Abstract
Herpesviruses characteristically transmit infection from immune hosts. Although their success in escaping neutralization by pre-formed antibody is indisputable, the underlying molecular mechanisms remain largely unknown. Glycoprotein B (gB) is the most conserved component of the herpesvirus entry machinery and its N terminus (gB-NT) is a common neutralization target. We used murid herpesvirus-4 to determine how gB-NT contributes to the virus–antibody interaction. Deleting gB-NT had no obvious impact on virus replication, but paradoxically increased virion neutralization by immune sera. This reflected greater antibody access to neutralization epitopes on gH/gL, with which gB was associated. gB-NT itself was variably protected against antibody by O-linked glycans; on virions from epithelial cells it was protected almost completely. gB-NT therefore provides a protective and largely protected cover for a vulnerable part of gH/gL. The conservation of predicted glycosylation sites in other mammalian herpesvirus gB-NTs suggests that this evasion mechanism is widespread. Interestingly, the gB-NT glycans that blocked antibody binding could be targeted for neutralization instead by a lectin, suggesting a means of therapeutic counterattack.
Keywords: antibody, glycoprotein, immune evasion, virus
Introduction
Neutralizing antibody is the sine qua non of anti-viral vaccines (Zinkernagel and Hengartner, 2006). Persistent viruses present a major challenge to vaccine development because they have evolved to coexist with antibody. Developing new strategies to control their spread means understanding why neutralization normally fails (Burton et al, 2005). Herpesviruses are among the most sophisticated of all persistent viruses and provide a template for understanding some of the limits viruses impose on antibody function. Once a herpesvirus has entered its host, latency and cell–cell spread (Peeters et al, 1993; Dingwell et al, 1994) offer few opportunities for neutralization; antibody must act instead through mechanisms such as cytotoxicity (Sissons and Oldstone, 1980). α- and β-Herpesviruses counteract this with viral IgG Fc receptors (Johnson and Feenstra, 1987; Nagashunmugam et al, 1998; Atalay et al, 2002). γ-Herpesviruses may not need to because their host colonization depends more on latency-associated lymphoproliferation than on lytic spread (Stevenson et al, 1999; Coleman et al, 2003). In contrast to cell–cell spread within hosts, herpesviruses transmit between hosts as cell-free virions. These are potentially vulnerable to neutralization. However, herpesviruses enter and exit immune hosts (Sitki-Green et al, 2003) without even much selection of antigenic variants (Xu et al, 1996). It is difficult to know exactly how much antibody each virion encounters, but antibody is abundant in the mucosal sites from where γ-herpesviruses are shed (Yao et al, 1985), and any antigen excess would presumably just elicit more antibody. An antibody excess therefore seems likely. Other mucosal infections make it clear that even quite low antibody amounts can significantly reduce infectivity if neutralization is efficient (Mozdzanowska et al, 2003). γ-Herpes virions must therefore resist neutralization.
Reconciling reports of in vitro γ-herpesvirus neutralization (Thorley-Lawson and Poodry, 1982; Stevenson and Doherty, 1998; Dialyna et al, 2004) with the evident lack of in vivo neutralization raises two important issues. First, neutralization aimed at cell binding may be cell type-specific. Thus, the Epstein–Barr virus gp350 is a neutralization target for B-cell infection (Thorley-Lawson and Poodry, 1982) but not for epithelial infection (Janz et al, 2000), which gp350-specific antibodies even promote (Turk et al, 2006). Similarly, immune sera block fibroblast binding by murid herpesvirus-4 (MuHV-4) (Gill et al, 2006), but fail to block and even enhance its infection of IgG Fc receptor-bearing cells (Rosa et al, 2007). Second, neutralization often reflects reduced rather than ablated infectivity. The requirements for each may be qualitatively distinct, for example if not all the copies of a virion glycoprotein are equally susceptible to antibody binding.
MuHV-4 provides a means to identify key, common themes in γ-herpesvirus antibody evasion. Robust neutralization generally targets essential virion proteins, and MuHV-4 is no exception: its only mAb-defined neutralization targets are glycoprotein B (gB) and gH/gL (Gill et al, 2006; Gillet et al, 2006). Both are conserved in all mammalian herpesviruses and essential for infectivity (Forrester et al, 1992; Heldwein et al, 2006). The gB N terminus (gB-NT) is a neutralization target for many herpesviruses, including MuHV-4 (Ohlin et al, 1993; Holloway et al, 1998; Akula et al, 2002; Gillet et al, 2006; Okazaki et al, 2006). The basis for this neutralization is not clear. The herpes simplex virus gB-NT has a non-essential heparin-binding function (Laquerre et al, 1998) and the Kaposi's Sarcoma-associated herpesvirus gB-NT binds to integrins (Akula et al, 2002), but gB-NT-directed MuHV-4 neutralization blocks infection at a post-binding step close to membrane fusion (Gillet et al, 2006). To understand how gB-NT contributes to the virus–antibody interaction, we have deleted it from MuHV-4.
Results
gB-NT is non-essential for MuHV-4 infection
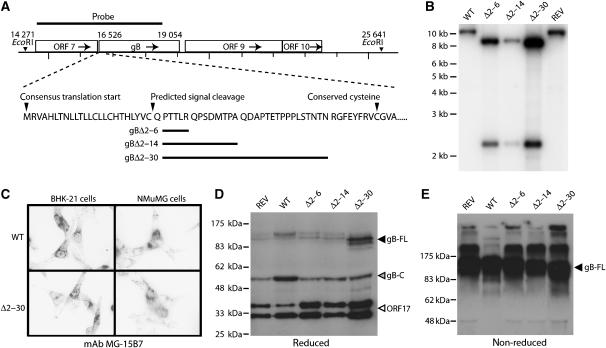
To establish the functional importance of gB-NT, we introduced one of three deletions, removing amino-acid residues 2–6, 2–14 or 2–30 after the predicted MuHV-4 gB signal sequence (Figure 1A). (Its first conserved cysteine is residue 39.) All these mutants retained infectivity. Southern blots confirmed their predicted genomic structures (Figure 1B). Immunofluorescence showed no obvious effect of gB-NT deletion on gB expression in infected cells (Figure 1C) and immunoblots established that the gB content of each mutant was comparable to that of wild-type virus (Figure 1D and E).
Figure 1.
Generation of MuHV-4 mutants lacking gB-NT. (A) The indicated regions of the gB coding sequence were removed and replaced with an EcoRI site. (B) Southern blotting showed the predicted introduction of an EcoRI restriction site for each deletion mutant (gBΔ2–6, gBΔ2–14, gBΔ2–30). REV, revertant of the gBΔ2–30 mutant. (C) Immunofluorescence of cells infected with the largest deletion mutant (gBΔ2–30) showed no effect on gB expression in either BHK-21 fibroblasts or NMuMG epithelial cells. gB-specific staining is black in this reversed image. Similar staining was observed with the gBΔ2–6 and gBΔ2–14 mutants. (D) Immunoblot of wild-type (WT), gBΔ2–6, gBΔ2–14 and gBΔ2–30 virions with the gB-C-specific mAb MG-4D11. REV, revertant of the 2–30 deletion mutant; gB-FL, uncleaved gB; gB-C, C-terminal half of furin-cleaved gB (Lopes et al, 2004), which contains the MG-4D11 epitope. The ORF17-specific mAb 150-7D1 was included in the same blot to give a loading control. The two ORF17 bands correspond to a post-translational cleavage, as described for the homologous herpes simplex virus UL26. (E) Unreduced gB immunoblotted with mAb MG-4D11 shows a single major band (gB-FL) since the furin-cleaved gB fragments are held together by disulfide bonds. Higher molecular weight forms are presumably gB multimers or aggregates.
