic GA acts as an ADP mimic that specifically binds into the nucleotide pocket of Hsp90 (61) and thus blocks ATP binding and the ATP hydrolysis-mediated Hsp90 chaperoning cycle (23). GA has been reported to suppress DHBV replication in transfected cells and to interfere, though at 100- to 1000-fold higher concentrations, with priming in reticulocyte lysate by in vitro translated RT (12,13) or recombinant RT (32). This may suggest that Hsp90, while dispensable in the reconstitution system, could be essential for viral DNA synthesis under physiological conditions, although inhibition of RT activation is but one out of very many alternative explanations for the inhibitory effects of GA. In addition, GA is now known to be a potent heat-shock inducer, mainly via the Hsp90-controlled heat-shock transcription factor 1 (62), and is rather cytotoxic at higher concentrations. Two improved GA derivatives are 17-AAG and 17-DMAG (63); the latter has the added advantage of being water-soluble. To shed more light on the role of Hsp90 on DHBV replication in cells, we re-examined the effects of pharmacological Hsp90 inhibition on DHBV synthesis in the chicken hepatoma cell line LMH which, though not infectable by DHBV, supports efficient DHBV replication and virus formation when transfected with appropriate DHBV expression vectors (3).
Preliminary dose-finding experiments (17-DMAG and 17-AAG varied from 1 ng/ml to 1 µg/ml) showed, after 24 h of treatment with 10 ng/ml or more of both drugs, a strong increase in the relative amounts of cytoplasmic proteins with apparent masses of around 90, 70 and 25 kDa (Supplementary Figure S2); by western blotting they were confirmed to be Hsp90 and Hsc/Hsp70 (Figure 7); the 25 kDa band contained, as identified by mass spectroscopy, Hsp27 related chicken proteins. Incubation for 48 h or longer led to a massive loss of viable cells at the higher drug doses.
Figure 7.
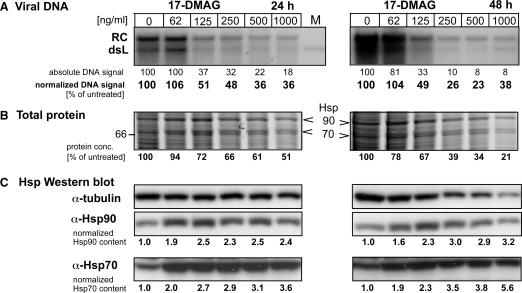
Effects of the Hsp90 inhibitor 17-DMAG on DHBV replication in transfected LMH cells. LMH cells were incubated with the indicated inhibitor concentrations 6 h prior to and during transfection with the DHBV expression vector pCD16. Cells were kept in the presence of drug for 24 h (left panels) or 48 h (right panels), then cytoplasmic lysates were prepared for viral DNA and cellular protein analysis. (A) Southern blot for viral DNA. Viral DNAs isolated from cytoplasmic nucleocapsids were detected using a 32P-labeled DHBV-specific DNA probe. RC denotes the typical 3 kb relaxed circular, and dsL the double-stranded linear DNA product of reverse transcription. Lane M contained a plasmid-derived 3.0 kb DHBV marker fragment. Band intensities were quantified by phosphorimaging, and the raw data are given below each lane relative to the untreated control set at 100% (designated absolute DNA signal). Correcting the raw data for total protein content of the corresponding lysates yielded the values designated normalized DNA signal. (B) Impact of 17-DMAG on total protein content. Aliquots from the same lysates used for viral DNA isolation were analyzed by SDS–PAGE analysis and Coomassie Blue staining. Only the section of the gel covering the molecular mass range between 60 and 100 kDa is shown (for the full range see Supplementary Figures S2 and S3). Note the relative increase of bands at around the 90 and 70 kDa position, shown in (C) to represent Hsp90 and Hsp70. Total protein concentrations determined by the BCA assay are given below each lane relative to the untreated sample set at 100%. (C) 17-DMAG upregulates Hsp70 and Hsp90 expression. Equal aliquots of the lysates were analyzed for Hsp90 and Hsc70/Hsp70 by western blotting, using chemiluminescent substrates; signals were quantified using an LAS3000 imaging system. Normalized values for Hsp90 and Hsp70 content were derived by correction for the total protein content in each lysate. Tubulin was simultaneously detected as a loading control. The decreasing tubulin signals with increasing drug concentration parallel the decrease in total protein content.
