Abstract
Bloom syndrome (BS) is an autosomal recessive disorder characterized by genomic instability and the early development of many types of cancer. Missense mutations have been identified in the BLM gene (encoding a RecQ helicase) in affected individuals, but the molecular mechanism and the structural basis of the effects of these mutations remain to be elucidated. We analysed five disease-causing missense mutations that are localized in the BLM helicase core region: Q672R, I841T, C878R, G891E and C901Y. The disease-causing mutants had low ATPase and helicase activities but their ATP binding abilities were normal, except for Q672, whose ATP binding activity was lower than that of the intact BLM helicase. Mutants C878R, mapping near motif IV, and G891E and C901Y, mapping in motif IV, displayed severe DNA-binding defects. We used molecular modelling to analyse these mutations. Our work provides insights into the molecular basis of BLM pathology, and reveals structural elements implicated in coupling DNA binding to ATP hydrolysis and DNA unwinding. Our findings will help to explain the mechanism underlying BLM catalysis and interpreting new BLM causing mutations identified in the future.
INTRODUCTION
DNA helicases are important in DNA metabolism and are involved in genome replication, DNA repair, recombination, transcription and telomere maintenance (1,2). Helicases function as molecular motors. They use the energy from nucleotide triphosphate (NTP) hydrolysis to translocate along a nucleic acid strand, and separate complementary strands of a nucleic acid duplex (3). The conversion of the energy derived from NTP hydrolysis into unwinding of double-stranded nucleic acids is coordinated by seven sequence motifs (I, Ia, II, III, IV, V and VI); these sequence motifs are features of superfamily-1 and -2 helicases (4). Motif 0, N-terminal to motif I, is an additional motif that is also conserved in proteins of the RecQ family (5). A similar element called a ‘Q motif’ has also been characterized in DEAD-box RNA helicases where it is important for NTP binding and hydrolysis (6). Crystal structures of a catalytic core fragment of Escherichia coli RecQ helicase show that motifs 0, I and II are important in ATP binding and hydrolysis by RecQ helicases (7). Studies of SF-1 helicases (PcrA and Rep) have identified several highly conserved residues in the seven motifs as being involved directly or indirectly in linking DNA binding to ATP hydrolysis. The crystal structures of HCV, PcrA, Rep and DEAD-box protein Vasa revealed that motifs Ia, IV and V bind the phosphate backbone of DNA (8–11). Several highly conserved residues in motif IV, including two Arg residues and one Asn residue in PcrA and Rep, have been observed to interact directly with ds or ssDNA (10,12). Although no crystal structure of the RecQ helicase–DNA complex is available, it is widely thought that RecQ helicases bind DNA in a similar manner to other helicases (SF-1 and SF-2). Most RecQ family proteins have a conserved RecQ-Ct domain immediately downstream from the conserved seven signature motifs that is unique to the RecQ family helicases (13). The RecQ-Ct subdomain, as detailed in the E. coli RecQ crystal structure, contains a platform of alpha helices with four conserved cysteine residues that bind a Zn2+ ion and winged helix motif (7). The helicase and RecQ-Ct domains combine to form the catalytic ‘helicase core’ domain of E. coli RecQ, containing the sequence motifs necessary for its ATPase and DNA unwinding activities.
The RecQ family of DNA helicases has received much attention in the past few years because mutations in three RecQ helicases are associated with genome instability and cancer susceptibility, and give rise to the human disorders: Bloom syndrome (BS) (14), Werner syndrome (WRN) (15) and Rothmund–Thomson syndrome (16,17).
Bloom syndrome is a rare autosomal recessive genetic disorder. It is characterized by growth deficiency, unusual facies, immunodeficiency, male sterility/female subfertility and an increased risk of a broad spectrum of cancers that develop at a young age. Although Werner syndrome and Rothmund–Thomson syndrome individuals also develop cancers, BS is the only RecQ-related disorder that causes cancers of the types observed in elderly general populations (18). Cells from BS patients exhibit chromosomal instability characterized by elevated rates of sister chromatid exchanges (SCEs), loss of heterozygosity and quadriradials (19). BS cells exhibit hyper-recombination and abnormalities in DNA replication involving an extended S phase and accumulation of an abnormal profile of replication intermediates. Thus, the BLM protein is expected to play a central role in one or more DNA metabolic pathways (20).
BLM, the gene mutated in BS, encodes a 1417 amino acid protein which has amino acid sequence similarities with the RecQ family of DNA helicases (Figure 1) (14). The natural DNA substrates of BLM in the cell have not been identified. However, in vitro studies demonstrate that BLM unwinds the canonical Watson–Crick duplex, and recognizes and disrupts alternative DNA structures including Holliday junctions, triple helices and the highly stable G-quadruplex (21–25). In addition to its DNA unwinding activity, BLM displays a strand annealing activity (26). BLM's helicase activity is necessary for the correction of the genomic instability of BLM cells (27). Like other RecQ family helicases, BLM interacts physically and functionally with numerous proteins in the cell to perform its diverse functions (20).
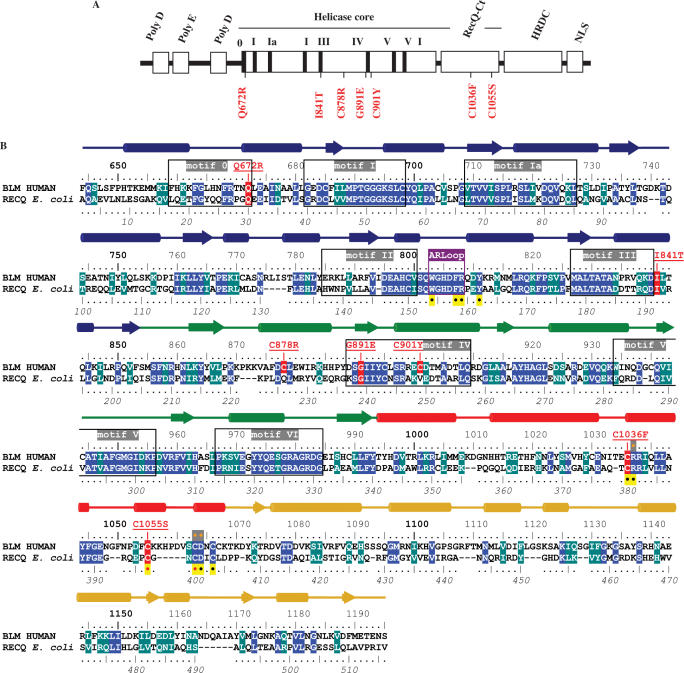
Figure 1.
(A) Schematic presentation of distribution of the identified mutant alleles in BLM patients (red). Domain organization of the BLM protein is shown above the mutations. (B) Mutation map of helicase catalytic cores of BLM and RecQ from E. coli. Alignment was performed using CLUSTALW and refined manually. Conserved residues are highlighted in blue and similar residues are highlighted in marine green. The secondary structure elements of RecQ from E. coli are indicated with alpha-helices appearing as cylinders and strands as arrows. Lobes 1 and 2 of the helicase domain are respectively coloured blue and green, the zinc binding domain red and the WH domain gold. The seven conserved helicase motifs are boxed and labelled in grey. The conserved aromatic-rich loop sequence is boxed in purple under the label ARLoop. The upper line numbering indicates residue positions in the sequence of BLM and the underside line numbering, in italics, refers to the sequence of E. coli RecQ. Sites of all the missense mutations causing Bloom syndrome are highlighted in red with the observed mutations underlined in red above the sequence. Other mutation sites tested in BLM are indicated by an orange point inside a grey box. All the residues that have been mutated and studied to date in E. coli RecQ are indicated by a black point in a yellow box.
Various disease-linked mutations have been identified in the BLM gene. The most frequent mutations include stop codons, insertions and deletions that generate frameshift mutations and missense mutations. Seven disease-causing missense mutations have been described: Q672R, I841T, C878R, G891E, C901Y, C1036F and C1055S (14,28–31). Five of these mutations have been mapped to the seven sequence motifs that are conserved (Figure 1A). The seminal work of the Keck laboratory on the atomic structure of RecQ helicase has provided new insight into the structural consequences of some missense mutants (7). Biochemical and structural characterizations of the mutations C1036F and C1055S, localized in the RecQ-Ct domain, have shown that both mutations affect sites directly involved in the formation and stability of the zinc binding domain; the zinc binding domain plays an essential role in protein folding and DNA substrate recognition and discrimination (32). Q672R and I841T, mutations localized in the helicase core domain, have been characterized using partially purified enzymes (27,33): both mutants have been shown to be deficient in ATPase and helicase activities, but the molecular basis of their mutagenic effects remains to be determined. In this study, we analysed the five disease-causing mutations found in the helicase core, using molecular modelling and biochemical and biophysical approaches. Studies of these mutants help elucidate the possible molecular bases of the BLM disease-causing mutations, and also to provide information on the conserved mechanisms employed by the RecQ family helicases.