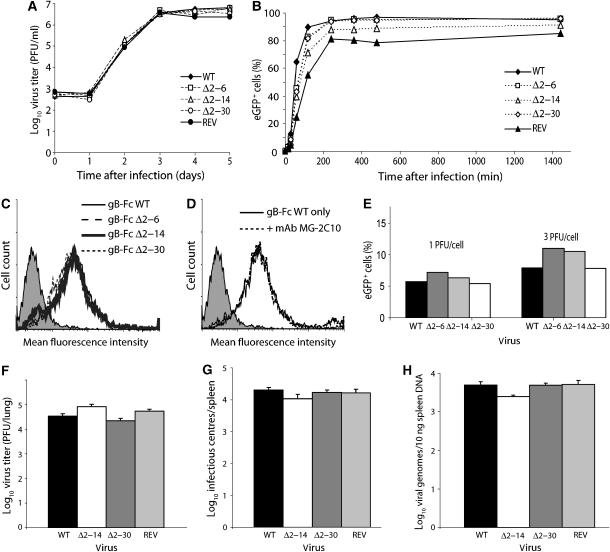
Growth curves (Figure 2A) established that gB-NT was not required for MuHV-4 lytic propagation. Nor did any of the mutants show reduced cell binding (Figure 2B). gB is a cell-binding protein of MuHV-4 (Gillet et al, 2007a), but gB-NT deletions did not affect cell binding by the N-terminal half of gB fused to IgG Fc (Figure 2C). Nor did a gB-NT-specific neutralizing mAb block cell binding by gB-Fc (Figure 2D), consistent with the idea that these mAbs do not act by blocking binding (Gillet et al, 2006). We considered that gB-NT might instead be important for B-cell infection. However, the gB-NT deletion mutants infected NS0 myeloma cells much like wild-type MuHV-4 (Figure 2E). The 2–14 and 2–30 deletion mutants (the 2–6 mutant was not tested) also showed no deficit in colonizing the lungs and spleens of mice after intranasal infection (Figure 2F–H). gB-NT was therefore not important for entry.
Figure 2.
Propagation and cell binding of gB-NT deletion mutants. (A) BHK-21 cells were infected (0.01 p.f.u./cell) with wild-type (WT) MuHV-4, each deletion mutant or a revertant of the gBΔ2–30 mutant (REV). Virus growth with time was then monitored by plaque assay. (B) EGFP+ version of the same viruses as in (A) was added to BHK-21 cells (1 p.f.u./cell) for the time indicated, then washed with PBS to remove unbound virions. The cells were then cultured overnight and infection was quantitated the next day by flow cytometric assay of viral eGFP expression. Equivalent data were obtained in two further experiments. (C) NMuMG cells were stained with gB-Fc fusions recovered from supernatants of transfected 293T cells. The shaded histogram is staining with secondary antibody only. None of the gB-NT deletions affected cell binding. (D) The gB-Fc fusion protein was pre-incubated with the neutralizing gB-NT-specific mAb MG-2C10 before being used to stain NMuMG cells. No difference in staining was observed. (E) NS0 myeloma cells were exposed (18 h) to eGFP+ wild-type (WT) MuHV-4 or gB-NT deletion mutants. Infection was then quantitated by flow cytometric assay of viral eGFP expression. (F) Mice were infected intranasally with wild-type (WT) MuHV-4, the gBΔ2–14 or gBΔ2–30 mutant or a revertant of the gBΔ2–30 mutant (REV). Lungs were harvested at 6 days post-infection and infectious virus quantitated by plaque assay. Each bar shows the mean±s.e.m. titers of five mice per group. (G) At 14 days after infection as for (B), spleens were harvested and virus load measured by infectious center assay. Each bar shows the mean±s.e.m. titers of five mice per group. (H) DNA was extracted from the same spleens as in (C) and assayed for viral genome load by real-time PCR. Each bar shows the mean±s.e.m. of five mice per group.
Altered antigenicity of MuHV-4 lacking gB-NT
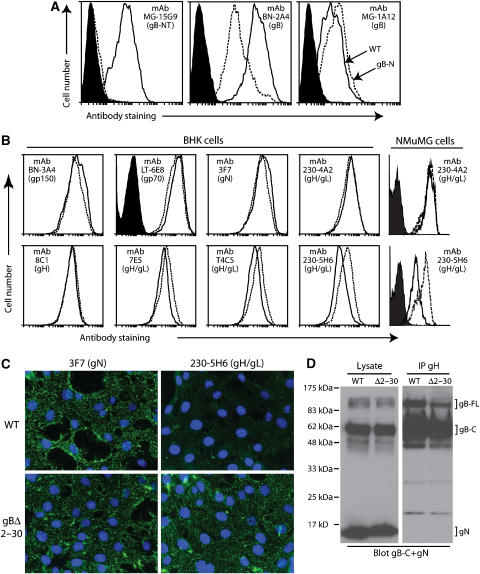
Our subsequent analysis focused on the largest gB-NT deletion (gBΔ2–30). The virions found on infected cell surfaces (de Lima et al, 2004) provide a means of probing antigenic differences between MuHV-4 glycoprotein mutants. As expected, gB-NT-specific neutralizing mAbs—previously mapped to gB residues 1–40 (Gillet et al, 2006)—did not recognize cells infected with gBΔ2–30 MuHV-4 (Figure 3A). gB recognition by several other mAbs was also affected. Thus, mAb MG-1A12 recognized the gBΔ2–30 mutant slightly better than wild type (mean fluorescence intensity (MFI) 352 versus 192), whereas mAb BN-2A4 recognized the mutant noticeably less well (MFI 63 versus 495). These epitopes may be close to gB-NT. For example, the BN-2A4 epitope could partly incorporate gB-NT and the MG-1A12 epitope could be partly obscured by it. The normal infectivity of the gBΔ2–30 mutant made it unlikely that gBΔ2–30 was substantially misfolded.
Figure 3.
Antigenicity of the gBΔ2–30 fusion complex. (A) BHK-21 cells were infected (2 p.f.u./cell, 18 h) with wild-type (solid lines) or gBΔ2–30 mutant (dotted lines) MuHV-4, and then analyzed by flow cytometry. The filled histogram shows uninfected cells. MAb 15G9 recognizes gB-NT, mAb BN-2A4 recognizes an epitope elsewhere in the N-terminal half of gB and mAb MG-1A12 recognizes a complex gB epitope. (B) BHK-21 or NMuMG cells were infected as for (A) and analyzed by flow cytometry for the expression of other MuHV-4 glycoproteins. Again, the dotted line corresponds to the gBΔ2–30 mutant and the solid line corresponds to wild type. The filled histogram shows secondary antibody only staining. Each histogram shows 30 000 events. The reduction in gH/gL staining for T4C5, 7E5 and 230-5H6 were all statistically significant (P<0.001 by Student's t-test). Equivalent data were obtained in four further experiments. (C) NMuMG cells were exposed to wild-type (WT) or gBΔ2–30 virions (2 h, 4°C), then washed with PBS and fixed with 2% paraformaldehyde. The cells were then stained for gN with mAb 3F7 and for gH/gL with mAb 230-5H6. Nuclei were counter-stained with DAPI. (D) Wild-type (WT) and gBΔ2–30 virions were lysed in 1% digitonin. gH was immunoprecipitated with mAb 8C1 (IgG2b). The original lysates and the precipitates (IP gH) were then immunoblotted for gB-C with mAb MG-4D11 and for gN with mAb 3F7 (both IgG2a) May et al (2005). Detection was with an IgG2a-specific secondary antibody. The positions of gN and full-length (gB-FL) and cleaved (gB-C) gB are marked. While both gB and gN were present in the lysates, only gB was co-precipitated with gH. No difference in co-precipitate was observed between the gBΔ2–30 mutant and wild type. Equivalent data were obtained in two further experiments.