In the subsequent transfection experiments, we used 17-DMAG at concentrations of 62.5, 125, 250, 500 and 1000 ng/ml. Addition of the drug immediately after the ∼16 h transfection period (see Materials and Methods section for details) revealed a modest reduction (by 30–40%) in viral DNA signal intensity at the two highest drug doses after 48 h, and a strong reduction (by 70–80%) after 72 h (Supplementary Figure S3). However, there was a parallel loss of viable cells, manifest by a strong reduction in the total protein content of the lysates from which the virus DNA was isolated. Because chaperone-mediated RT activation for pgRNA encapsidation is an early event in replication and because nucleocapsids are highly stable (64), we next kept the cells in the presence of inhibitor already 1 h before and during transfection. This time viral DNA was prepared 24 and 48 h post-transfection. No specific inhibition was seen after the short treatment period, and the reduction in absolute DNA signal strengths at the two highest drug concentrations after 48 h (∼60 and 70%, respectively) correlated again with a concomitant reduction in total protein content of the lysates (Supplementary Figure S4). Because binding of GA to Hsp90 has recently been suggested to occur with very slow kinetics (65), we finally started treatment of the cells 6 h prior to transfection. Under these conditions, drug dose-dependent reductions in DNA signal intensity from ∼50–65% after 24 h, and 50% to about 80% after 48 h were observed after correction for total protein content of the source lysates (Figure 7A). However, not even the highest dose of 17-DMAG was able to completely block viral replication although cell viability was already strongly affected (Figure 7B). While compatible with a similar auxiliary but non-essential role for Hsp90 in RT activation in vivo as in vitro, all drug concentrations tested led to a significant upregulation (Figure 7C) of Hsp90 (up to 2.8-fold) and Hsc70/Hsp70 (1.8- to more than 4-fold). Hence the intricate networking between the chaperone systems in the cell precludes clear assignments on whether or not Hsp90 is absolutely required for DHBV replication.
We also tested the impact of 17-DMAG on the in vitro reconstitution system, with and without Hsp90/Hop (Supplementary Figure S5). No inhibition was detectable at 1 µg/ml (the highest dose tested in vivo); at 10 µg/ml Hsp90/Hop stimulation was substantially reduced from ∼4- to 1.6-fold whereas the reaction without Hsp90/Hop was only slightly affected (∼25% reduction). At 100 µg/ml, the drug essentially abolished the priming signal in the Hsp90/Hop supplemented reaction, similarly as previously reported for in vitro priming in reticulocyte lysate (12,13); however, the reaction without Hsp90/Hop was also nearly completely inhibited. Preincubation of Hsp90 with the drug for 1.5 h to account for the potentially low on-rate of Hsp90–inhibitor complex formation did not enhance the inhibitory effect (data not shown). Hence these data are in line with specific inhibition of the stimulatory activity of Hsp90 at intermediate drug concentration whereas the nearly complete inhibition at the highest dose cannot be attributed to Hsp90-specific effects.
DISCUSSION
The crucial role of chaperones for virus propagation, though at diverse levels, is increasingly acknowledged (19). However, obligate chaperone dependence, even in vitro, remains a distinctive feature of the hepadnaviral P protein, probably shared by telomerase (66–68), the cellular RT maintaining chromosome end integrity (69). Here, we exploited in vitro reconstitution of priming-active DHBV RT to dissect the roles of the Hsp70 and Hsp90 systems in activation. The data firmly establish the fundamental role of the Hsp70 system in generating a metastable state, P*, which is autonomously capable of Dε RNA binding and authentic initiation of reverse transcription. This mode of activation is different from that of typical Hsp90 clients such as the steroid hormone receptors, which do not reach ligand-binding competence without the Hsp90 system. Clearly, however, different from typical Hsp70 clients such as clathrin, the Hsp90 system can substantially stimulate RT activity. This stimulation, mechanistically distinct from that exerted by the Hsp70 nucleotide exchange factor Bag-1, requires Hsp70-activated P* as substrate, and it depends on Hop as an adaptor between both chaperone systems. These data support a model for RT activation (Figure 8) whereby Hsp70 induces the basic conformational changes distinguishing P* from P, whereas Hsp90 increases the quantity, not the quality, of activated RT molecules. The GA inhibition data are compatible with, but do not prove, a similar stimulatory but non-essential role of Hsp90 for virus replication in intact cells; importantly, they neither provide unambiguous evidence that Hsp90 is essential.
Figure 8.