MATERIALS AND METHODS
Plasmid construction and site-directed mutagenesis
A plasmid for producing the BLM helicase core consisting of amino acid residues 642–1290 was generated by inserting the corresponding gene between the NdeI and XhoI sites of the expression plasmid pET15b (Novagen). The resulting plasmid, pET-BLM642–1290, was used as the target for site-directed mutagenesis. All point mutations were constructed by ‘splicing by overlap extension’ as described (34) with the desired mutations in the internal mutagenic primers (Table 1). To avoid undesired mutations, PCR fragments were sequenced by the dideoxy method performed by MWG (MWG Biotech, Germany).
Table 1.
Sequences of oligonucleotides used for site-directed mutagenesis and DNA substrates
| BLM | Recombinant or mutagenic PCR primer (5′-3′) | |
|---|---|---|
| WT642–1290 | F- GGAATTCATATGGAGCGTTTCCAAAGTCTTAGTTTTCCT R- CCGCTCGAGTTACGATGTCCATTCAGAGTATTTCTGTAA | |
| Q672R | F- TTTAGAACTAATCGGCTAGAGGCGATC R- GATCGCCTCTAGCCGATTAGTTCTAAA | |
| I841T | F- GTACAGAAGGACACCCTGACTCAGCTG R- CAGCTGAGTCAGGGTGTCCTTCTGTAC | |
| C878R | F- AAGGTGGCATTTGATCGCCTAGAATGGATCAGA R- TCTGATCCATTCTAGGCGATCAAATGCCACCTT | |
| G891E | F- CCATACGATTCAGAGATAATTTACTGC R- GCAGTAAATTATCTCTGAATCGTATGG | |
| C901Y | F- TCCAGGCGAGAATATGACACCATGGCT R- AGCCATGGTGTCATATTCTCGCCTGGA |
| DNA substrate | Length | DNA substrate sequence |
|---|---|---|
| HJ1 | 40 | CCGTGATCACCAATGCAGATTGACGAACCTTTGCCCACGT |
| HJ2 | 40 | ACGTGGGCAAAGGTTCGTCAATGGACTGACAGCTGCATGG |
| HJ3 | 40 | CCATGCAGCTGTCAGTCCATTGTCATGCTAGGCCTACTGC |
| HJ4 | 40 | GCAGTAGGCCTAGCATGACAATCTGCATTGGTGATCACGG |
| A | 44 | GCACTGGCCGTCGTTTTACGGTCGTGACTGGGAAAACCCTGGCG |
| B | 45 | TTTTTTTTTTTTTTTTTTTTTTAGCCGTAAAACGACGGCCAGTGC |
| F21 | 21 | Fluo- GGGTTAGGGTTAGGGTTAGGG |
Protein production and purification
All proteins (wild type and mutants) were purified by the following protocol. A single colony of the E. coli strain (BL21(DE3)-condonplus) producing the protein was grown overnight in 10 ml of LB containing 80 µg/ml ampicillin and 34 µg/ml chloramphenicol at 37°C. An aliquot of 0.1 ml of this culture was diluted into 1 l of pre-warmed LB, and the cells were grown to the mid-exponential phase (A600 of 0.5–0.6) at 37°C. Protein production was induced by the addition of isopropyl-1-thio-α-d-galactopyranoside to a final concentration of 0.25 mM, and the culture was incubated with shaking at 18°C for 18 h. The cells were harvested by centrifugation and suspended in a final volume of 25 ml of the lysis buffer (50 mM Tris–HCl, pH 7.5, 500 mM NaCl, 0.1% Triton X-100, PMSF 0.1 µM and 10% glycerol). Cells were lysed by passage through a French pressure cell and the samples were sonicated to reduce viscosity. To remove any insoluble materials, the cell lysate was centrifuged twice at 15 000 g for 45 min. The soluble extract was applied to a column containing 20 ml of Ni-NTA resin (Qiagen), and the subsequent purification procedures were performed with FPLC system (ÄKTA Purifier) at 18°C. The column was washed with lysis buffer until the UV absorbance at 280 nm became stable. Bound proteins were eluted with 300 ml linear gradient of imidazole (0.02–0.4 M). Fractions containing the proteins were identified by SDS-polyacrylamide gel electrophoresis. Pooled fractions were concentrated and further purified by FPLC size exclusion chromatography (Superdex 200, Amersham Bioscience). The purified proteins were electrophoresed on SDS-polyacrylamide gels and visualized with Coomassie Brilliant Blue. The concentrations of the purified proteins were determined by the Bio-Rad dye method using bovine serum albumin as the standard.
DNA substrates preparation
PAGE-purified oligonucleotides listed in Table 1 were purchased from Proligo (France). Duplex DNA substrates were prepared as described previously (32). Briefly, 250 µM DNA substrates were denatured in 1× TE containing 1 M NaCl or 1 M KCl by heating at 95°C for 10 min. The denatured DNA was then annealed at 37°C for 48 h. The annealed products were separated by 8% non-denaturing PAGE with buffer containing 10 mM KCl and at 4°C for 12 h with constant current of 20 mA.
ATPase assay
ATPase activity was assayed by measuring the release of free phosphate during ATP hydrolysis (35,36). The reaction was carried out in ATPase reaction buffer (50 mM Tris–HCl, pH 8.0, 3 mM MgCl2, 0.5 mM DTT) at 37°C in a volume of 100 µl. The reactions were initiated by the addition of enzymes into a reaction mixture containing 0.5 µM ssDNA (nt, 60-mer oligonucleotide) and the indicated concentration of ATP. The reaction was stopped by transferring 80 µl aliquots from the reaction mixture every 30 s into a hydrochloric solution of ammonium molybdate. The liberated radioactive γ32Pi was extracted with a solution of 2-butanol:benzene:acetone:ammonium molybdate (750:750:15:1) saturated with water. An aliquot (60 µl) was removed from the organic phase and the radioactivity was quantified using a liquid scintillation counter.
Helicase assay
Radiometric assay
DNA helicase reactions were carried out at 37°C in reaction mixtures containing 25 mM HEPES-NaOH, pH 7.5, 25 mM CH3CO2Na, 7.5 mM (CH3CO2)2Mg, 2 mM ATP, 1 mM DTT, 0.1 mg/ml BSA and the appropriate 32P-labelled partial duplex DNA substrate (10 fmol, 3000 c.p.m./fmol). Reactions were initiated by addition of the indicated concentration of BLM proteins and incubated at 37°C for 30 min. Reactions were terminated by the addition of 5 µl of 5 × loading buffer (50 mM EDTA, 0.5% SDS, 0.1% xylene cyanol, 0.1% bromophenol blue and 50% glycerol). The products of helicase reactions were resolved on 12% (w/v) polyacrylamide gel (acylamide to bis-acrylamide ratios 19:1). The gel was run in TBE buffer (90 mM Tris, 90 mM boric acid and 1 mM EDTA, pH 8.3) at 100 V for 2 h at 4°C.
Fluorometric assay
Stopped-flow DNA-unwinding assays were performed as described (37). Briefly, a Bio-logic SFM-400 mixer with a 1.5 mm × 1.5 mm cell (Bio-Logic, FC-15) and a Bio-Logic MOS450/AF-CD optical system equipped with a 150-W mercury-xenon lamp were used. They were performed in two-syringe mode, where helicase and duplex DNA substrates were preincubated in syringe 1 for 5 min and ATP in syringe 4. Each syringe contained unwinding reaction buffer (25mM Tris–HCl, pH 7.5 at 25°C, 50 mM NaCl, 1 mM MgCl2 and 0.1 mM DTT) and the unwinding reaction was initiated by rapid mixing. The sequences of the two strands, labelled with hexachlorofluorescein (H) and fluorescein (F), respectively, of the 56:16-mer DNA substrates were 5′H-AATCCGTCGAGCAGAG(dT40)-3′ and 3′F-TTAGGCAGCTCGTCTC-5′. To convert the output data from volts to percent unwinding, another experiment was performed in the four-syringe mode, where helicase in syringe 1, H-labelled ss oligonucleotides in syringe 2 and F-labelled ss oligonucleotides in syringe 3 were incubated in unwinding reaction buffer, the solution in syringe 4 being the same as in the above unwinding experiment. The fluorescent signal of the mixed solution from the four syringes was used to define 100% unwinding. The standard reaction temperature was 25°C and all concentrations listed were after mixing unless noted otherwise. Data were fitted with Equation (1)
| 1 |
where A1 (A2) and kobs,1 (kobs,2) represent, respectively, the unwinding amplitude and rate of the fast (slow) phase.