A second and more surprising effect of gB-NT deletion was greater accessibility of gH/gL-neutralizing epitopes (Figure 3B). Thus, while mAbs LT-6E8 (anti-gp70), 3F7 (anti-gN) and BN-3A4 (anti-gp150) stained the gBΔ2–30 mutant no more than wild type, the gH/gL-specific mAbs 7E5, T4C5 and 230-5H6 all stained the mutant slightly better (MFI 334 versus 233, 334 versus 209 and 225 versus 109, respectively for BHK-21 cells). 7E5 and T4C5 define two gH/gL neutralization epitopes (Gill et al, 2006). MAb 230-5H6 always showed the greatest difference, but all the differences were highly reproducible and statistically significant. They did not reflect greater gH/gL expression, since the gH-specific mAb 8C1 and another gH/gL-specific mAb, 230-4A2, showed equivalent expression between the mutant and wild type. Instead it appeared that neutralization epitopes on gH/gL were selectively exposed by gB-NT deletion. Increased gH/gL accessibility on gBΔ2–30 virions was confirmed by immunofluorescence (Figure 3C). Here, we bound virions to cells rather than using infected cells, so as to present a more defined antigen. Both wild-type and gBΔ2–30 virions were readily visualized by staining for gN, but the 230-5H6 epitope of gH/gL was poorly displayed unless gB-NT was deleted.
An effect of gB on gH seemed plausible because these proteins are associated in MuHV-4 virions (Figure 3D). This association was independent of gB-NT, but provided a physical basis for gB-NT contacting gH/gL.
Increased susceptibility of gBΔ2–30 MuHV-4 to gH/gL-directed neutralization
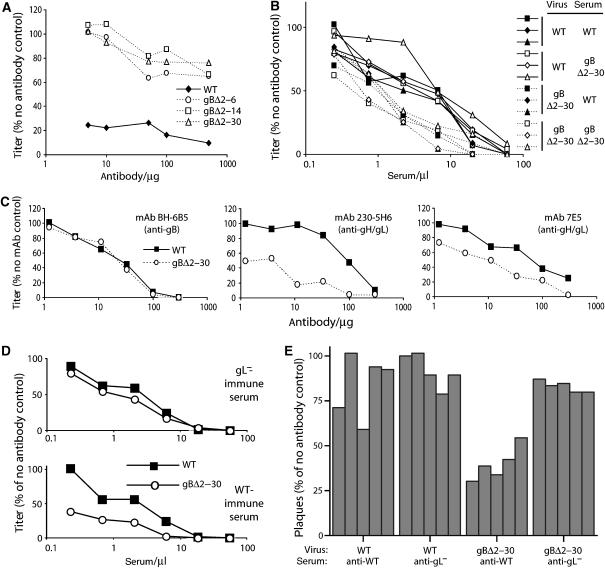
As expected, the gBΔ2–30 mutant was no longer susceptible to gB-NT-specific neutralizing mAbs (Figure 4A). However, despite the removal of this neutralization epitope, immune sera neutralized the gBΔ2–30 mutant better than wild type (Figure 4B). There was no obvious difference in neutralization titer between wild-type-immune and gBΔ2–30-immune sera, arguing that neutralization did not depend on a new epitope created by the gB-NT deletion, but rather on an epitope that could also be presented by wild-type MuHV-4.
Figure 4.
Neutralization of the gBΔ2–30 mutant. (A) Wild-type (WT) and gB-NT deletion virions were incubated with the gB-NT-specific neutralizing mAb MG-2C10, then assayed for infectivity on BHK-21 cells. Equivalent data were obtained in two further experiments. (B) Sera from mice infected with wild-type (WT) or gBΔ2–30 MuHV-4 were used to neutralize WT or gBΔ2–30 virions for BHK-21 cell infection. Each of the six serum samples is from one mouse. Equivalent data were obtained in four further experiments, using different immune sera. We compared titers by χ2 test (scoring as more or less than the interpolated median) for all the sera at each dilution. In the experiment shown, five of six serum dilutions showed a significant difference in neutralization (P<0.05) between WT and gBΔ2–30 viruses. Each repeat experiment showed significant differences at more than half the dilutions tested. (C) Wild-type (WT) and gBΔ2–30 virions were exposed to different neutralizing mAbs, then assayed for infectivity on BHK-21 cells. BH-6B5 is gB specific but not gB-NT specific. Scoring the titer for each virus to the average for that dilution and then comparing the values for all the dilutions of an assay by χ2 test showed that gH/gL-specific mAbs neutralized the gBΔ2–30 mutant significantly better than the wild type (P<0.002). Equivalent data were obtained in four further experiments. (D) Wild-type (WT) and gBΔ2–30 virions were exposed to sera from mice infected with WT or gL-deficient (gL−) MuHV-4. Both showed greater neutralization of the gBΔ2–30 mutant, but the difference was greater for wild-type-immune sera. (E) Equivalent results to (D) for five different immune sera in each group at a single serum dose (2.5 μl/1000 p.f.u. virus). Wild-type-immune sera showed a significant difference in neutralization between wild-type and gBΔ2–30 virions (P<0.002 by Student's t-test). The gL−-immune sera did not (P=0.14).
We then selected mAbs from wild-type MuHV-4-immune mice by their capacity to neutralize the gB-NT mutant. All (n=10) recognized gH/gL. Previously established gH/gL-specific mAbs also neutralized the gBΔ2–30 mutant better than wild type (Figure 4C). This was consistent with the greater accessibility of gH/gL epitopes observed for gBΔ2–30-infected cells (Figure 3B) and gBΔ2–30 virions (Figure 3C). Notably, mAb 230-5H6 showed the greatest difference between the gBΔ2–30 mutant and wild type in both staining and neutralization. In contrast to the improved gH/gL-specific neutralization, a gB-specific neutralizing mAb (BH-6B5) that recognizes a region of gB outside gB-NT showed no difference between the gBΔ2–30 mutant and wild type (Figure 4C). We tested further the gH/gL dependence of gBΔ2–30 virus neutralization by comparing sera from mice infected with gL+ and gL− MuHV-4 (Figure 4D and E). The latter make no gH/gL-specific antibody response, since gL− MuHV-4 does not display gH/gL epitopes (Gillet et al, 2007c). While wild-type-immune sera showed better neutralization of the gBΔ2–30 mutant, gL knockout-immune sera did not. gB-NT deletion therefore conferred a greater susceptibility to neutralization by exposing gH/gL.
This neutralization also operated in vivo (Table I). Thus, immune sera blocked the entry of gBΔ2–30 MuHV-4 into new hosts much more effectively than that of the wild type. Immune sera normally block intranasal MuHV-4 infection poorly because they poorly inhibit membrane fusion; this can be remedied by boosting anti-gH/gL immunity (Gillet et al, 2007d). Thus, the increased susceptibility of the gBΔ2–30 mutant to in vivo neutralization was consistent with its greater vulnerability to gH/gL-specific antibodies.