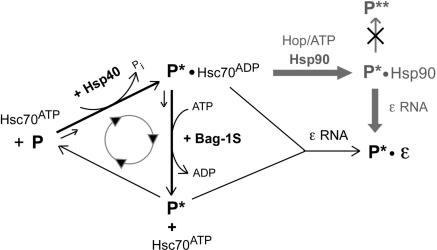
Model for chaperone contributions to P protein activation. As in the general Hsp70 chaperoning cycle, Hsp40 stimulates the ATPase activity of Hsc70, converting Hsc70ATP into high substrate affinity Hsc70ADP which binds P and produces P*-Hsc70ADP. Spontaneous recycling of P*-bound Hsc70ADP into free Hsc70ATP is rate limiting. Proper concentrations of Bag-1S promote ATP for ADP exchange on Hsc70 and substrate release, enabling faster cycling and faster production of P*, increasing its steady-state concentration. Over-acceleration by excess Bag-1S reduces the time span Hsc70ADP can act on P, reducing P to P* conversion and thus the steady-state P* concentration. Dε RNA may react with either free P* or P*-Hsc70ADP. While the increased release of P* by Bag-1S would favor free P* as binding partner, the simultaneously generated Hsc70ATP becomes available for a new round of P*-Hsc70ADP production. The cyclic nature of this process thus prevents a clear distinction between the two options. Hsp90 (pathway in gray) does not act directly on P but requires prior Hsc70 action; the Hop dependence strongly suggests the P*–Hsc70ADP complex, not free P*, as Hsp90 substrate. Hsp90 increases the steady-state levels of P*, by slowing down decay or promoting production, but does not induce formation of a more active form P** (crossed-out pathway). The exact compositions of the chaperone-containing complexes are not known and may be subject to dynamic changes. Similarly, not each single interaction between Hsc70 and P may be productive in P* generation.
Essential versus non-essential role of the Hsp90 system in hepadnavirus RT activation
Given the evidence supporting hepadnaviral P protein being a bona fide Hsp90 client (12,13,19,33), our previous finding that Hsc70 plus Hsp40 plus ATP are sufficient to activate P protein (31) was surprising. One important aspect of the current work is therefore that it fully corroborates this conclusion. That Hsp90/Hop substantially stimulate this basal Hsc70/Hsp40 activation could therefore well explain why at low P protein concentrations Hsp90/Hop were seemingly essential to generate a detectable priming signal (33). Notably, occasional activation of DHBV RT in the absence of Hsp90 has been noted in the same report (33), and in vitro binding of HBV P protein to its cognate ε RNA, though not yielding any DNA products, was at least weakly possible with just Hsc70/Hsp40 (50). Hence activatability without Hsp90 may be a general property of hepadnaviral RTs.
The previously observed suppression of DHBV replication in intact cells by GA (12,13) might have suggested a strict requirement for Hsp90 in RT activation in vivo, questioning the physiological relevance of Hsp90 dispensability in vitro. However, our thorough re-examination of the impact of Hsp90 inhibition on virus replication revealed severe experimental limitations of this pharmacological approach. First, although we used the less toxic 17-DMAG derivative, substantial inhibition of viral DNA synthesis required drug concentrations that severely affected cell viability. That inhibition required several hours of pre-incubation with the drug is consistent with a role of Hsp90 at a very early replication step, including pgRNA packaging and initiation of reverse transcription (12,13); that a 1-h pretreatment was insufficient may relate to the kinetics of drug uptake and binding to intracellular Hsp90 which is still poorly understood (65,70). However, even the highest, barely tolerated drug concentrations did not completely block viral DNA synthesis. This would be expected if, as in vitro, the Hsp70 system alone were able to bring about a basal activation of the RT. However, several issues call for a very cautious interpretation. It is unclear whether all Hsp90 molecules in the cell were blocked, and possibly such blockage would be lethal. Second, the large number of cellular Hsp90 clients and genes affected by GA treatment (71) leaves many options for the inhibitory effect of GA other than preventing Hsp90-mediated RT activation. Most importantly with regard to this study, both 17-DMAG and 17-AAG induced a strong upregulation of Hsp70/Hsc70, Hsp27 related proteins and Hsp90 itself; this is also seen in various other cell types and probably mediated by the major heat-shock transcription factor HSF-1 which is itself regulated by Hsp90 (72). This interdependence of the chaperone systems in the cell underscores the importance of defined in vitro systems for understanding mechanistic aspects of client activation by chaperones.
Nucleotide exchange as rate-limiting step in Hsc70/Hsp40-mediated RT activation in vitro
The Hsp70-specific co-chaperone Bag-1S, at optimal concentration, stimulated Hsc70/Hsp40-driven P activation to a similar extent as Hsp90/Hop. That Bag-1S acts genuinely on Hsc70 was demonstrated by the lack of stimulation by a mutant Bag-1S unable to interact with Hsc70, and by the Bag-1S-like stimulation by just the Bag domain (construct Bag-1C) which suffices for Hsc70 binding (37). Given the dependence of Hsc70-mediated P activation on Hsp40 and on ATP hydrolysis (31,33), this is fully in line with the general chaperoning cycle of Hsp70 in which Bag-1 accelerates ADP for ATP exchange (35) and thus the frequency of cycling (Figure 7). Different from general refolding, however, P protein is not converted from an unfolded into a stable, native conformation that continuously accumulates; rather, the activated product is a metastable intermediate, P*, which in the absence of Dε RNA decays with a half-life of minutes as soon as the chaperoning activity of Hsc70 is interrupted by ATP depletion (31). The resulting steady-state level of P* will thus be determined by the rates of generation and decay of P*.