DNA binding assay
Electrophoretic mobility shift assay
Binding reactions (20 µl) were conducted in standard binding buffer (40 mM Tris–HCl, pH 7.0, 1 mM EDTA, 20 mM NaCl, 8% glycerol and 20 µg/ml bovine serum albumin). Protein and DNA substrate concentrations are indicated in the figure legends. Reactions were incubated for 30 min at room temperature. Non-denaturing loading dye (4 µl of 0.25% bromphenol blue in 30% glycerol) was added to reaction mixes and the samples were loaded onto 6% non-denaturating polyacrylamide gels (19:1). Electrophoresis was carried out at a constant voltage of 14 V/cm at 4°C in 1× TAE (40 mM Tris acetate, 1 mM EDTA, pH 8.0) for 3 h. The gels were dried and processed for autoradiography.
Fluoresence polarization assay
DNA binding was studied by fluorescence polarization as described previously (37). The assays were performed using a Bio-logic auto-titrator (TCU-250) and a Bio-Logic optical system (MOS450/AF-CD) in fluorescence anisotropy mode. Various amounts of proteins were added to 1 ml of binding buffer containing 1 nM DNA substrate. Each sample was allowed to equilibrate in solution for 1.5 min, and fluorescence polarization was then measured. Titrations were performed in a temperature-controlled cuvette at 25°C. The solution was stirred continuously by a small magnetic stir bar throughout the titration process. The binding isotherms were determined and fit by Equation (2):
| 2 |
where A is the fluorescence anisotropy at a given concentration of RecQ, Amax is the anisotropy at saturation, Amin is the initial anisotropy, Δ = DT + NPT + KD, DT is the total concentration of DNA, PT is the concentration of the enzyme in the binding solution and KD is the apparent dissociation constant.
ATP binding assay
The ATP binding affinity of BLM642–1290 and the mutants was measured by nitrocellulose filter binding as described in reference (38). The assays were performed at 4°C with a fixed amount of ATP and various concentrations of the proteins. Nitrocellulose filters (25 mm) were washed with 0.5 M NaOH for 10 min, rinsed with double-distilled water, and then equilibrated in wash buffer (40 mM Tris–HCl, pH 7.5/10 mM MgCl2/50 mM potassium glutamate). The proteins (from 5 to 30 μM) were mixed with ATP (200 μM) and [γ-32P]ATP in the absence of DNA in 40 mM Tris–HCl (pH 7.5)/10 mM MgCl2/10 mM DTT/50 mM potassium glutamate/10% glycerol in a total volume of 20 μl. The reaction mixtures were incubated for 30 min on ice, and 15 μl aliquots were filtered through nitrocellulose filters. The membranes were washed twice with 2 ml of ice-cold wash buffer. The radioactivity bound to the nitrocellulose membrane was measured in a liquid scintillation counter. The stoichiometry of ATP binding was calculated from the radioactivity count.
Strand annealing assay
DNA strand annealing activity was measured according to Cheok et al. with some modifications (26). Briefly, the assay was performed using fully complementary oligonucleotides, one of which was 5′−32P-end-labelled. Reactions were carried out in a reaction buffer (20 µl) containing 20 mM Tris–acetate, pH 7.9, 50 mM KOAc, 10 mM Mg(OAc)2, 1 mM DTT, 50 µg/ml BSA and the indicated protein concentration. The reaction was initiated by adding the unlabelled oligonucleotide, immediately followed by incubation at 37°C for 15 min and was stopped by the addition of stop buffer (50 mM EDTA, 1% SDS and 0.1 mg/ml of proteinase K). The resulting DNA products were analysed as described for the helicase assays.
RESULTS
Structural model of the BLM helicase core-DNA complex
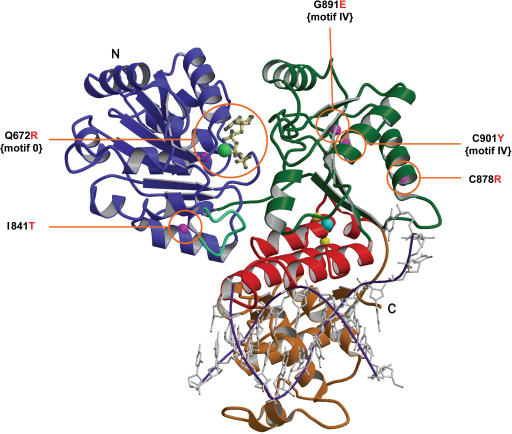
We have previously modelled the atomic structure of the BLM helicase core of the apo-BLM enzyme. We further modelled the 3D structure of the BLM helicase core by homology modelling using both amino acid sequence alignment (Figure 1B), and the template structure of E. coli RecQ helicase in complex with ATP-γ-S (7). Bernstein and colleagues compared several 3D structures of helicases in complexes with ATP/DNA, and the E. coli RecQ atomic structure. Using these comparisons, they proposed a model for the RecQ-DNA complex: (i) dsDNA is bound in a depression formed between the zinc finger and WH domains and (ii) ssDNA is bound both to the top of the RecA-like lobe 1, where ATP also binds, and lobe 2 (7,39). This DNA binding topology is different from that of SF1 helicases PcrA and Rep (10,12). The recently published structure of the HRDC domain of E. coli RecQ, and the study of the aromatic-rich loop of RecQ mutants are consistent with this model (40). We introduced a heteroduplex DNA into the BLM structure, made adjustments to our structure using Bernstein and colleagues’ model for DNA binding to E. coli RecQ, and obtained a model of BLM in complex with DNA and ATP (Figure 2). Most of the disease-linked mutations are localized in or near the ATP and DNA binding sites (Figure 2). The structural environment of each mutated residue is indicated in Figure 3. The molecular structure of BLM–DNA complex provides an excellent model for elucidating the biophysical basis of known BLM-causing mutations.
Figure 2.
Mutation sites in the modelled BLM structure. Ribbon drawing of the BLM helicase catalytic core, obtained after refinement of the structure generated by the Modeller computation (see text for details) with a trace of a partially unwound DNA molecule in a possible orientation, shown in purple and grey. This model is used to consider possible relationships between structure and enzyme activities. The N-terminal domains 1A (lobe 1) and 2A (lobe 2) are respectively coloured blue and green, the zinc binding domain is red, and the WH domain, gold. The conserved aromatic-rich loop is shown in blue-green. Labels N and C refer to the N-terminus and C-terminus, respectively. ATPγS is drawn in cream with a ball-and-stick representation, the manganese ion is shown as a light green ball and the zinc ion in cyan. Magenta spheres are the Cα positions of the five mutations studied in this article and observed in patients suffering from Bloom syndrome. Yellow spheres are the Cα positions of the two mutations in the zinc binding domain studied previously.
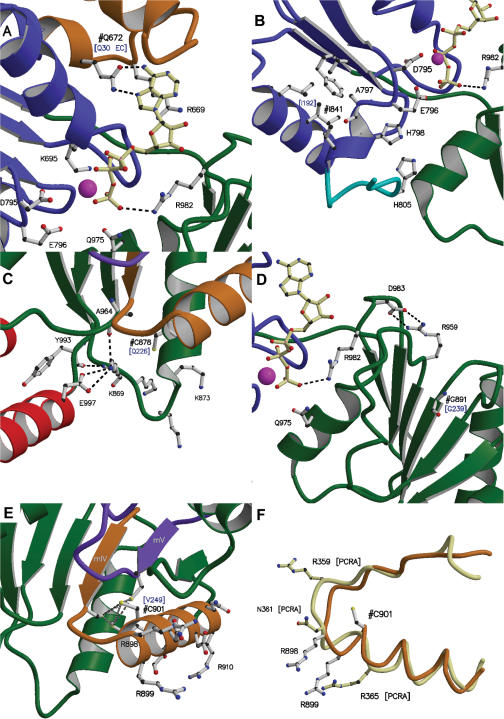
Figure 3.