Table 1. gB-NT protects against neutralization in vivo.
| Challenge | Neutralization | Mice infected | |
|---|---|---|---|
| Wild type | gBΔ2–30 | ||
| 100 p.f.u. | Nil | 10/10 | 10/10 |
| 100 p.f.u. | BALB/c immune serum | 4/10 | 0/10 |
| 100 p.f.u. | C57BL/6 immune serum | 2/10 | 0/10 |
| 1000 p.f.u. | Nil | 10/10 | 10/10 |
| 1000 p.f.u. | BALB/c immune serum | 10/10 | 0/10 |
| 1000 p.f.u. | C57BL/6 immune serum | 10/10 | 0/10 |
| Wild-type and gBΔ2–30 virions were incubated (2 h, 37°C) with or without immune sera (5 μl) pooled from mice 3-6 months post-infection with wild-type MuHV-4. They were then inoculated intranasally into naive mice. The mice were assayed for infection or not 7 days later by titering infectious virus in lungs. Significant differences were observed from the pooled BALB/c immune sera at 100 p.f.u. (P<0.05 by χ2 test) and for both serum pools at 1000 p.f.u. (P<10−5). No significant difference was observed between sera from C57BL/6 and BALB/c mice. | |||
Cell type-specific susceptibility to gB-NT-directed neutralization
Since gB-NT itself can be a neutralization target, why should exchanging a gB-NT target for one on gH/gL affect overall virion susceptibility to neutralization? The obvious explanation would be that these targets are not equivalent. We have noted before that mAbs specific for gB-NT neutralize MuHV-4 only to a threshold minimum, not to zero (Gillet et al, 2006). Moreover, while plaque formation on BHK-21 fibroblasts is blocked quite well, plaque formation on NMuMG epithelial cells is blocked hardly at all. These data suggested that gB-NT-directed neutralization might be less effective than it appeared from assays using fibroblasts, and so might be less important to MuHV-4 than gH/gL-directed neutralization. We therefore sought to define how gB-NT might itself evade antibody.
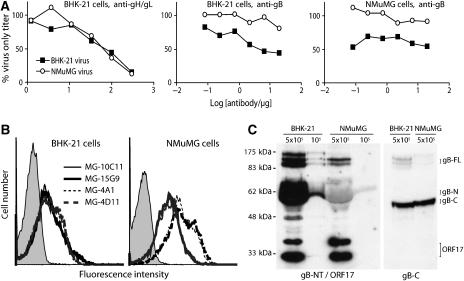
One question raised by the difference in gB-NT-directed neutralization between NMuMG epithelial cells and BHK-21 fibroblasts was whether this reflected different entry requirements or differences in the virions produced, once an initial infection had occurred. To answer this, we compared NMuMG-derived and BHK-derived virions for their susceptibility to neutralization (Figure 5A). Each was similarly susceptible to gH/gL-directed neutralization, but NMuMG cell-derived virions were much less susceptible to gB-NT-directed neutralization for both BHK-21 and NMuMG cell infections. Thus, the difference in neutralization reflected mainly a difference between the virions each cell type produced.
Figure 5.
MuHV-4 virions derived from BHK-21 fibroblasts and NMuMG epithelial cells differ in gB-NT antigenicity and in susceptibility to gB-NT-directed neutralization. (A) Virions from BHK-21 or NMuMG cells were incubated with gH/gL-specific or gB-NT-specific neutralizing mAbs, then titered on BHK-21 or NMuMG cells. The difference in gB-NT-directed neutralization between BHK-21-derived and NMuMG-derived virions was significant by χ2 test for both BHK-21 and NMuMG cells (P<0.002). Equivalent data were also obtained in two further experiments. (B) BHK-21 or NMuMG cells were infected with MuHV-4 and 18 h later analyzed for cell surface gB expression by flow cytometry. MAbs MG-10C11 and MG-15G9 are gB-NT-specific and neutralizing, MG-4A1 and MG-4D11 are not gB-NT-specific and non-neutralizing. The difference in MFI between MG-10C11/MG-15G9 and MG-4A1/MG-4D11 was significantly greater for NMuMG cells (>4-fold) than for BHK-21 cells (<2-fold). Equivalent data were obtained in two further experiments. (C) BHK-21- and NMuMG-derived virions were immunoblotted for gB-NT (mAb MG-2C10) plus ORF17 as a loading control (mAb 150-7D1), or for gB-C (mAb MG-4D11). The positions of uncleaved gB (gB-FL), its cleavage products (gB-N, gB-C) and ORF17 are marked.
As before, we examined gB antigenicity by flow cytometry of virus-infected cells (Figure 5B). MuHV-4-infected NMuMG cells showed more total gB-specific staining than infected BHK-21 cells (MFI 135 versus 32 for mAb MG-4A1 and 161 versus 40 for MG-4D11), probably reflecting greater virion accumulation on the plasma membrane. But they displayed relatively less of the gB-NT neutralization epitope (MFI 77 versus 51 for mAbs MG-10C11 and 79 versus 46 for MG-15G9). Immunoblots (Figure 5C) showed a similar gB content for NMuMG- and BHK-derived virions based on staining with the gB-C-specific mAb MG-4D11, but gB-NT of NMuMG-derived virions was recognized substantially less well. The 65 kDa band in Figure 5C is cleaved N-terminal gB, which is abundant in mature virions. The less abundant 120 kDa band is uncleaved gB. The different bands around 120 kDa probably correspond to different N-linked gB glycoforms (Lopes et al, 2004). The poor recognition of these bands of NMuMG-derived virions by mAb MG-2C10 indicated that gB-NT was antigenically different.
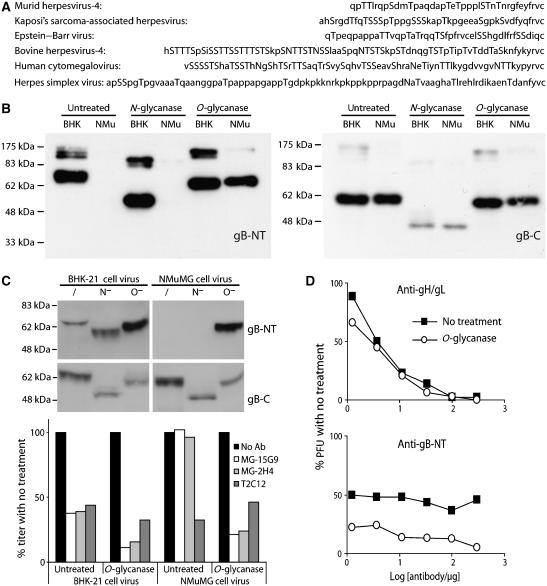
O-linked glycans protect gB against neutralization
The MuHV-4 gB-NT contains nine potential O-glycosylation sites (Figure 6A). Despite considerable sequence diversity between the gB-NTs of different herpesviruses, multiple predicted O-glycosylation sites are preserved. Human cytomegalovirus and bovine herpesvirus-4 provide particularly striking examples. Our gB-NT-specific neutralizing mAbs do not require O-glycans for recognition, since they recognize bacterially expressed gB fragments (Gillet et al, 2006), but O-glycans could potentially inhibit their recognition. We tested this by digesting NMuMG cell-derived virion lysates with sialidase A plus O-glycanase (Figure 6B). This gave a marked improvement in gB detection. In contrast, removing N-linked glycans had no effect.
Figure 6.
O-linked glycans mask gB-NT of NMuMG cell-derived virions. (A) Herpesvirus gB alignment from the predicted signal sequence cleavage site to the first conserved cysteine residue, with potential glycosylation sites, both N- and O-linked, in upper case. (B) Reduced MuHV-4 virion lysates derived from BHK-21 (BHK) or NMuMG cells (NMu) were either left untreated or deglycosylated as indicated, then immunoblotted for gB-NT with mAb MG-2C10 or for gB-C with mAb MG-4D11. (C) Intact BHK-21 or NMuMG cell-derived virions were deglycosylated without denaturation. /, no enzyme, N−, protein N-glycanase, O−, sialidase A+O-glycanase. Each was then immunoblotted for gB-NT/gB-C and tested for neutralization. MAbs MG-15G9 and MG-2H4 are gB-NT-specific, T2C12 is gH/gL-specific. Equivalent data were obtained in two further experiments. Comparing results for all the experiments by Student's t-test, T2C12 neutralization was not significantly different between untreated and deglycosylated samples, whereas MG-15G9 and MG-2H4 showed a significant improvement in neutralization with deglycosylation for both BHK-21 cells (P<0.02) and NMuMG cells (P<0.001). (D) MuHV-4 virions were recovered from infected mouse lungs, deglycosylated (O-glycanase) or not (no treatment) as in (C), then exposed to gH/gL-specific (T2C12) or gB-NT-specific (MG-2C10)-neutralizing mAbs. The improvement in gB-NT-directed neutralization by deglycosylation was significant by χ2 test (P<0.002). Equivalent data were obtained in a repeat experiment.