The clear stimulation by active Bag-1S, but not inactive Bag-1S-R182A, demonstrates that in the non-supplemented reaction exchange of ADP for ATP on Hsc70 is rate limiting for P* generation. Maximal stimulation occurred at around 1:1 stoichiometry of Bag-1S to Hsc70. The decline at higher Bag-1S to Hsc70 ratios is, again, in accord with the cycling model of Hsp70 chaperoning; in this case, the life span of the RT-Hsc70ADP complex would be so much reduced (39) that Hsc70-mediated restructuring from P to P* cannot occur. Hence Bag-1S stimulation is likely based on tuning the kinetics of the Hsp70 cycle such that the maximal number of P* molecules per time is generated, thus increasing their steady-state concentration (Figure 7), as corroborated by RNA gel shift assays (Figure 6B).
Not mutually exclusively, Hsc70 might have to be released from P* before Dε RNA can bind. Whether the chaperone and the RNA occupy overlapping sites on the RT is not known. Hsp70-binding sites on substrates conform to general, rather than specific sequence motifs (73); interestingly, one such candidate motif, predicted by an algorithm developed for DnaK (73), overlaps with a C terminal region in the TP domain that contains essential residues for Dε RNA binding (M.S., J.B. and M.N., manuscript submitted), and with a region termed T3 that appears to act as an intra- or intermolecular contact point (74). While the functional relevance of such predictions remains to be determined, the current data support a model (Figure 8) whereby the Hsp70 system acts on the RT as it does on its general substrates; the product is, however, a specific, metastable, Dε RNA binding-competent intermediate.
Predictions on the in vivo role of Bag-1 in hepadnavirus RT activation are currently as problematic as outlined above for Hsp90, given the plethora of isoforms (36) with apparently opposing effects (40,42,75,76), the importance of Bag1 to Hsp70 stoichiometry [see above and ref. (39)], and the presence of many other co-operating and competing co-chaperones that affect the Hsp70 cycle (16). Of practical importance, the enhancement of in vitro priming signals by Bag-1C, which is small and easily accessible in recombinant form, should facilitate use of the in vitro reconstitution system for screening for drugs that affect P protein activity.
Stimulatory activity of the Hsp90 system
Hsp90 did not promote any RT activation in the absence of Hsc70 or Hsp40; furthermore, in their presence, stimulation of P activation depended on the presence of Hop, and on the ability of Hsp90 to interact with Hop. Clearly, therefore, Hsp70-activation must precede Hsp90 binding. Furthermore, the established adaptor role of Hop indicates that P* complexed with Hsc70, not free P*, is the Hsp90 target (Figure 7). One way of exerting a stimulatory effect would be that Hsp90 induces qualitative changes in P* that promote its affinity for Dε RNA, or enhance its capacity to use Dε as template for DNA synthesis initiation. Our data do not support such an explanation. The same Dε RNA concentration (∼0.3 µM) was required for half-maximal priming activity of the Hsp70 system activated and the Hsp70 plus Hsp90 system activated RT. Second, direct monitoring of P–Dε complex formation by RNA gel shift assays, though not as accurately quantifiable as the priming signals, showed comparable increases upon addition of Hsp90/Hop, as well as Bag-1S, and a combination of the two, as those seen in the priming assays. Combined with the RNA titration data, this strongly suggests that the Hsp90 system acts by increasing the steady-state concentration of P* generated by the Hsp70 system.
As with Bag-1S, this could be achieved by accelerating P* production, or by slowing down P* decay, i.e. by P* stabilization. Hsp90 stimulation depended on its ability to interact with Hop but not on ATP hydrolysis, as shown by the substantial stimulatory activity of the variant yHsp90Δ16 which still binds ATP but is unable to mediate dimerization of the N terminal Hsp90 domains (47) that precedes ATP hydrolysis (23). Notably, this result is highly reminiscent of the Hop-dependent stimulatory activity of Hsp90 on Hsp70/Hsp40-mediated refolding of luciferase (56); there another ATP-binding competent but ATPase-deficient Hsp90 variant (E46A) could also partially substitute for wild-type Hsp90. Intriguingly, the mutation did not prevent Hsp90 loading on pregesteron receptor complexes but blocked mature complex formation and gain of ligand-binding competence. By contrast, yHsp90Δ16 stimulated Dε RNA binding by P protein nearly as good as wild-type Hsp90 (75%). Hence different from steroid hormone receptors, RT activation is neither strictly dependent on Hsp90, nor does the stimulatory activity of Hsp90 require the full Hsp90 chaperoning cycle. However, whether Hsp90 binding increases production or decreases decay of P* remains to be determined.
