Although genetic linkage analysis is a vital tool for identifying disease genes, further study is often hindered by the lack of a known function for the corresponding gene products. In the case of hereditary multiple exostoses (HME), a dominantly inherited genetic disorder characterized by the formation of multiple cartilaginous tumors, extensive genetic analyses of affected families linked HME to mutations in two members of a novel family of putative tumor suppressor genes, EXT1 and EXT2. The biological function of the corresponding proteins, exostosin-1 (EXT1) and exostosin-2 (EXT2), has emerged in part by way of a serendipitous discovery made in the study of herpes simplex virology, which revealed that the pathogenesis of HME is linked to a defect in heparan sulfate (HS) biosynthesis. Biochemical analysis shows that EXT1 and EXT2 are type II transmembrane glycoproteins and form a Golgi-localized hetero-oligomeric complex that catalyzes the polymerization of HS. In this Perspective we will review the identification and characterization of the EXT family, with a particular focus on the biology of the EXT proteins in vivo, and we will explore their possible role(s) in both normal bone development and the formation of exostoses.
Hereditary multiple exostoses
Hereditary multiple exostoses (HME), an autosomal dominant bone disorder, is the most common type of benign bone tumor, with an estimated occurrence of 1 in 50,000–100,000 in Western populations. It is characterized by cartilage-capped tumors, known as osteochondromas or exostoses, which develop primarily on the long bones of affected individuals from early childhood until puberty (1). Individuals with HME are often short in stature with varying degrees of orthopedic deformity. Although exostoses are in themselves benign, surgery may be required to alleviate secondary complications such as joint pain and restricted movement. Also, recent evidence suggests that early removal of exostoses may ameliorate the growth retardation that typically leads to shortened bones in these individuals (2). While classical histopathology identifies exostoses as benign tumors, the histological organization of an exostosis shows remarkable similarities with the parent bone from which it arises. The cartilage cap of an exostosis often shows the same structural organization as that of growth plate cartilage, and the trabecular and periosteal bone are continuous with that of the underlying bone (1). A neoplastic model of pathogenesis for exostosis development and progression has recently been put forward as an alternative to the traditional skeletal dysplasia theory (3). According to this model, which is founded on the molecular biology of exostoses, aberrant bone growth would arise through clonal expansion of a single mutant chondrocyte, and not due to a defect in all cells of the growth-inducing tissue. Moreover, some studies have provided evidence that individuals with HME might have a significantly higher risk than the general population, 0.5–3%, of developing subsequent malignancies such as chondrosarcomas or osteosarcomas.
A highly conserved family of putative tumor suppressor genes
Although the pathogenesis of HME was first described nearly 200 years ago (1), it is only in the past few years that elegant genetic analyses have determined that both hereditary and sporadic cases of exostoses are linked to two main loci, EXT1 on chromosome 8q24.1 (4) and EXT2 on chromosome 11p11-p12 (5), with rare linkage to another locus, EXT3, on chromosome 19p (6). Because exostosis formation is associated with loss of heterozygosity at one or several EXT loci, it has been suggested that the EXT proteins have tumor suppressor function (4). The human EXT1 and EXT2 genes have both been cloned (7, 8), as have homologues in mice (9–11), the nematode Caenorhabditis elegans (9), and the fruit-fly Drosophila melanogaster (12). In addition, database searches have revealed the existence of three EXT-like (EXTL) genes, EXTL1 (13), EXTL2 (14), and EXTL3 (15, 16), whose products share amino acid homology with EXT1 and EXT2, particularly at their carboxy-termini. While none of the EXTL proteins are associated with HME, genetic analysis of various tumor types suggests that the EXTL proteins may also possess tumor suppressor activity. Altogether, the accumulating data suggest the existence of an EXT family of tumor suppressor genes.
EXT tumor suppressor proteins and HS biosynthesis
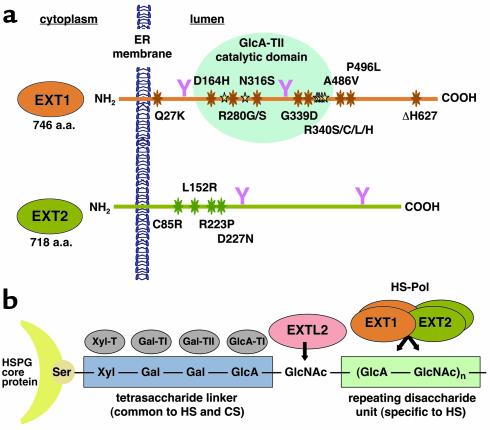
Human EXT1 and EXT2 (illustrated in Figure 1a) are ubiquitously expressed proteins of 746 and 718 amino acids, respectively, sharing about 70% similarity at the amino acid level (7, 8). EXT1 and EXT2 both have a predicted type II transmembrane glycoprotein structure: a short N-terminal cytoplasmic tail, a transmembrane domain, a stalk region, and a globular lumenal C-terminal tail (17, 18); and each protein localizes predominantly to the endoplasmic reticulum when overexpressed in cell culture (10, 17, 19).
Figure 1.