We then removed O-glycans from intact, NMuMG cell-derived virions. This conferred susceptibility to gB-directed neutralization (Figure 6C). The neutralization of BHK-21 cell-derived virions also improved, consistent with the previously noted failure of gB-NT-specific mAbs to eliminate their infectivity completely (Gillet et al, 2006). Thus, gB-NT on NMuMG cell-derived virions was strongly protected against neutralization by O-glycans, and the greater susceptibility of BHK-21 cell-derived virions to gB-NT-directed neutralization correlated with less O-glycan-dependent protection. gB-NT O-glycosylation also explained why deleting just a few amino-acid residues noticeably reduced the gB apparent molecular weight (Figure 1D and E).
O-glycans protect ex vivo virions against neutralization
The γ-herpes virions shed by infected hosts are probably derived from epithelial cells (Jiang et al, 2006). These characteristically produce extensively O-glycosylated mucins (Patton et al, 1995). They may therefore produce extensively O-glycosylated virions. Virions recovered from lung homogenates of MuHV-4-infected mice showed substantial O-glycan-dependent resistance to gB-directed neutralization (Figure 6D). Similar results were obtained with virions recovered by bronchoalveolar lavage (data not shown). Thus, the resistance of NMuMG cell-derived virions to neutralization was consistent with the situation in vivo. In contrast, the susceptibility of BHK-21 cell-derived virions to gB-NT-directed neutralization probably underestimates the obstacles that antibody normally faces.
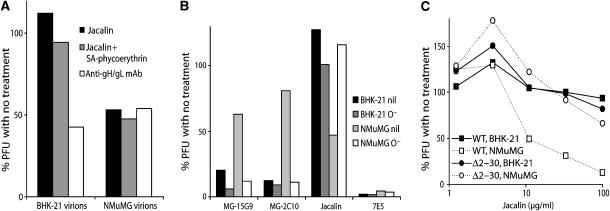
gB-NT O-glycans can be a target for lectin-mediated neutralization
Since gB-NT was non-essential for MuHV-4 replication, gB-NT-directed neutralization probably operates by steric hindrance, for example by keeping fusion loops away from host membranes. This suggested that any bulky molecule in the same site might inhibit infection equally well. We therefore tested the capacity of jacalin, an O-glycan-specific lectin, to inhibit MuHV-4 infection (Figure 7A). Inhibition was evident for NMuMG cell-derived but not for BHK-21 cell-derived virions, the converse of antibody-mediated neutralization. We used biotinylated jacalin so as to also attach streptavidin-conjugated phycoerythrin (molecular weight 300 kDa), but biotinylated jacalin alone (60 kDa) was also effective. Removing O-glycans from MuHV-4 propagated in NMuMG cells, the same treatment that conferred susceptibility to neutralization by mAbs, abolished the inhibition by jacalin (Figure 7B). Probably multiple virion glycoproteins are differentially glycosylated between NMuMG and BHK-21 cells. However, the gBΔ2–30 mutant escaped inhibition by jacalin when propagated in NMuMG cells (Figure 7C). Lectin-mediated neutralization was therefore specific to gB-NT.
Figure 7.
O-linked glycans on the gB N terminus allow virus neutralization by jacalin. (A) BHK-21 or NMuMG cell-derived virions were left untreated or incubated with biotinylated jacalin (10 μg/ml), biotinylated jacalin+phycoerythrin-conjugated streptavidin (10 μg/ml) or the gH/gL-specific mAb T2C12. Each sample was then compared by plaque assay. Equivalent data were obtained in a repeat experiment. The reduction in titer by jacalin was significantly greater for NMuMG-derived virions than for BHK-21-derived virions (P<0.02 by Student's t-test). (B) BHK-21 or NMuMG-derived virions were left untreated (nil) or deglycosylated with sialidase A+O-glycanase (O−), then neutralized with gB-NT-specific mAbs (MG-15G9, MG-2C10), biotinylated jacalin/phycoerythrin-conjugated streptavidin (jacalin) or a gH/gL-specific mAb (7E5). Equivalent data were obtained in a repeat experiment. In contrast to intact virions, there was no significant difference in gB-NT-directed mAb neutralization between deglycosylated NMuMG- and BHK-21-derived virions. Jacalin showed significantly better neutralization of intact virions from NMuMG cells (P<0.03 by Student's t-test) but not of deglycosylated virions. (C) Wild-type (WT) and gBΔ2–30 MuHV-4 were grown in BHK-21 cells or NMuMG cells and then tested for susceptibility to neutralization by jacalin. Equivalent data were obtained in a repeat experiment. Unlike wild-type virions, the cell type of origin had no significant effect on jacalin-mediated neutralization for gBΔ2–30 virions.
Discussion
Epidemiology tells us that herpesviruses evade neutralization by antibody (Xu et al, 1996). There is no evidence for weak immunogenicity, indeed persistent infection potently elicits both serum and mucosal antibody responses (Gyselink et al, 1978; Tamura et al, 1980; Yao et al, 1991). But while most viruses cannot transmit from immune hosts without antigenic variation, herpes virions readily do so. Most neutralizing antibodies block receptor binding (Knossow and Skehel, 2006). A key part of herpesvirus antibody evasion may therefore be a capacity to infect via alternative uptake pathways such as IgG Fc receptors (Inada et al, 1985; Maidji et al, 2006; Rosa et al, 2007; Gillet et al, 2007b). But even if virions evade a block to cell binding, they remain potentially susceptible to a block to membrane fusion. The data presented here provide the first insights into how γ-herpesviruses can protect their fusion machinery against antibody.
MAbs directed against the MuHV-4 fusion complex are strikingly inefficient at neutralization: those binding to gB cannot ablate infectivity completely (Gillet et al, 2006), and those binding to gH/gL require a very large antibody excess (Gill et al, 2006). gB-NT played a central role in both aspects of antibody resistance: it protected a critical region of gH/gL, and was itself protected by O-linked glycans. Removing gB-NT made a subset of gH/gL epitopes more accessible, implying that gB-NT covers a crucial part of gH/gL. The redundancy of gB-NT for the gB/gH/gL interaction suggested that its interaction with gH/gL was weak; the partial availability of the 230-5H6 epitope on wild-type virions suggested that it might even be reversible. gB-NT is presumably displaced from gH/gL after virions engage the cell surface. This may stop antibodies binding to neutralization epitopes on gH/gL unless they are co-endocytosed, which would explain why gH/gL-directed neutralization requires a large antibody excess.