Mechanistic implications
Non-activated hepadnaviral RT is unable to bind its cognate Dε RNA. Given that chaperones are folding enzymes, and the ATP dependence of P activation and stimulation, both are expected to involve transient structural alterations in the RT that make the RNA-binding site accessible; we therefore agree with a previously proposed conceptual model (34), except that we ascribe the fundamental structuring role to Hsc70 rather than Hsp90. Possibly, chaperone activity extends to enabling the relative domain movements that must accompany DNA synthesis. For instance, the priming Tyr residue in the TP domain has to be properly oriented in the polymerase active site to allow attachment of the 5′ nt, and then must move out as the primer oligo is growing, similar to what was recently shown for the protein-priming DNA polymerase from bacteriophage phi29 (77). Obtaining active P–ε complexes in sufficient amounts for such crystallographic studies will be difficult. However the in vitro reconstitution system, combined with the data from the current study, should help to further elucidate how the chaperones achieve P protein activation. Limited proteolysis of DHBV RT produced in RL has already provided evidence for structural differences between non-activated, and Dε RNA containing P protein (10,11). The defined in vitro system will now, aided by the development of site-specific monoclonal anti-RT antibodies (74), allow to sequentially follow structural alterations induced by the individual chaperone systems, by the bound Dε RNA, and/or by primer synthesis. Ongoing experiments suggest, indeed, that the same basic chaperone components as required for RT activation also induce significant rearrangements in the TP domain, including a region that is crucial for Dε RNA binding (M.S., J.B., and M.N., manuscript submitted).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Harald Wegele and Johannes Buchner for providing recombinant human Hsp90 and Hop protein; David Smith, Jörg Höhfeld and Reinhild Prange for providing chaperone and co-chaperone encoding plasmids; Jianming Hu for sharing RNA gel shift protocols; Sacha Baginsky and Daniel Mayer for mass spectrometric analyses; and Kerstin Semmler and Christine Rösler for expert technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft (DFG Na 14/7-2) and is part of the activities of the VIRGIL European Network of Excellence on Antiviral Drug Resistance sponsored by the European Commission (LSHM-CT-2004-503359). Funding to pay the Open Access publication charges for this article was provided by the DFG.
Conflict of interest statement. None declared.
REFERENCES
- 1.Beck J, Nassal M. Hepatitis B virus replication. World J. Gastroenterol. 2007;13:48–64. doi: 10.3748/wjg.v13.i1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganem D, Schneider R. Hepadnaviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 4th. Vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 2923–2969. [Google Scholar]
- 3.Schultz U, Grgacic E, Nassal M. Duck hepatitis B virus: an invaluable model system for HBV infection. Adv. Virus Res. 2004;63:1–70. doi: 10.1016/S0065-3527(04)63001-6. [DOI] [PubMed] [Google Scholar]
- 4.Wang GH, Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992;71:663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- 5.Hu J, Boyer M. Hepatitis B virus reverse transcriptase and epsilon RNA sequences required for specific interaction in vitro. J. Virol. 2006;80:2141–2150. doi: 10.1128/JVI.80.5.2141-2150.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck J, Nassal M. Sequence- and structure-specific determinants in the interaction between the RNA encapsidation signal and reverse transcriptase of avian hepatitis B viruses. J. Virol. 1997;71:4971–4980. doi: 10.1128/jvi.71.7.4971-4980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck J, Nassal M. Formation of a functional hepatitis B virus replication initiation complex involves a major structural alteration in the RNA template. Mol. Cell. Biol. 1998;18:6265–6272. doi: 10.1128/mcb.18.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck J, Nassal M. Reconstitution of a functional duck hepatitis B virus replication initiation complex from separate reverse transcriptase domains expressed in Escherichia coli. J. Virol. 2001;75:7410–7419. doi: 10.1128/JVI.75.16.7410-7419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollack JR, Ganem D. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J. Virol. 1994;68:5579–5587. doi: 10.1128/jvi.68.9.5579-5587.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tavis JE, Ganem D. Evidence for activation of the hepatitis B virus polymerase by binding of its RNA template. J. Virol. 