Environments of the five studied Bloom Syndrome mutation sites in the BLM structure model. For all panels of the figure, domains 1A and 2A are in blue and green, respectively, and the zinc binding domain is in red. ATPγS is drawn as a ball-and-stick representation in cream, the manganese ion is shown as a magenta ball and the conserved aromatic-rich loop is in cyan. The conserved motifs are coloured as follows: motif 0 is drawn in orange, motif IV in gold and motif V in purple. The mutated residues are labelled ‘#’ and the one letter codes and positions of the analogous residues in RecQ from E. coli are given in blue and in brackets. Possible interactions between neighbour atoms are indicated with dashed lines. (A) Detailed view of the environment of the mutation site Q672 and the ATP-binding site. Residues believed to be involved in ATP binding or hydrolysis are indicated. (B) Detailed view of the environment of I841. The conserved hydrophobic sites and the ATP binding site are shown. (C) Site of C878 in the BLM structure model. Interactions between conserved residues, all involving LYS 869, are indicated as dashed lines. (D) Detailed view of the environment of G891. The major interactions between the conserved residues R959 and D983 and between R982 and the phosphate moiety of ATP are shown as dashed lines. (E) Detailed view of the environment of C901. Binding distances for the sulphur atoms of the cluster (C895, C901, M904 and C944) are indicated with dashed lines. Conserved charged or polar residues in motif IV are also indicated. (F) Comparison of motif IV in PcrA (cream) with that in BLM (gold). PcrA residues are highlighted in cream. Residue numbering for PcrA is indicated by the label ‘[PCRA]’. A least squares comparison gave a RMSD of 1.7 Å for 75 atoms superimposed.
Design, over-production and purification of BLM mutant proteins
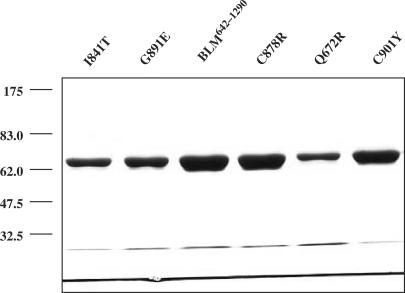
The recombinant helicase core of BLM protein made up of residues 642–1290 (BLM642–1290) displays similar biochemical properties, both in vitro and in vivo, to that of the full length BLM protein (41). The BLM642–1290 fragment, similar to the full-length protein, is a DNA-dependent ATPase and an ATP-dependent DNA helicase that displays a 3′-5′ polarity. The BLM642–1290 fragment efficiently unwinds Holliday junctions and suppresses spontaneous and UV-induced illegitimate recombination in E. coli (41). We used the BLM642–1290 protein fragment to study the functional consequences of the five disease-causing mutations (Q672R, I841T, C878R, G891E and C901Y). The mutants were produced by site-directed mutagenesis, and the mutated residues were identical to those identified in BS patients. The part of the BLM gene corresponding to amino acid residues 642–1290 (BLM642–1290), and the mutated genes, containing the five mutations, were expressed in an E. coli pET expression system. The recombinant proteins were purified using Ni-agarose affinity and gel filtration chromatography. All mutants and the BLM642–1290 fragment were over-produced and purified to homogeneity. The mutant protein preparations were between 90 and 95% pure (Figure 4). Mutant Q672R displayed a high level of aggregation, even in the presence of the chemical chaperone betaine (42) (data not shown). This suggests that Q672R is involved in protein folding (see below).
Figure 4.
SDS-PAGE analysis of the purified BLM642–1290 and various mutants. The proteins (indicated above the figure) were resolved in 10% SDS-polyacrylamide gel and stained with Coomassie blue. The amounts of the proteins used were 3.5 µg for BLM, C878R and C901Y and 1.5 µg for I841T, G891E and Q672R. The positions of marker proteins (in kDa) are indicated on the left.
Helicase activity assay
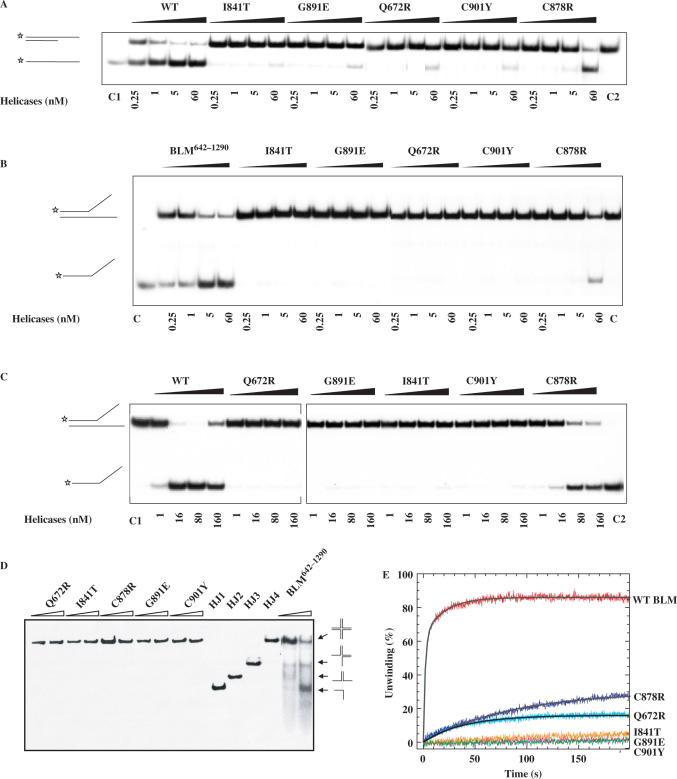
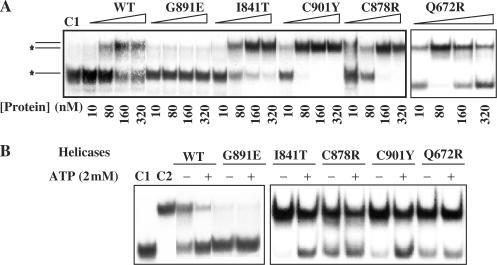
Wild type and mutants of the BLM642–1290 fragment were tested for their helicase activities by electrophoresis mobility-shift assay. Mutant C878R exhibited a detectable helicase activity but only at high protein concentrations (Figure 5A). All other mutants displayed no significant DNA unwinding activity at the protein concentrations used in this study (Figure 5A). As BLM helicase unwinds forked duplex DNA more efficiently than partial duplex DNA, we characterized the DNA unwinding activity of the five mutants with forked duplex DNA substrates. BLM642–1290 unwound forked DNA efficiently. C878R unwound DNA only at high protein concentrations, and the other mutants did not unwind partial duplex DNA (Figure 5A and B). We studied the DNA unwinding activity of these mutants with various protein concentrations. Under similar experimental conditions, only C878R was able to unwind DNA to similar levels as the BLM642–1290 protein fragment, and this required a 10-fold greater protein concentration (Figure 5C). This suggests that the mechanisms of impairment of DNA unwinding differ between C878R, which retain intrinsic DNA unwinding ability under certain conditions, and the other mutants. Previous studies have shown that the BLM can act in concert with topoisomerase IIIα to resolve recombination intermediates containing double Holliday junctions (21,24), a potential physiological DNA substrate of BLM. Since BLM helicase activity is required for dissolution of the double Holliday junction structure, we then compared the Holliday junction resolution activity of wild type and mutant BLM642–1290 proteins. Figure 5D shows that the intact BLM642–1290 was able to disrupt four-way junctions to the component single strands, whereas all the mutant BLM proteins failed to do so.
Figure 5.
DNA unwinding activity of wild-type and mutant BLM proteins. (A−C) Helicase activities revealed by radiometric assay using duplex DNA and forked DNA. 5′-32P-labelled DNA substrate (1 ng) was incubated at 37°C with wild-type and the various mutants in helicase assay buffer. The reactions were terminated after 30 min and the samples were analysed by electrophoresis in a 12% non-denaturing polyacrylamide gel. The concentrations of the proteins were between 0.25 and 60 nM as indicated at the bottom of the gel panel, where C1 and C2 represent labelled ssDNA and partial duplex DNA substrate, respectively. (D) Unwinding of the Holliday junction by BLM642–1290 and the all mutants. Reaction mixtures (20 µl) in unwinding buffer contain 1 nM DNA substrate, 1 mM ATP and the indicated amounts of the proteins. The four-way DNA junction was constructed from oligonucleotides HJ1, HJ2, HJ3 and HJ4. Reactions were carried out and analysed as described for A. HJ1, HJ2, HJ3 and HJ4 represent single-stranded, two-stranded, three-stranded and four-way junction substrate, respectively. (E) DNA unwinding activity by stopped-flow DNA unwinding assay. As described in the ‘Material and Methods’ section, 1 nM 56:16-mer DNA substrate was pre-incubated with 20 nM helicase for 5 min at 25°C. The unwinding reaction was initiated by mixing with 1 mM ATP. The fluorescence emission of fluorescein at 520 nm (excited at 492 nm) was monitored. The fraction of DNA unwound was obtained by normalization of the fluorescence signal. The solid curves are the best fit of the data to Equation (1) and the values of A1 (A2) and kobs,1 (kobs,2) are given in Table 2.