The protection of gB-NT itself by O-linked glycans illustrates two important points: susceptibility to neutralization may depend on the cell type producing the virus; and infectivity reduction rather than ablation may overestimate the ease of infection control. Although gB-NT-specific mAbs could largely neutralize fibroblast-derived virions, they almost completely failed to neutralize epithelial cell-derived virions. Ex vivo virions also showed substantial resistance. The conservation of predicted O-glycosylation sites in other herpesvirus gB-NTs suggests that this evasion mechanism is widespread; indeed, much as N-linked glycans protect conformational epitopes (Burton et al, 2005; Knossow and Skehel, 2006), O-glycans could protect linear epitopes on many viral glycoprotein N termini. Despite O-glycosylation, gB-NTs may eventually be targeted well enough to reduce viral transmission. This could explain what appears to be weak selection for herpesvirus gB-NT amino-acid diversity (Basgoz et al, 1992; Xu et al, 1996; Auerbach et al, 2000). The recorded amino-acid diversity by itself would seem insufficient for antibody evasion—HIV and influenza vary much more—but could act by modulating O-glycosylation (Mardberg et al, 2004). Bovine herpesvirus-4, which is particularly rich in potential O-linked glycan attachment sites, encodes a mucin-type β-1,6-N-acetylglucosaminyltransferase (Vanderplasschen et al, 2000), which could modify gB-NT further. Thus, while herpesviruses generally pursue a faithful replication strategy, more subtle forms of antigenic variation may not be beyond them.
Although O-glycan binding mAbs can occur (Burton et al, 2005), T-cell tolerance makes them rare. In essence, O-glycans like N-glycans provide an antibody-proof shield. However, glycans provide potential binding sites for lectins (Wilson et al, 1999; Rudiger and Gabius, 2001). The extensive N-glycosylation of the HIV gp120 makes it a target for N-glycan-binding lectins (Balzarini, 2006). Our data show that an O-glycan-binding lectin can reduce herpesvirus infectivity. A combination of lectins and antibody could be particularly effective, since the evasion of one predisposed to recognition by the other. A possible application might be reducing perinatal infection.
Materials and methods
Mice
Female BALB/c mice were purchased from Harlan UK Ltd (Bicester, UK), housed in the Cambridge University Department of Pathology, and infected intranasally with MuHV-4 (2 × 104 p.f.u.) when 6–8 weeks old (Home Office Project Licence 80/1992). Immune sera were collected at least 3 months post-infection. Ex vivo virus was recovered from lung homogenates 5 days post-infection. Cell debris was pelleted by centrifugation (1000 g, 5 min) three times. Supernatant virus was then recovered by ultracentrifugation (38 000 g, 90 min).
Cells
BHK-21 cells, NMuMG cells, NIH-3T3-CRE cells (Stevenson et al, 2002), 293T cells and NS0 cells were grown in Dulbecco's modified Eagle's medium (Invitrogen, Paisley, UK) supplemented with 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin and 10% fetal calf serum (PAA Laboratories, Linz, Austria). Medium for murine embryonic fibroblasts was further supplemented with 50 μM 2-mercaptoethanol. Cells were transfected where indicated using Fugene-6 (Roche Diagnostics, Lewes, UK).
Viruses
All viruses were derived from a cloned MuHV-4 BAC (Adler et al, 2000). The coding sequence for amino-acid residues 2–6 (PTTLR), 2–14 (PTTLRQPSDMTPA) or 2–30 (PTTLRQPSDMTPAQDAPTETPPPLSTNTN) of the mature gB were replaced with an EcoRI restriction site (encoding EF). We first PCR-amplified (Phusion DNA polymerase; New England Biolabs, Hitchin, UK) coordinates 15 001–16 600 of the MuHV-4 genome (Virgin et al, 1997), including KpnI and EcoRI restriction sites in the respective 5′ and 3′ primers, and cloned the PCR product into the KpnI/EcoRI sites of pSP73 (Promega Corporation, Southampton, UK). We then amplified genomic coordinates 16 616–18 025, 16 640–18 025 or 16 688–18 025 as EcoRI/BglII-restricted fragments and cloned these into the corresponding sites of the same vector. This generated the relevant deletions between genomic flanks of approximately 1.6 kb. Each construct was then subcloned as a BglII/KpnI fragment into the BamHI/KpnI sites of the pST76K-SR shuttle vector, and recombined into the MuHV-4 BAC (Adler et al, 2000). The correct construction of each mutant was confirmed by DNA sequencing. Infectious virus was reconstituted by transfecting each BAC into BHK-21 cells. Except where indicated, the loxP-flanked BAC/eGFP cassette was removed by virus passage in NIH-3T3-CRE cells, and virus stocks were grown in BHK-21 cells and titered by plaque assay in BHK-21 cells. gL-deficient MuHV-4 has been described (Gillet et al, 2007c).
Plasmids
A fusion of the N-terminal half of gB to human IgG1-Fc has been described (Gillet et al, 2007a). Deletion mutants of this construct corresponding to the gB 2–6, 2–14 and 2–30 virus mutants were made by PCR amplification of the same region using DNA from each mutant virus as a template. As with the wild type (Gillet et al, 2007a), each PCR product was cloned into the XbaI/NotI sites of pTORSTEN, using AvrII and NotI restriction sites in the respective 5′ and 3′ primers. Fusion proteins were generated by transfecting each plasmid into 293T cells and harvesting the supernatant 48 h later.
Southern blotting
DNA was extracted from virions (Wizard genomic DNA purification kit; Promega Corporation), digested with EcoRI, electrophoresed through 0.8% agarose in Tris acetate buffer and transferred to positively charged nylon membranes (Roche Diagnostics) (de Lima et al, 2004). A 32P-dCTP-labeled probe (APBiotech, Little Chalfont, UK) corresponding to genomic coordinates 15 001–18 025 was generated by random primer extension (Nonaprimer kit; Qbiogene, Bingham, UK). Membranes were hybridized with probe (65°C, 18 h), washed to a stringency of 0.2% SSC, 0.1% SDS and exposed to X-ray film.
Viral genome quantitation
Viral genome loads were measured by real-time PCR (Bennett et al, 2005). Genomic coordinates 24 832–25 071 were amplified by PCR (Rotor Gene 3000; Corbett Research, Cambridge, UK) from 10 ng of DNA extracted (Wizard genomic DNA purification kit; Promega Corporation) from spleens. The PCR products were quantitated with Sybr green (Invitrogen) and the copy number was calculated by comparison with a standard curve of cloned MK3 template, serially diluted in control spleen DNA and amplified in parallel. Amplified products were distinguished from paired primers by melting curve analysis, and the correct size of the amplified products was confirmed by electrophoresis and staining with ethidium bromide.
Virus titrations
For plaque assays, 10-fold virus dilutions were plated onto BHK-21 cell, NMuMG cell or murine embryonic fibroblast monolayers for 2 h, then overlaid with 0.3%. carboxymethylcellulose. The monolayers were fixed in 10% formaldehyde after 4 days for BHK-21 cells, after 5 days for murine embryonic fibroblasts and after 6 days for NMuMG cells. The fixed cells were stained with 0.1% toluidine blue and plaques were counted with a plate microscope. To titer infectious virus in lungs, the lungs were homogenized in complete medium, frozen, thawed and sonicated. Tissue debris was pelleted by brief centrifugation (1000 g, 1 min). Infectious virus in homogenate supernatants was then measured by plaque assay. Latent virus in spleens was measured by infectious center assay (de Lima et al, 2004). Single spleen cell or mediastinal lymph node cell suspensions were cultured on murine embryonic fibroblast monolayers, which were fixed and stained for plaque counting after 6 days. Pre-formed infectious virus—that forming plaques even after freeze–thawing—contributed <1% of the infectivity recoverable from lymphoid tissue, so the infectious center assay essentially measured reactivating latent virus.
Neutralization assays
Viruses were pre-incubated (2 h, 37°C) with dilutions of monoclonal antibody (mAb), immune serum or biotinylated jacalin (Vector Laboratories, Burlingame, CA) plus streptavidin-phycoerythrin (BD Biosciences, Oxford, UK). The virus–antibody or virus–lectin mixtures were then added to cell monolayers for plaque assay.