1996;70:5741–5750. doi: 10.1128/jvi.70.9.5741-5750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tavis JE, Massey B, Gong Y. The duck hepatitis B virus polymerase is activated by its RNA packaging signal, epsilon. J. Virol. 1998;72:5789–5796. doi: 10.1128/jvi.72.7.5789-5796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J, Seeger C. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl Acad. Sci. USA. 1996;93:1060–1064. doi: 10.1073/pnas.93.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu J, Toft DO, Seeger C. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 1997;16:59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 15.Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell. Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 16.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamer J, Multhaup G, Tomoyasu T, McCarty JS, Rüdiger S, Schonfeld HJ, Schirra C, Bujard H, Bukau B. A cycle of binding and release of the DnaK, DnaJ and GrpE chaperones regulates activity of the Escherichia coli heat shock transcription factor sigma32. EMBO J. 1996;15:607–617. [PMC free article] [PubMed] [Google Scholar]
- 18.Guisbert E, Herman C, Lu CZ, Gross CA. A chaperone network controls the heat shock response in E. coli. Genes Dev. 2004;18:2812–2821. doi: 10.1101/gad.1219204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer MP. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 2005;153:1–46. doi: 10.1007/s10254-004-0025-5. [DOI] [PubMed] [Google Scholar]
- 20.Fotin A, Cheng Y, Grigorieff N, Walz T, Harrison SC, Kirchhausen T. Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating. Nature. 2004;432:649–653. doi: 10.1038/nature03078. [DOI] [PubMed] [Google Scholar]
- 21.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 22.Young JC, Moarefi I, Hartl FU. Hsp90: a specialized but essential protein-folding tool. J. Cell. Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearl LH, Prodromou C. Structure and mechanism of the hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 24.Picard D. Chaperoning steroid hormone action. Trends Endocrinol. Metab. 2006;17:229–235. doi: 10.1016/j.tem.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Smith DF. Tetratricopeptide repeat cochaperones in steroid receptor complexes. Cell Stress Chaperones. 2004;9:109–121. doi: 10.1379/CSC-31.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wegele H, Wandinger SK, Schmid AB, Reinstein J, Buchner J. Substrate transfer from the chaperone Hsp70 to Hsp90. J. Mol. Biol. 2006;356:802–811. doi: 10.1016/j.jmb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Wegele H, Müller L, Buchner J. Hsp70 and Hsp90 – a relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 2004;151:1–44. doi: 10.1007/s10254-003-0021-1. [DOI] [PubMed] [Google Scholar]
- 28.Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 29.Cintron NS, Toft DO. Defining the requirements for Hsp40 and Hsp70 in the Hsp90 chaperone pathway. J. Biol. Chem. 2006;281:26235–26244. doi: 10.1074/jbc.M605417200. [DOI] [PubMed] [Google Scholar]
- 30.Riggs DL, Cox MB, Cheung-Flynn J, Prapapanich V, Carrigan PE, Smith DF. Functional specificity of co-chaperone interactions with Hsp90 client proteins. Crit. Rev. Biochem. Mol. Biol. 2004;39:279–295. doi: 10.1080/10409230490892513. [DOI] [PubMed] [Google Scholar]
- 31.Beck J, Nassal M. Efficient Hsp90-independent in vitro activation by Hsc70 and Hsp40 of duck hepatitis B virus reverse transcriptase, an assumed Hsp90 client protein. J. Biol. Chem. 2003;278:36128–36138. doi: 10.1074/jbc.M301069200. [DOI] [PubMed] [Google Scholar]
- 32.Hu J, Anselmo D. In vitro reconstitution of a functional duck hepatitis B virus reverse transcriptase: posttranslational activation by Hsp90. J. Virol. 2000;74:11447–11455. doi: 10.1128/jvi.74.24.11447-11455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu J, Toft D, Anselmo D, Wang X. In vitro reconstitution of functional hepadnavirus reverse transcriptase with cellular chaperone proteins. J. Virol. 2002;76:269–279. doi: 10.1128/JVI.76.1.269-279.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Qian X, Guo HC, Hu J. Heat shock protein 90-independent activation of truncated hepadnavirus reverse transcriptase. J. Virol. 2003;77:4471–4480. doi: 10.1128/JVI.77.8.4471-4480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brehmer D, Rüdiger S, Gässler CS, Klostermeier D, Packschies L, Reinstein J, Mayer MP, Bukau B. Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat. Struct. Biol. 2001;8:427–432. doi: 10.1038/87588. [DOI] [PubMed] [Google Scholar]