To measure the unwinding amplitude, and the unwinding rate, we used rapid stopped-flow fluorescence assays. The assays were based on fluorescence resonance energy transfer (FRET) (43) to measure the helicase activity of BLM642–1290 and BLM mutants under similar experimental conditions. Multiple turnover kinetic studies were performed with all the mutants and BLM642–1290. BLM642–1290 -catalyzed unwinding of a 16 bp duplex was biphasic, and we could best fit time courses using the sum of two exponential terms (Figure 5E). The observed rate constants of the fast and slow phases were 0.52 and 0.05 s−1, respectively, for BLM. Under similar experimental conditions, mutants E841T, G891E and C901Y did not unwind DNA, whereas Q672R clearly displayed helicase activity. C878R displayed DNA unwinding activity at high protein concentrations, consistent with results obtained by radiometric assay (Figure 5E and Table 2).
Table 2.
ATPase, helicase, DNA binding and ATP binding activities of the wild-type and mutant BLM proteins
| Helicases | ATPase | Helicase | DNA binding | ATP binding | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Vm | KM | kcat | kcat/KM | A1 | kobs,1 | A2 | kobs,2 | ssDNA | dsDNA | [ATP]/[Protein] | |
| µM·min−1 | µM | s−1 | s−1·mM−1 | % | s−1 | % | s−1 | nM | nM | ||
| BLM642–1290 | 164.2 ± 9.3 | 80.7 ± 2.0 | 13.7 | 169.5 | 62.8 ± 0.6 | 0.52 ± 0.01 | 23.2 ± 0.6 | 0.05 ± 0.01 | 7.2 ± 0.4 | 9.8 ± 1.1 | 1.02 ± 0.23 |
| Q672R | 127.6 ± 11.4 | 1755 ± 127 | 10.6 | 6.0 | 16.2 ± 0.1 | 0.03 ± 0.01 | 10.9 ± 0.8 | 15.3 ± 1.9 | 0.22 ± 0.04 | ||
| I841T | 39.8 ± 7.4 | NDa | 3.3 | ND | 12.1 ± 1.2 | 14.8 ± 1.5 | 1.01 ± 0.15 | ||||
| C878R | 196.5 ± 11.5 | 3460 ± 693 | 16.3 | 4.7 | 30.2 ± 0.1b | 0.01 ± 0.01 | 168 ± 10 | 327 ± 27 | 0.97 ± 0.23 | ||
| G891E | 4.6 ± 0.9 | ND | 0.38 | ND | 352 ± 12 | 349 ± 12 | 0.98 ± 0.14 | ||||
| C901Y | 5.8 ± 0.8 | ND | 0.48 | ND | 475 ± 25 | 122 ± 18 | 0.89 ± 0.27 | ||||
aND: cannot be determined precisely. The ATP saturation curves of the BLM642–1290 and its corresponding variant were fitted by Michaelis–Menten, Lineweaver–Burk and Eadie–Hofstee equations. The ATP saturation curves of BLM642–1290, Q672R and C878R were fitted well by the three equations, yielding similar Vm and Km values, whatever the fitting procedure. In contrast, for poorly active mutants, I841, G891E and C901Y, the Lineweaver–Burk and Eadie–Hofstee plots were not linear. For these mutants, only the Vm values were precisely recovered by fitting with the Michaelis–Menten equation.
bData obtained with 150 nM protein.
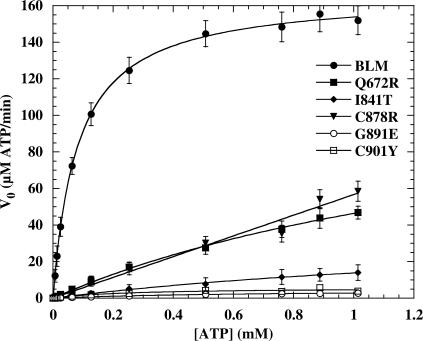
ATPase activity assay
BLM is a DNA-stimulated ATPase and ATP-dependent helicase, so we next investigate the biochemical basis for the reduced helicase activity of each of the mutant proteins by assaying for the various sub-activities that together constitute the overall DNA unwinding activity. The ATPase activities of the five disease-causing BLM mutant proteins were measured as described in ‘Materials and Methods’ section. The rate constant (kcat), KM values and the ATPase catalytic efficiencies (kcat/KM) for intrinsic and DNA-stimulated ATPase activity were assayed by varying the concentration of ATP substrate. The resulting curves were fitted using the Michaelis–Menten equation (Figure 6, Table 2). We did not detect ATPase activity in mutant proteins in the absence of a DNA cofactor (data not shown). The kcat values of the G891E and C901Y were 36-fold lower than that of BLM642–1290, indicating that for both mutant proteins, the turnover rates for ATP hydrolysis were significantly lower. The kcat values of Q672R, I841T and C878R were between 3.3 and 16.3 s−1 (Table 2). However, their KM values were higher than that of BLM642–1290 (Table 2). The ATPase activity of the BLM protein is stimulated by DNA. Thus, the observed low apparent kcat values of these mutant proteins may have resulted from a low affinity for either ATP or DNA. To determine the structural basis for the lower ATPase activity of these mutants, ATP binding affinities of BLM642–1290 and mutant proteins were qualitatively assayed by nitrocellulose filter binding. All mutant proteins bound ATP with similar stoichiometry to that of BLM642–1290, except for Q672R (Table 2). Most mutant proteins bound ATP normally, but were unable to hydrolyze ATP efficiently. Thus, the low levels of ATP hydrolysis by other mutant proteins cannot be due to a failure of ATP binding. Q672R had a significantly poorer ATP binding activity (Table 2), suggesting that Q672 is involved in ATP binding.
Figure 6.
ATP hydrolysis activity of the wild-type and mutant BLMs as a function of ATP concentration. Experiments were performed in ATPase assay buffer at 37°C with 0.5 µM ssDNA (nt, 60-mer oligonucleotide) and 0.1 or 0.2 µM of each enzyme. The ATP hydrolysis was quantified as described in ‘Material and Methods’ section. Solid lines represent the best fit of the data to the Michaelis–Menten equation. The apparent kcat and KM values obtained are summarized in Table 2.
DNA binding assay
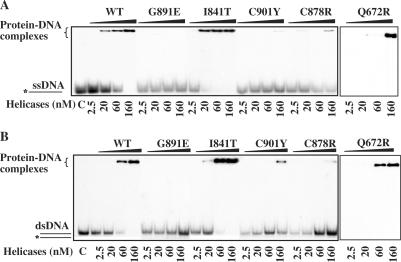
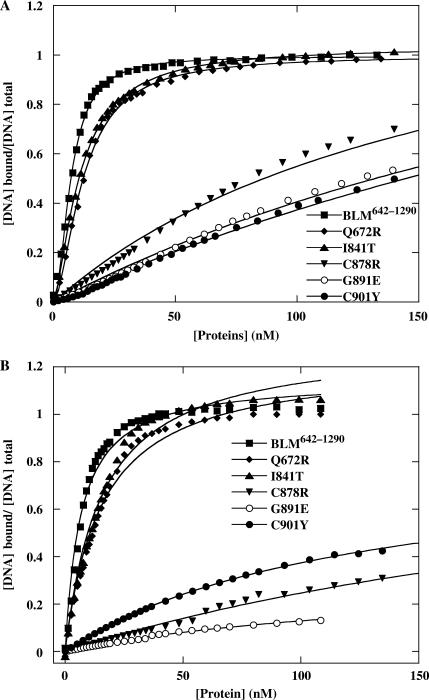
Helicases are DNA-stimulated ATPases, so we investigated the DNA binding ability of these mutants to determine whether the lower ATPase and helicase activities were a result of failure to bind DNA. We used electrophoretic mobility-shift assays to probe the ability of BLM proteins to bind ssDNA and dsDNA. Mutants Q672R and I841T bound ssDNA and dsDNA with affinities similar to those of BLM642–1290. DNA binding by C878R, G891E and C901Y was significantly weaker than that by BLM642–1290 (Figure 7). C901Y had a lower affinity for ssDNA than for dsDNA, but C878R had a lower affinity for dsDNA than for ssDNA (Figure 7). The difference in ssDNA and dsDNA binding for C901Y and C878R may reflect intrinsic energetic properties of these mutants for DNA binding. To confirm the differences in ssDNA and dsDNA binding by these mutants and measure their dissociation constants for DNA binding, we used a fluorescence anisotropy assay: we measured the apparent Kd values under equilibrium conditions (44). We titrated fluorescein-labelled 36-mer ssDNA and dsDNA with various concentrations of BLM642–1290 and mutant proteins. The resulting binding isotherms were fitted with Equation (2) (Figure 8). The apparent dissociation constants determined from the titration curves are consistent with the results obtained with the radiometric assay. The binding affinity of C901Y for ssDNA was significantly lower than that for dsDNA with dissociation constants of 475 nM for ssDNA and 122 nM for dsDNA. The mutant protein C878R had significantly lower dsDNA binding than ssDNA binding. Mutants Q672R and I841T displayed DNA binding affinity similar to that of the BLM642–1290.