Monoclonal antibodies
All mAbs were derived from MuHV-4-infected BALB/c mice by fusion with NS0 cells (Köhler and Milstein, 1975). The mAbs used in this study are listed in Supplementary Table 1.
Immunoblotting and immunoprecipitation
Virions were lysed in Laemmli's buffer, denatured (95°C, 5 min), resolved by SDS–PAGE and transferred to PVDF membranes (Boname et al, 2004). The membranes were blocked with 10% non-fat milk in PBS/0.1% Tween-20, then incubated with MuHV-4-specific mAbs. Bound antibody was detected with horseradish peroxidase-conjugated rabbit anti-mouse IgG pAb (Dako Corporation, Ely, UK), followed by washing in PBS/0.1% Tween-20, development with ECL substrate (APBiotech) and exposure to X-ray film. To look for protein associations, virions were lysed (30 min, 4°C) in 1% digitonin/50 mM Tris–Cl pH 7.4/150 mM NaCl/5 mM MgCl2/5 mM CaCl2/1 mM PMSF plus complete protease inhibitors (Roche Diagnostics). Insoluble debris was removed by centrifugation (13 000 g, 15 min). gH was then immunoprecipitated with mAb 8C1 (IgG2b) plus protein A-sepharose (Sigma Chemical Co., Poole, UK). The beads were washed 5 × in lysis buffer. The proteins were then eluted and immunoblotted as above, using IgG2a primary mAbs and a horseradish peroxidase-conjugated goat anti-mouse IgG2a secondary antibody (Southern Biotech, Birmingham, AL).
Deglycosylation
For SDS–PAGE analyzing, virions were lysed, reduced with 2-mercaptoethanol, denatured in 1% SDS and deglycosylated with protein N-glycanase or with sialidase A+O-glycanase according to the manufacturer's instructions (Prozyme, San Leandro, CA). For neutralization assays, N- or O-linked glycans were removed with the same enzymes, but without reduction or denaturation. Thus, intact virions were incubated with either protein N-glycanase or sialidase A+O-glycanase (3 h, 37°C) in PBS/5% fetal calf serum buffered to pH 6.
Immunofluorescence
Adherent MuHV-4-infected cells were washed in PBS, fixed in 4% paraformaldehyde, then permeabilized with 1% Triton X-100. Viral glycoproteins were detected with murine mAbs plus Alexa 488-conjugated goat anti-mouse IgG (Invitrogen). Nuclei were counterstained with DAPI. Fluorescence was visualized with an Olympus IX70 microscope plus a Retiga 2000R camera line (QImaging) or with a Leica confocal microscope.
Flow cytometry
Cells exposed to eGFP+ viruses were washed in PBS and analyzed directly for green channel fluorescence. For surface staining, cells were incubated (1 h, 4°C) with MuHV-4 glycoprotein-specific mAbs followed by fluorescein-conjugated rabbit anti-mouse IgG pAb (Dako Cytomation) or Alexa 633-conjugated goat anti-mouse pAb (Invitrogen). Fc fusion protein binding was detected with phycoerythrin-conjugated goat anti-human pAb (Sigma). All cells were washed 2 × in PBS after each antibody and analyzed on a FACS Calibur (BD Biosciences).
Supplementary Material
Supplementary data Table 1
Acknowledgments
We thank Janet May and Susanna Colaco for outstanding technical support. Laurent Gillet is a Postdoctoral Researcher of the Fonds National Belge de la Recherche Scientifique (FNRS). Philip Stevenson is a Wellcome Trust Senior Clinical Fellow (GR076956MA). This work was also supported by Medical Research Council grants G0400427 and G9800903 and by Cancer Research UK grant C19612/A6189.
References
- Adler H, Messerle M, Wagner M, Koszinowski UH (2000) Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J Virol 74: 6964–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akula SM, Pramod NP, Wang FZ, Chandran B (2002) Integrin alpha3beta1 (CD 49c/29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108: 407–419 [DOI] [PubMed] [Google Scholar]
- Atalay R, Zimmermann A, Wagner M, Borst E, Benz C, Messerle M, Hengel H (2002) Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcgamma receptor homologs. J Virol 76: 8596–8608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach MR, Czajak SC, Johnson WE, Desrosiers RC, Alexander L (2000) Species specificity of macaque rhadinovirus glycoprotein B sequences. J Virol 74: 584–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzarini J (2006) Inhibition of HIV entry by carbohydrate-binding proteins. Antiviral Res 71: 237–247 [DOI] [PubMed] [Google Scholar]
- Basgoz N, Qadri I, Navarro D, Sears A, Lennette E, Youngblom J, Pereira L (1992) The amino terminus of human cytomegalovirus glycoprotein B contains epitopes that vary among strains. J Gen Virol 73: 983–988 [DOI] [PubMed] [Google Scholar]
- Bennett NJ, May JS, Stevenson PG (2005) Gamma-herpesvirus latency requires T cell evasion during episome maintenance. PLoS Biol 3: e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boname JM, de Lima BD, Lehner PJ, Stevenson PG (2004) Viral degradation of the MHC class I peptide loading complex. Immunity 20: 305–317 [DOI] [PubMed] [Google Scholar]
- Burton DR, Stanfield RL, Wilson IA (2005) Antibody versus HIV in a clash of evolutionary titans. Proc Natl Acad Sci USA 102: 14943–14948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman HM, de Lima B, Morton V, Stevenson PG (2003) Murine gammaherpesvirus 68 lacking thymidine kinase shows severe attenuation of lytic cycle replication in vivo but still establishes latency. J Virol 77: 2410–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima BD, May JS, Stevenson PG (2004) Murine gammaherpesvirus 68 lacking gp150 shows defective virion release but establishes normal latency in vivo. J Virol 78: 5103–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dialyna IA, Graham D, Rezaee R, Blue CE, Stavrianeas NG, Neisters HG, Spandidos DA, Blackbourn DJ (2004) Anti-HHV-8/KSHV antibodies in infected individuals inhibit infection in vitro. AIDS 18: 1263–1270 [DOI] [PubMed] [Google Scholar]
- Dingwell KS, Brunetti CR, Hendricks RL, Tang Q, Tang M, Rainbow AJ, Johnson DC (1994) Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol 68: 834–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T (1992) Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol 66: 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill MB, Gillet L, Colaco S, May JS, de Lima BD, Stevenson PG (2006) Murine gammaherpesvirus-68 glycoprotein H-glycoprotein L complex is a major target for neutralizing monoclonal antibodies. J Gen Virol 87: 1465–1475 [DOI] [PubMed] [Google Scholar]
- Gillet L, Adler H, Stevenson PG (2007a) Glycosaminoglycan interactions in murine gammaherpesvirus-68 infection. PLoS ONE 2: e347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet L, Gill MB, Colaco S, Smith CM, Stevenson PG (2006) Murine gammaherpesvirus-68 glycoprotein B presents a difficult neutralization target to monoclonal antibodies derived from infected mice. J Gen Virol 87: 3515–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet L, May JS, Colaco S, Stevenson PG (2007b) The murine gammaherpesvirus-68 gp150 acts as an immunogenic decoy to limit virion neutralization. PLoS ONE 2: e705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet L, May JS, Colaco S, Stevenson PG (2007c) Glycoprotein L disruption reveals two functional forms of the murine gammaherpesvirus 68 glycoprotein H. J Virol 81: 280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet L, May JS, Stevenson PG (2007d) Post-exposure vaccination improves gammaherpesvirus neutralization. PLoS ONE 2: e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyselink R, Coles D, Ash RJ, Fritz ME (1978) Salivary neutralizing activity against herpes simplex virus type 1. J Infect Dis 137: 583–586 [DOI] [PubMed] [Google Scholar]
- Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC (2006) Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313: 217–220 [DOI] [PubMed] [Google Scholar]