- 36.Takayama S, Reed JC. Molecular chaperone targeting and regulation by BAG family proteins. Nat. Cell. Biol. 2001;3:E237–E241. doi: 10.1038/ncb1001-e237. [DOI] [PubMed] [Google Scholar]
- 37.Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291:1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- 38.Briknarova K, Takayama S, Brive L, Havert ML, Knee DA, Velasco J, Homma S, Cabezas E, Stuart J, et al. Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein. Nat. Struct. Biol. 2001;8:349–352. doi: 10.1038/86236. [DOI] [PubMed] [Google Scholar]
- 39.Gässler CS, Wiederkehr T, Brehmer D, Bukau B, Mayer MP. Bag-1M accelerates nucleotide release for human Hsc70 and Hsp70 and can act concentration-dependent as positive and negative cofactor. J. Biol. Chem. 2001;276:32538–32544. doi: 10.1074/jbc.M105328200. [DOI] [PubMed] [Google Scholar]
- 40.Lüders J, Demand J, Papp O, Höhfeld J. Distinct isoforms of the cofactor BAG-1 differentially affect Hsc70 chaperone function. J. Biol. Chem. 2000;275:14817–14823. doi: 10.1074/jbc.275.20.14817. [DOI] [PubMed] [Google Scholar]
- 41.Kanelakis KC, Morishima Y, Dittmar KD, Galigniana MD, Takayama S, Reed JC, Pratt WB. Differential effects of the hsp70-binding protein BAG-1 on glucocorticoid receptor folding by the hsp90-based chaperone machinery. J. Biol. Chem. 1999;274:34134–34140. doi: 10.1074/jbc.274.48.34134. [DOI] [PubMed] [Google Scholar]
- 42.Takayama S, Bimston DN, Matsuzawa S, Freeman BC, Aime-Sempe C, Xie Z, Morimoto RI, Reed JC. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 1997;16:4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Höhfeld J, Jentsch S. GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohtsuka K. Cloning of a cDNA for heat-shock protein hsp40, a human homologue of bacterial DnaJ. Biochem. Biophys. Res. Commun. 1993;197:235–240. doi: 10.1006/bbrc.1993.2466. [DOI] [PubMed] [Google Scholar]
- 45.Honore B, Leffers H, Madsen P, Rasmussen HH, Vandekerckhove J, Celis JE. Molecular cloning and expression of a transformation-sensitive human protein containing the TPR motif and sharing identity to the stress-inducible yeast protein STI1. J. Biol. Chem. 1992;267:8485–8491. [PubMed] [Google Scholar]
- 46.Buchner J, Bose S, Mayr C, Jakob U. Purification and characterization of prokaryotic and eukaryotic Hsp90. Methods Enzymol. 1998;290:409–418. doi: 10.1016/s0076-6879(98)90034-9. [DOI] [PubMed] [Google Scholar]
- 47.Richter K, Reinstein J, Buchner J. N-terminal residues regulate the catalytic efficiency of the Hsp90 ATPase cycle. J. Biol. Chem. 2002;277:44905–44910. doi: 10.1074/jbc.M208457200. [DOI] [PubMed] [Google Scholar]
- 48.Richter K, Muschler P, Hainzl O, Buchner J. Coordinated ATP hydrolysis by the Hsp90 dimer. J. Biol. Chem. 2001;276:33689–33696. doi: 10.1074/jbc.M103832200. [DOI] [PubMed] [Google Scholar]
- 49.Beck J, Nassal M. In vitro reconstitution of epsilon-dependent duck hepatitis B virus replication initiation. Methods Mol. Med. 2004;95:315–325. doi: 10.1385/1-59259-669-X:315. [DOI] [PubMed] [Google Scholar]
- 50.Hu J, Flores D, Toft D, Wang X, Nguyen D. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J. Virol. 2004;78:13122–13131. doi: 10.1128/JVI.78.23.13122-13131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beck J, Vogel M, Nassal M. dNTP versus NTP discrimination by phenylalanine 451 in duck hepatitis B virus P protein indicates a common structure of the dNTP-binding pocket with other reverse transcriptases. Nucleic Acids Res. 2002;30:1679–1687. doi: 10.1093/nar/30.7.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Protzer U, Nassal M, Chiang PW, Kirschfink M, Schaller H. Interferon gene transfer by a hepatitis B virus vector efficiently suppresses wild-type virus infection. Proc. Natl Acad. Sci. USA. 1999;96:10818–10823. doi: 10.1073/pnas.96.19.10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rösler C, Köck J, Kann M, Malim MH, Blum HE, Baumert TF, von Weizsäcker F. APOBEC-mediated interference with hepadnavirus production. Hepatology. 2005;42:301–309. doi: 10.1002/hep.20801. [DOI] [PubMed] [Google Scholar]
- 54.Prodromou C, Panaretou B, Chohan S, Siligardi G, O’Brien R, Ladbury JE, Roe SM, Piper PW, Pearl LH. The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J. 2000;19:4383–4392. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grenert JP, Johnson BD, Toft DO. The importance of ATP binding and hydrolysis by hsp90 in formation and function of protein heterocomplexes. J. Biol. Chem. 1999;274:17525–17533. doi: 10.1074/jbc.274.25.17525. [DOI] [PubMed] [Google Scholar]