Figure 7.
Analysis of DNA binding activity of the intact and mutant BLMs using electrophoretic mobility shift assay. One nanomole of 5′-labelled ssDNA or dsDNA was incubated at room temperature for 20 min with 2.5 to 160 nM protein in unwinding buffer. Bound and free DNA were separated by electrophoresis through a non-denaturing 15% polyacrylamide and revealed by autoradiography.
Figure 8.
The anisotropy-based DNA binding isotherms of wild-type and mutant BLM proteins. Fluorescence anisotropy values were determined as a function of BLM concentration for 36 mer ssDNA (A) and dsDNA (B) substrates. A total of 5 nM fluorescein-labelled DNA was titrated with increasing amounts of BLM as described in the ‘Materials and Methods’ section. The solid lines represent the best fits of the data to the Equation (2). The apparent Kd, values are summarized in Table 2.
Strand annealing assay
Recently, a novel strand annealing activity of BLM was reported and it has been suggested to play an important role in replication fork regression (26). To compare the BLM-mediated strand annealing activity between the BLM642–1290 and mutant proteins, we incubated two partially complementary single strand oligonucleotides with increasing concentrations of purified proteins and analysed the products on native polyacrylamide gels. The results in Figure 9A showed that BLM642–1290 and most missense mutants, except for G891E, displayed significant strand annealing activity at protein concentrations above 80 nM. More interestingly, some mutants such as C901Y and Q672R exhibited stronger annealing activity than that of BLM642–1290. In accordance with the previous studies, the strand annealing activities of the wild-type and the mutants proteins are partially inhibited by ATP (Figure 9B), except Q672R. This is consistent with the fact that Q672R is deficient in ATP binding.
Figure 9.
Strand annealing activity of BLM642–1290 and mutant proteins. (A) Strand annealing activity of BLM proteins was assayed by the formation of a 23 bp forked duplex. Two partially complementary oligonucleotides (0.5 nM oligonucleotide A and B), one of which was radiolabelled at the 5′-end and the other was unlabelled, were incubated with the indicated concentrations of BLM protein for 15 min at 37°C in the helicase reaction buffer in the absence of ATP. The reaction products were separated by 12% non-denaturing PAGE and revealed by autoradiography. (B) Effect of nucleotides on the BLM-mediated ssDNA annealing. The reactions contained 100 nM proteins and were carried under the same conditions as in A. ATP concentration was 2 mM, as indicated in the left of the figure.
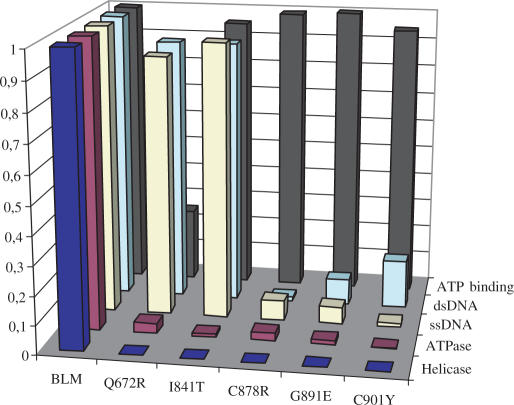
Thus, we have characterized the enzymatic activities of all the mutants. ATPase, helicase, ATP binding and DNA binding activities of these proteins were expressed as ratios relative to the wild-type enzyme in Figure 10.
Figure 10.
Graphic representation of the relative enzymatic activity of BLM mutants. Data points are the means of two to three experiments.
DISCUSSION
In this study, we describe the molecular consequences of five mutations that cause Bloom syndrome. Combination of the biochemical study and the molecular modelling approaches improve our understanding of the molecular basis of how these mutations affect BLM enzymatic functions, thereby perturbing the integrity and stability of chromosomes. These analyses have also provided new insight into how the RecQ family helicases couple DNA binding to ATP hydrolysis and DNA unwinding, and how this coupling is impaired in the BLM misense mutants.
Residue Q672 is involved in ATP binding
The BLM missense mutation, Q672R, is naturally occurring and causes Bloom's syndrome, and has been mapped to motif 0 (14). Several additional mutations found in various species have indicated that this residue is crucial for enzyme function. The E. coli RecQ in complex with ATPγS structure shows that motif 0 forms a loop connecting two helices creating a pocket that accommodates the adenine base of the ATPγS molecule. OE1 and NE2 atoms of the C-terminal Gln residue 672 (Q672 in BLM is equivalent to Q30 in E. coli RecQ) of motif 0 form hydrogen bonds with the N6 and N7 atoms of the adenine, specifically binding ATP. Consistent with the previous results obtained with the partial purified full length BLM (27,33), our data indicates that Q672R displays normal DNA binding activity, but its ATPase activity is significantly impaired. This implicates Q672 in either ATP binding or hydrolysis. The KM and kcat values obtained from the ATP saturation curves show that the Q672R turnover rate for ATP hydrolysis is similar to that of the BLM642–1290, but that its ATP binding affinity is significantly lower. Our direct ATP binding experiment confirms that the ATP binding ability of Q672R is dramatically decreased. These findings are consistent with the data from the crystal structure of E. coli RecQ in complex with ATPγS (7) and our molecular modelling structure of the BLM helicase core (Figures 2 and 3A). The Q672R mutant protein tended to aggregate. This suggests that either residue Q672 or ATP binding is important in protein folding as the mutant protein Q672R is deficient in ATP binding.
Residue I841 contributes to stabilize the residues involved in ATP hydrolysis
Motifs I and II (Walker A and B motifs, respectively) are highly conserved in SF-1 and SF-2 helicases, and have residues that interact with MgATP/MgADP. Our molecular BLM model, consistent with the crystal structures of PcrA, Rep and E. coli RecQ helicases, shows that the carboxyl of the highly conserved D795 coordinates the Mg2+ ion of Mg2+-ATP through outer sphere interactions, and that E796 may act as a catalytic base in ATP hydrolysis (Figure 3B). Residues D795, E796, A797 and H798 map in a loop connecting a β-strand and a α-helix in our model. The spatial positions of the crucial residues D795 and E796 are further stabilized by a conserved hydrophobic pocket involving residues A797, and I841 (Figure 3B). Replacement of residue I with T in position 841 inserts a polar group that may disrupt the coherence of this pocket, and affect the interactions involving residues D795, E796, A797 and H798 in motif II (Figure 3B). These structural modifications may have direct consequences on the orientation of motif II with regard to the γ-phosphate of ATP. The modifications may disturb the position of E796, (equivalent to E147 in RecQ from E. coli) which acts as the catalytic base that polarizes a water molecule for attack on the γ-phosphate of ATP. The modifications may also affect the position of D795 (equivalent to D146 in RecQ from E. coli) that coordinates the catalytic ion Mg2+ (7). Thus, the mutation I841T may have an effect on the positions of D795 and E796, which are both predicted to be directly involved in ATP binding and hydrolysis. This is in agreement with our biochemical data showing that this mutation impairs the ATPase and helicase activities whereas DNA and ATP binding activities remain unchanged.