- Holloway SA, Studdert MJ, Drummer HE (1998) Characterization of glycoprotein B of the gammaherpesvirus equine herpesvirus-2. J Gen Virol 79: 1619–1629 [DOI] [PubMed] [Google Scholar]
- Inada T, Chong KT, Mims CA (1985) Enhancing antibodies, macrophages and virulence in mouse cytomegalovirus infection. J Gen Virol 66: 871–878 [DOI] [PubMed] [Google Scholar]
- Janz A, Oezel M, Kurzeder C, Mautner J, Pich D, Kost M, Hammerschmidt W, Delecluse HJ (2000) Infectious Epstein–Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J Virol 74: 10142–10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Scott RS, Hutt-Fletcher LM (2006) Epstein–Barr virus shed in saliva is high in B-cell-tropic glycoprotein gp42. J Virol 80: 7281–7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Feenstra V (1987) Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J Virol 61: 2208–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knossow M, Skehel JJ (2006) Variation and infectivity neutralization in influenza. Immunology 119: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler G, Milstein C (1975) Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256: 495–497 [DOI] [PubMed] [Google Scholar]
- Laquerre S, Argnani R, Anderson DB, Zucchini S, Manservigi R, Glorioso JC (1998) Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J Virol 72: 6119–6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes FB, Colaco S, May JS, Stevenson PG (2004) Characterization of the MuHV-4 glycoprotein B. J Virol 78: 13370–13375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidji E, McDonagh S, Genbacev O, Tabata T, Pereira L (2006) Maternal antibodies enhance or prevent cytomegalovirus infection in the placenta by neonatal Fc receptor-mediated transcytosis. Am J Pathol 168: 1210–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardberg K, Nystrom K, Tarp MA, Trybala E, Clausen H, Bergstrom T, Olofsson S (2004) Basic amino acids as modulators of an O-linked glycosylation signal of the herpes simplex virus type 1 glycoprotein gC: functional roles in viral infectivity. Glycobiology 14: 571–581 [DOI] [PubMed] [Google Scholar]
- May JS, Colaco S, Stevenson PG (2005) Glycoprotein M is an essential lytic replication protein of the murine gammaherpesvirus 68. J Virol 79: 3459–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdzanowska K, Feng J, Gerhard W (2003) Virus-neutralizing activity mediated by the Fab fragment of a hemagglutinin-specific antibody is sufficient for the resolution of influenza virus infection in SCID mice. J Virol 77: 8322–8328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashunmugam T, Lubinski J, Wang L, Goldstein LT, Weeks BS, Sundaresan P, Kang EH, Dubin G, Friedman HM (1998) In vivo immune evasion mediated by the herpes simplex virus type 1 immunoglobulin G Fc receptor. J Virol 72: 5351–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlin M, Sundqvist VA, Mach M, Wahren B, Borrebaeck CA (1993) Fine specificity of the human immune response to the major neutralization epitopes expressed on cytomegalovirus gp58/116 (gB), as determined with human monoclonal antibodies. J Virol 67: 703–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K, Fujii S, Takada A, Kida H (2006) The amino-terminal residue of glycoprotein B is critical for neutralization of bovine herpesvirus 1. Virus Res 115: 105–111 [DOI] [PubMed] [Google Scholar]
- Patton S, Gendler SJ, Spicer AP (1995) The epithelial mucin, MUC1, of milk, mammary gland and other tissues. Biochim Biophys Acta 1241: 407–423 [DOI] [PubMed] [Google Scholar]
- Peeters B, Pol J, Gielkens A, Moormann R (1993) Envelope glycoprotein gp50 of pseudorabies virus is essential for virus entry but is not required for viral spread in mice. J Virol 67: 170–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa GT, Gillet L, Smith CM, de Lima BD, Stevenson PG (2007) IgG fc receptors provide an alternative infection route for murine gamma-herpesvirus-68. PLoS ONE 2: e560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger H, Gabius HJ (2001) Plant lectins: occurrence, biochemistry, functions and applications. Glycoconj J 18: 589–613 [DOI] [PubMed] [Google Scholar]
- Sissons JG, Oldstone MB (1980) Killing of virus-infected cells: the role of antiviral antibody and complement in limiting virus infection. J Infect Dis 142: 442–448 [DOI] [PubMed] [Google Scholar]
- Sitki-Green D, Covington M, Raab-Traub N (2003) Compartmentalization and transmission of multiple Epstein–Barr virus strains in asymptomatic carriers. J Virol 77: 1840–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson PG, Belz GT, Castrucci MR, Altman JD, Doherty PC (1999) A gamma-herpesvirus sneaks through a CD8(+) T cell response primed to a lytic-phase epitope. Proc Natl Acad Sci USA 96: 9281–9286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson PG, Doherty PC (1998) Kinetic analysis of the specific host response to a murine gammaherpesvirus. J Virol 72: 943–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson PG, May JS, Smith XG, Marques S, Adler H, Koszinowski UH, Simas JP, Efstathiou S (2002) K3-mediated evasion of CD8(+) T cells aids amplification of a latent gamma-herpesvirus. Nat Immunol 3: 733–740 [DOI] [PubMed] [Google Scholar]
- Tamura T, Chiba S, Chiba Y, Nakao T (1980) Virus excretion and neutralizing antibody response in saliva in human cytomegalovirus infection. Infect Immun 29: 842–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorley-Lawson DA, Poodry CA (1982) Identification and isolation of the main component (gp350–gp220) of Epstein–Barr virus responsible for generating neutralizing antibodies in vivo. J Virol 43: 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk SM, Jiang R, Chesnokova LS, Hutt-Fletcher LM (2006) Antibodies to gp350/220 enhance the ability of Epstein–Barr virus to infect epithelial cells. J Virol 80: 9628–9633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderplasschen A, Markine-Goriaynoff N, Lomonte P, Suzuki M, Hiraoka N, Yeh JC, Bureau F, Willems L, Thiry E, Fukuda M, Pastoret PP (2000) A multipotential beta-1,6-N-acetylglucosaminyl-transferase is encoded by bovine herpesvirus type 4. Proc Natl Acad Sci USA 97: 5756–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, Speck SH (1997) Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol 71: 5894–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Chen C, Ratcliffe NA (1999) Innate immunity in insects: the role of multiple, endogenous serum lectins in the recognition of foreign invaders in the cockroach, Blaberus discoidalis. J Immunol 162: 1590–1596 [PubMed] [Google Scholar]
- Xu J, Lyons PA, Carter MD, Booth TW, Davis-Poynter NJ, Shellam GR, Scalzo AA (1996) Assessment of antigenicity and genetic variation of glycoprotein B of murine cytomegalovirus. J Gen Virol 77: 49–59 [DOI] [PubMed] [Google Scholar]
- Yao QY, Rickinson AB, Epstein MA (1985) A re-examination of the Epstein–Barr virus carrier state in healthy seropositive individuals. Int J Cancer 35: 35–42 [DOI] [PubMed] [Google Scholar]
- Yao QY, Rowe M, Morgan AJ, Sam CK, Prasad U, Dang H, Zeng Y, Rickinson AB (1991) Salivary and serum IgA antibodies to the Epstein–Barr virus glycoprotein gp340: incidence and potential for virus neutralization. Int J Cancer 48: 45–50 [DOI] [PubMed] [Google Scholar]
- Zinkernagel RM, Hengartner H (2006) Protective ‘immunity' by pre-existent neutralizing antibody titers and preactivated T cells but not by so-called ‘immunological memory'. Immunol Rev 211: 310–319 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data Table 1