- 57.Johnson BD, Chadli A, Felts SJ, Bouhouche I, Catelli MG, Toft DO. Hsp90 chaperone activity requires the full-length protein and interaction among its multiple domains. J. Biol. Chem. 2000;275:32499–32507. doi: 10.1074/jbc.M005195200. [DOI] [PubMed] [Google Scholar]
- 58.Brinker A, Scheufler C, Von Der Mulbe F, Fleckenstein B, Herrmann C, Jung G, Moarefi I, Hartl FU. Ligand discrimination by TPR domains. Relevance and selectivity of EEVD-recognition in Hsp70 x Hop x Hsp90 complexes. J. Biol. Chem. 2002;277:19265–19275. doi: 10.1074/jbc.M109002200. [DOI] [PubMed] [Google Scholar]
- 59.Sondermann H, Ho AK, Listenberger LL, Siegers K, Moarefi I, Wente SR, Hartl FU, Young JC. Prediction of novel Bag-1 homologs based on structure/function analysis identifies Snl1p as an Hsp70 co-chaperone in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:33220–33227. doi: 10.1074/jbc.M204624200. [DOI] [PubMed] [Google Scholar]
- 60.Beck J, Bartos H, Nassal M. Experimental confirmation of a hepatitis B virus (HBV) epsilon-like bulge-and-loop structure in avian HBV RNA encapsidation signals. Virology. 1997;227:500–504. doi: 10.1006/viro.1996.8329. [DOI] [PubMed] [Google Scholar]
- 61.Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 62.Neckers L. Chaperoning oncogenes: Hsp90 as a target of geldanamycin. Handb. Exp. Pharmacol. 2006:259–277. doi: 10.1007/3-540-29717-0_11. [DOI] [PubMed] [Google Scholar]
- 63.Jez JM, Chen JC, Rastelli G, Stroud RM, Santi DV. Crystal structure and molecular modeling of 17-DMAG in complex with human Hsp90. Chem. Biol. 2003;10:361–368. doi: 10.1016/s1074-5521(03)00075-9. [DOI] [PubMed] [Google Scholar]
- 64.Yao E, Tavis JE. Kinetics of synthesis and turnover of the duck hepatitis B virus reverse transcriptase. J. Biol. Chem. 2003;278:1201–1205. doi: 10.1074/jbc.M208895200. [DOI] [PubMed] [Google Scholar]
- 65.Gooljarsingh LT, Fernandes C, Yan K, Zhang H, Grooms M, Johanson K, Sinnamon RH, Kirkpatrick RB, Kerrigan J, et al. A biochemical rationale for the anticancer effects of Hsp90 inhibitors: slow, tight binding inhibition by geldanamycin and its analogues. Proc. Natl Acad. Sci. USA. 2006;103:7625–7630. doi: 10.1073/pnas.0602650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Forsythe HL, Jarvis JL, Turner JW, Elmore LW, Holt SE. Stable association of hsp90 and p23, but Not hsp70, with active human telomerase. J. Biol. Chem. 2001;276:15571–15574. doi: 10.1074/jbc.C100055200. [DOI] [PubMed] [Google Scholar]
- 67.Keppler BR, Grady AT, Jarstfer MB. The biochemical role of the heat shock protein 90 chaperone complex in establishing human telomerase activity. J. Biol. Chem. 2006;281:19840–19848. doi: 10.1074/jbc.M511067200. [DOI] [PubMed] [Google Scholar]
- 68.Toogun OA, Zeiger W, Freeman BC. The p23 molecular chaperone promotes functional telomerase complexes through DNA dissociation. Proc. Natl Acad. Sci. USA. 2007;104:5765–5770. doi: 10.1073/pnas.0701442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 70.Kamal A, Thao L, Sensintaffar J, Zhang L, Boehm MF, Fritz LC, Burrows FJ. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 71.Maloney A, Clarke PA, Naaby-Hansen S, Stein R, Koopman JO, Akpan A, Yang A, Zvelebil M, Cramer R, et al. Gene and protein expression profiling of human ovarian cancer cells treated with the heat shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2007;67:3239–3253. doi: 10.1158/0008-5472.CAN-06-2968. [DOI] [PubMed] [Google Scholar]
- 72.Neckers L. Heat shock protein 90: the cancer chaperone. J. Biosci. 2007;32:517–530. doi: 10.1007/s12038-007-0051-y. [DOI] [PubMed] [Google Scholar]
- 73.Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cao F, Badtke MP, Metzger LM, Yao E, Adeyemo B, Gong Y, Tavis JE. Identification of an essential molecular contact point on the duck hepatitis B virus reverse transcriptase. J. Virol. 2005;79:10164–10170. doi: 10.1128/JVI.79.16.10164-10170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bimston D, Song J, Winchester D, Takayama S, Reed JC, Morimoto RI. BAG-1, a negative regulator of Hsp70 chaperone activity, uncouples nucleotide hydrolysis from substrate release. EMBO J. 1998;17:6871–6878. doi: 10.1093/emboj/17.23.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Terada K, Mori M. Human DnaJ homologs dj2 and dj3, and bag-1 are positive cochaperones of hsc70. J. Biol. Chem. 2000;275:24728–24734. doi: 10.1074/jbc.M002021200. [DOI] [PubMed] [Google Scholar]
- 77.Kamtekar S, Berman AJ, Wang J, Lazaro JM, de Vega M, Blanco L, Salas M, Steitz TA. The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition. EMBO J. 2006;25:1335–1343. doi: 10.1038/sj.emboj.7601027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.