Residue C878 may connect the zinc binding motif and lobe 2, and to enhance DNA binding
The C878R mutant can unwind partial and forked duplex DNA at high protein concentration and displays low, but detectable ATPase activity. However, its DNA binding ability is substantially below that of the BLM62–1290 fragment. The modelled BLM structure reveals two structural features related to C878 (Figure 3C): (i) residue C878 is close to the hydrogen bonds between K869 and D997, and its sulphur atom is 5.22 Å from the side chain of K869. The hydrogen bond interactions between K869 and D997 are highly conserved from bacteria to human, and may contribute to the stabilization of the zinc binding motif relative to the lobe 2 of the helicase domain, thereby stabilizing DNA binding. (ii) C878 is also part of an α-helix connected to a lysine-rich loop (residues 869–873), which is unique to BLM. The lysine-rich loop869–873, comprising four lysine residues and a proline, constitutes a flexible and positively charged surface and is totally accessible to solvent. This loop is very close to the dsDNA binding sites in both the E. coli RecQ helicase model and our BLM model, and may contribute to DNA binding and substrate specificity. The mutation C878R, involving a very large and very positively charged residue, may interfere with the K869–D997 hydrogen bond linking the zinc binding motif to lobe 2, and thus displace the zinc binding motif from lobe 2, thereby affecting DNA binding. Indeed, mutant C878R displays a detectable unwinding activity at high protein concentration indicating that its intrinsic ATPase and helicase activities are not abolished, but that a high protein concentration is required to enhance DNA binding. We also measured ATPase activity of C878R with increasing DNA concentration as additional indication of the ability of the mutant enzyme to bind DNA. We found that the mutant C878R needs 12-fold higher ssDNA concentration to achieve the same extent of ATPase stimulation by the BLM642–1290 (data not shown). These various findings suggest that residue C878 may link the zinc binding motif and lobe 2 to enhance DNA binding. The BLM protein preferentially unwinds or migrates recombination intermediates, including Holliday junctions and D loops; possibly, the molecular basis for the pathological effects of the C878R mutant is that it fails to recognize, bind and unwind its substrate in cells during the DNA transaction process.
Residue G891 is involved in coupling DNA binding and ATP hydrolysis
The ATPase and DNA helicase activities and DNA binding are severely compromised in G891R although its ATP binding activity appears essentially normal. This implicates G891 in the coupling of DNA binding and ATP hydrolysis. Residue G891 is distant from the ATP binding/hydrolysis sites, so it is reasonable to assume that the mutation G891R causes long-distance structural modifications, thereby affecting ATP hydrolysis. Residue G891 of motif IV contributes to a hydrophobic site with several conserved residues that constitute parallel-strand beta-sheets (Figure 3D). These hydrophobic interactions probably greatly stabilize the spatial position of beta strand 12. Residue R959 is on top of the beta strand 12 and forms a salt bridge with D983 which may spatially restrict the position of the arginine residue 982, the only residue of lobe 2 expected to be involved in ATP hydrolysis (Figure 3D) (7,45). Replacement of the small apolar glycine residue 891 with a large and positively charged arginine residue undoubtedly destabilizes the local conformation near position 891, particularly beta strand 12 where R959 is implicated in stabilization of the putative arginine finger. R959 is only 5 Å from residue G891, so the side chain of the mutated 891 may directly disturb R959 through charge repulsion. As a consequence, R959 may not be properly oriented in the mutant to interact with D983.
Replacement of G891 with an arginine not only impairs ATP hydrolysis and DNA unwinding activities, but also results in DNA binding defects. At first glance, it is surprising that G891 in motif IV contributes to DNA binding. However, the crystal structures of Rep bound to ssDNA, PcrA bound to a hybrid DNA with both single-stranded and duplex segments, HCV helicase bound to ssDNA and DEAD-box helicase Vasa in complex with ssRNA indicate that motifs Ia and IV contribute substantially to the specific interactions with oligonucleotides. Several conserved residues in motif IV, including Asn, Arg and Lys, are directly involved in oligonucleotide binding (Table 3). No crystal structure of RecQ helicase in complex with DNA is available, so it is hard to ascertain whether G891 interacts directly with the oligonucleotide or stabilizes the relevant β strands through hydrophobic interactions, thereby ensuring a suitable conformation for DNA binding (Figure 3D). In the light of our findings, it is reasonable to suppose that G891 contributes to coupling DNA binding to ATP hydrolysis. DNA binding may result in a subtle change in the conformation of several β strands around G891 and in particular β strand 12. R959 is on top of the β strand 12 and interacts with D983. The spatial orientation of R959 may be altered upon DNA binding, affecting the arginine finger position through the D983–R959 salt bridge and activating ATP hydrolysis.
Table 3.
Helicase motif IV is specifically involved in oligonucleotide binding
| Residue numbers in Motif IV | Sequences | Interactions |
|---|---|---|
| PcrA (355–365) | AVLYRTNAQSR | R359 binds DNA and forms a salt bridge to E600; N361 interacts with ssDNA and R365 close to dsDNA |
| Rep (346–356) | AILYRGNHQSR | R350, N352, H353 and R356 interact with ssDNA |
| RecQ (237–258) | KSGIIYCNSRAKVEDTAAR | R246 equivalent to R365 of PcrA |
| BLM (898–907) | DSGIILCLSRRECDTMADT | R898 and R899 equivalent to N361 and R365 of PcrA, respectively |
| HCV(365–372) | LIFCHSKK | K371 interacts with ssDNA |
| DEAD-box Vasa (493–500) | IVFVETKR | E497 and K499 interact with ssRNA |
This possibility is in good agreement with our biochemical data showing that this mutation impairs ATPase, DNA helicase and DNA binding activities. As the mutation does not significantly disturb ATP binding, the loss of helicase activity is presumably the consequence of a coupling between DNA binding and ATP hydrolysis.
Residue C901 stabilizes the α-helix of motif IV which is probably involved in DNA binding
Residue C901 is in the central position of motif IV which is composed by a strand connected to an α-helix (Figure 3E). Crystal structures of several helicases in complex with oligonucleotides have shown that some conserved residues in motif IV are involved in DNA binding (Table 3). PcrA and BLM display a similar fold of motif IV. Residues R898 and R899 (equivalent to N361 and R365 from PcrA) of motif IV (Figure 3F) may play a key role in ssDNA binding (Figure 3E). Moreover, the α-helix of motif IV may swing as a rigid body due to thermodynamic movement (Figure 3E). Such a mobile α-helix may be unfavourable for DNA binding and must be further restricted to a spatial position appropriate for DNA binding. In our BLM molecular model, residue C901 is associated with an intriguing cluster of 3 cysteines and 1 methionine (C895, C901, M904 and C944). The distances between the cysteine or methionine side-chain sulphur groups are all 3.4 Å and close to the ideal distance (2.35 ± 0.09 Å) for structural metal-coordination sites (46). It is possible that the α-helices of the motif IV are maintained in the spatial orientation appropriate for DNA binding by this metal-binding motif including C901. Preliminary biochemical characterization indicates that Zn2+ is not involved in the formation of the putative metal-binding motif. The relevance of this putative metal-binding motif and the nature of the metal are currently under investigation in our laboratory. Alternatively, C901 may form disulfide bridges with one of its neighbour cysteines to stabilize the α-helix of motif IV.
In mutant C901Y a medium-sized polar cysteine residue (valine in RecQ from E. coli) is replaced with a large and planar tyrosine residue with an OH group. This is expected to disrupt the metal-binding motif or the putative disulfide bridges and consequently destabilize the helix of motif IV carrying residues that may be involved directly or indirectly in DNA binding. This model predicts that the residues in the helix are important for ssDNA binding, contributing to the formation of the BLM–DNA complex. Consistent with the model, mutant C901Y exhibited significant reduction in dsDNA binding and even greater loss of ssDNA binding (Figures 7 and 8).
The C901 involved in the putative metal-binding motif is not a conserved residue in RecQ family helicases. ATPase and helicase activities are common to all DNA helicases, so a fundamental question is how deficiency of a particular helicase gives rise to the characteristic biochemical, cellular, genetic and organismal consequences observed in helicase mutants. It is plausible that each helicase recognizes its own substrate in the cell: the structural feature involved in motif IV in BLM may have a physiological relevance in DNA substrate specificity recognition.
Although Cheok and colleagues (26) did not observe the strand annealing activity with BLM642–1290, we did detect the strand annealing activity with BLM642–1290 at protein concentrations above 80 nM (Figure 9). More interestingly, some mutants displayed stronger strand annealing activity than the intact BLM642–1290 fragment, demonstrating that the DNA unwinding and the strand annealing activities could be uncoupled. The same phenomenon was also observed in RECQL4 protein (47). The observations that the missense mutants lost helicase activity, but still possess the strand annealing activity indicated that the helicase activity or its coordination with the strand annealing is essential for the integrity and stability of chromosomes.
ACKNOWLEDGEMENTS
This research was supported by the grant from the Institut National du Cancer (France) to M.A.-G. and X.G.X., the National Natural Science Foundation of China, and the Innovation Project of the Chinese Academy of Sciences. Part of this work was performed by R.B.G., P.R., H.R. and X.G.X. in CNRS 8113, ENS de Cachan, France. We thank Dr Eric Deprez for helpful discussion. Funding to pay the Open Access publication charges for this article was provided by CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 2.Soultanas P, Wigley DB. DNA helicases: ‘inching forward’. Curr. Opin. Struct. Biol. 2000;10:124–128. doi: 10.1016/s0959-440x(99)00059-7. [DOI] [PubMed] [Google Scholar]
- 3.von Hippel PH, Delagoutte E. A general model for nucleic acid helicases and their ‘coupling’ within macromolecular machines. Cell. 2001;104:177–190. doi: 10.1016/s0092-8674(01)00203-3. [DOI] [PubMed] [Google Scholar]
- 4.Gorbalenya AE, Koonin EV. helicases: amino acid sequence comparisons and structure-function relationship. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 5.Bernstein DA, Keck JL. Domain mapping of Escherichia coli RecQ defines the roles of conserved N- and C-terminal regions in the RecQ family. Nucleic Acids Res. 2003;31:2778–2785. doi: 10.1093/nar/gkg376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanner NK, Cordin O, Banroques J, Doere M, Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E.coli RecQ helicase catalytic core. EMBO J. 2003;22:4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao N, Hesson T, Cable M, Hong Z, Kwong AD, Le HV, Weber PC. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 9.Subramanya HS, Bird LE, Brannigan JA, Wigley DB. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 10.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 11.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 12.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 13.Morozov V, Mushegian AR, Koonin EV, Bork P. A putative nucleic acid-binding domain in Bloom's and Werner's syndrome helicases. Trends Biochem. Sci. 1997;22:417–418. doi: 10.1016/s0968-0004(97)01128-6. [DOI] [PubMed] [Google Scholar]
- 14.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 15.Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 16.Vennos EM, Collins M, James WD. Rothmund-Thomson syndrome: review of the world literature. J. Am. Acad. Dermatol. 1992;27:750–762. doi: 10.1016/0190-9622(92)70249-f. [DOI] [PubMed] [Google Scholar]
- 17.Vennos EM, James WD. Rothmund-Thomson syndrome. Dermatol. Clin. 1995;13:143–150. [PubMed] [Google Scholar]
- 18.Amor-Gueret M. Bloom syndrome, genomic instability and cancer: the SOS-like hypothesis. Cancer Lett. 2006;236:1–12. doi: 10.1016/j.canlet.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 19.Mohaghegh P, Hickson ID. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum. Mol. Genet. 2001;10:741–746. doi: 10.1093/hmg/10.7.741. [DOI] [PubMed] [Google Scholar]
- 20.Hickson ID. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 21.Mohaghegh P, Karow JK, Brosh JR, Jr, Bohr VA, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karow JK, Chakraverty RK, Hickson ID. The Bloom's syndrome gene product is a 3′-5′ DNA helicase. J. Biol. Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 23.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 24.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 25.Brosh RM, Jr, Majumdar A, Desai S, Hickson ID, Bohr VA, Seidman MM. Unwinding of a DNA triple helix by the Werner and Bloom syndrome helicases. J. Biol. Chem. 2001;276:3024–3030. doi: 10.1074/jbc.M006784200. [DOI] [PubMed] [Google Scholar]
- 26.Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID. The Bloom's syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005;33:3932–3941. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neff NF, Ellis NA, Ye TZ, Noonan J, Huang K, Sanz M, Proytcheva M. The DNA helicase activity of BLM is necessary for the correction of the genomic instability of bloom syndrome cells. Mol. Biol. Cell. 1999;10:665–676. doi: 10.1091/mbc.10.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foucault F, Vaury C, Barakat A, Thibout D, Planchon P, Jaulin C, Praz F, mor-Gueret M. Characterization of a new BLM mutation associated with a topoisomerase II alpha defect in a patient with Bloom's syndrome. Hum. Mol. Genet. 1997;6:1427–1434. doi: 10.1093/hmg/6.9.1427. [DOI] [PubMed] [Google Scholar]
- 29.Barakat A, Ababou M, Onclercq R, Dutertre S, Chadli E, Hda N, Benslimane A, mor-Gueret M. Identification of a novel BLM missense mutation (2706T>C) in a Moroccan patient with Bloom's syndrome. Hum. Mutat. 2000;15:584–585. doi: 10.1002/1098-1004(200006)15:6<584::AID-HUMU28>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 30.Rong SB, Valiaho J, Vihinen M. Structural basis of Bloom syndrome (BS) causing mutations in the BLM helicase domain. Mol. Med. 2000;6:155–164. [PMC free article] [PubMed] [Google Scholar]
- 31.German J, Sanz MM, Ciocci S, Ye TZ, Ellis NA. Syndrome-causing mutations of the BLM gene in persons in the Bloom's syndrome registry. Hum. Mutat. 2007;28:743–753. doi: 10.1002/humu.20501. [DOI] [PubMed] [Google Scholar]
- 32.Guo RB, Rigolet P, Zargarian L, Fermandjian S, Xi XG. Structural and functional characterizations reveal the importance of a zinc binding domain in Bloom's syndrome helicase. Nucleic Acids Res. 2005;33:3109–3124. doi: 10.1093/nar/gki619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bahr A, De GF, Kedinger C, Chatton B. Point mutations causing Bloom's syndrome abolish ATPase and DNA helicase activities of the BLM protein. Oncogene. 1998;17:2565–2571. doi: 10.1038/sj.onc.1202389. [DOI] [PubMed] [Google Scholar]
- 34.McPherson MJ, Quirke P, Taylor GR. PCR: A Practical Approach. 1992. IRL, Oxford, pp. 207–209. [Google Scholar]
- 35.Avron M. Photophosphorylation by swiss-chard chloroplasts. Biochim. Biophys. Acta. 1960;40:257–272. doi: 10.1016/0006-3002(60)91350-0. [DOI] [PubMed] [Google Scholar]
- 36.Xu HQ, Deprez E, Zhang AH, Tauc P, Ladjimi MM, Brochon JC, Auclair C, Xi XG. The Escherichia coli RecQ helicase functions as a monomer. J. Biol. Chem. 2003;278:34925–34933. doi: 10.1074/jbc.M303581200. [DOI] [PubMed] [Google Scholar]
- 37.Zhang XD, Dou SX, Xie P, Hu JS, Wang PY, Xi XG. Escherichia coli RecQ is a rapid, efficient, and monomeric helicase. J. Biol. Chem. 2006;281:12655–12663. doi: 10.1074/jbc.M513089200. [DOI] [PubMed] [Google Scholar]
- 38.Crampton DJ, Guo S, Johnson DE, Richardson CC. The arginine finger of bacteriophage T7 gene 4 helicase: role in energy coupling. Proc. Natl Acad. Sci. USA. 2004;101:4373–4378. doi: 10.1073/pnas.0400968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zittel MC, Keck JL. Coupling DNA-binding and ATP hydrolysis in Escherichia coli RecQ: role of a highly conserved aromatic-rich sequence. Nucleic Acids Res. 2005;33:6982–6991. doi: 10.1093/nar/gki999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernstein DA, Keck JL. Conferring substrate specificity to DNA helicases: role of the RecQ HRDC domain. Structure. 2005;13:1173–1182. doi: 10.1016/j.str.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 41.Janscak P, Garcia PL, Hamburger F, Makuta Y, Shiraishi K, Imai Y, Ikeda H, Bickle TA. Characterization and mutational analysis of the RecQ core of the bloom syndrome protein. J. Mol. Biol. 2003;330:29–42. doi: 10.1016/s0022-2836(03)00534-5. [DOI] [PubMed] [Google Scholar]
- 42.Blackwell JR, Horgan R. A novel strategy for production of a highly expressed recombinant protein in an active form. FEBS Lett. 1991;295:10–12. doi: 10.1016/0014-5793(91)81372-f. [DOI] [PubMed] [Google Scholar]
- 43.Zhang XD, Dou SX, Xie P, Wang PY, Xi XG. RecQ helicase-catalyzed DNA unwinding detected by fluorescence resonance energy transfer. Acta Biochim. Biophys. Sin. (Shanghai) 2005;37:593–600. doi: 10.1111/j.1745-7270.2005.00084.x. [DOI] [PubMed] [Google Scholar]
- 44.Dou SX, Wang PY, Xu HQ, Xi XG. The DNA binding properties of the Escherichia coli RecQ helicase. J. Biol. Chem. 2004;279:6354–6363. doi: 10.1074/jbc.M311272200. [DOI] [PubMed] [Google Scholar]
- 45.Killoran MP, Keck JL. Sit down, relax and unwind: structural insights into RecQ helicase mechanisms. Nucleic Acids Res. 2006;34:4098–4105. doi: 10.1093/nar/gkl538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alberts IL, Nadassy K, Wodak SJ. Analysis of zinc binding sites in protein crystal structures. Protein Sci. 1998;7:1700–1716. doi: 10.1002/pro.5560070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst.) 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]