The EXT proteins and HS biosynthesis. (a) Schematic representations of the EXT1 and EXT2 putative tumor suppressor proteins (not drawn to scale). These endoplasmic reticulum–localized (ER-localized) type II transmembrane proteins have an N-terminal cytoplasmic tail, a single transmembrane domain, a stem region, and a long C-terminal lumenal tail. Amino acids (a.a.) mutated in HME patients are labeled and indicated by big stars, amino acids mutated in HS-deficient CHO cells (27) are indicated by small yellow stars, and N-linked glycosylation sites are represented by a pink Y. The proposed D-glucuronic acid glycosyltransferase (GlcA-TII) catalytic domain (27) is shaded in light blue. (b) A schematic representation of the HS biosynthesis pathway (recently reviewed in ref. 43). A tetrasaccharide unit, common to both HS and chondroitin sulfate (CS), is synthesized by sequential additions of xylose (Xyl), two galactose (Gal) residues, and a D-glucuronic acid (GlcA) residue, covalently linked to a serine residue on the HS proteoglycan (HSPG) core protein. HS biosynthesis is specifically initiated by the addition of an N-acetylglucosamine residue (GlcNAc), which is carried out by the glycosyltransferase (GlcNAc-TI) encoded by the EXTL2 gene. Next, the HS-polymerase (HS-Pol), a Golgi-localized hetero-oligomeric complex of which EXT1 and EXT2 are key components, elongates the nascent chain by adding alternating GlcA and GlcNAc residues.
Despite extensive genetic characterization, the function of the EXT proteins remained unknown until 1998, when the study of an HS-deficient cell line, sog9, revealed that EXT1 is involved in HS biosynthesis (17). Sulfated glycosaminoglycans (GAGs), including HS, are negatively charged oligosaccharide chains that decorate cell surface and ECM proteoglycans (PGs), playing important roles in ligand-binding, cell adhesion, and cell signaling (reviewed in ref. 20). Herpes simplex viruses (HSVs), like many other enveloped viruses, use HS as a primary receptor for attachment to the host cell (See Shukla and Spear, this Perspective series, ref. 21). Our laboratory screened for cDNAs capable of restoring susceptibility to HSV infection in the HS-deficient/HSV-resistant sog9 cell line. After several rounds of screening, a single cDNA was isolated that fully restored HS biosynthesis to the sog9 cell line, thereby rescuing HSV infectivity. Remarkably, DNA sequencing revealed that this cDNA encoded the putative tumor suppressor EXT1 (17) and that the HS-deficient sog9 cell line harbors a specific defect in the EXT1 gene (19). This cell line provides an in vivo functional assay for EXT1 function, because when a functional EXT1 gene is transfected into sog9 cells, the EXT1 defect is complemented, HS synthesis resumes, and the cells regain wild-type levels of susceptibility to HSV (17). This assay is specific for EXT1, because other EXT members are unable to complement the defect in HS biosynthesis (19).
Biochemical studies have confirmed that both EXT1 and its homologue, EXT2, possess the glycosyltransferase activities representative of an HS-polymerase (HS-Pol) in vitro, i.e., the ability to add single D-glucuronic acid (GlcA) and N-acetylglucosamine (GlcNAc) molecules to an artificial substrate molecule (18). Another member of the EXT gene family, EXTL2, encodes a functionally related enzyme, α1,4-N-acetylhexosaminyltransferase (22). As shown in Figure 1b, EXTL2 is proposed to initiate HS chain formation by transferring the first GlcNAc residue to a specific tetrasaccharide linker sequence on the HSPG core protein, thereby providing the substrate for polymerization by EXT1 and EXT2. By contrast, a study of C. elegans EXT homologues suggests that a single protein, Rib-2, which is most closely related to the human EXTL3 gene product, is able to carry out both the HS chain initiation and the polymerization steps (23), thereby demonstrating that the HS biosynthetic mechanism in C. elegans is distinct from that reported for the mammalian system. A recent report suggests that the human EXTL1 and EXTL3 genes also encode glycosyltransferases involved in HS biosynthesis (24), although another study has suggested that the human EXTL3 gene may encode a molecule with a strikingly different function, a cell surface receptor for the pancreatic β cell regeneration factor Reg (25).
One curious aspect of HME is that mutations in either EXT1 or EXT2 result in the formation of clinically indistinguishable exostoses, even though the two proteins do not appear to be functionally redundant in vivo (19). Insight into this problem came from two observations. First, EXT1 and EXT2 form a hetero-oligomeric complex in vivo that leads to an accumulation of both proteins in the Golgi apparatus (19). Second, Golgi-localized EXT1/EXT2 complexes possess substantially higher glycosyltransferase activity than either EXT1 or EXT2 alone (19). These results suggest a model (Figure 1b) in which a Golgi-localized EXT1/EXT2 heterocomplex represents the biologically relevant form of the HS-Pol enzyme. Solid support for this model has recently been provided by studies in yeast cells, which lack any endogenous HS-Pol activity (26). This model of the HS-Pol heterocomplex provides a convincing explanation of how inherited mutations in either of the two EXT genes might cause loss of HS biosynthesis activity, resulting in clinically identical HME.
More detailed information regarding the glycosyltransferase activity of EXT1 has come from a study involving HS-deficient mutant Chinese hamster ovary (CHO) cells, which were similarly rescued by human EXT1 expression (27). In this system, six reported missense mutations that clustered around a putative nucleotide sugar-binding domain were found to abolish GlcA-transferase (GlcA-T) but not GlcNAc-T activity, suggesting that the GlcA-T catalytic domain lies in the central region of the EXT1 protein.
Two-hybrid analysis has also been used to study the EXT proteins, and results indicate that fragments of the wild-type EXT proteins can interact with a chaperone protein and another glycosyltransferase enzyme. Significantly, these interactions are abrogated by a disease-causing HME mutation (28), suggesting that the EXT proteins may be components of a larger multienzyme GAG synthesis complex.
An important role for EXT and HS biosynthesis in vivo
The importance of HS, and therefore of EXT1 function, has been demonstrated in several in vivo models. Studies in Drosophila have identified an invertebrate homologue of EXT1 called tout-velu (Ttv), meaning “all-hairy” in reference to the corresponding mutant phenotype induced in fruit flies (12). Like human EXT1, Ttv appears to be a glycosylated type II integral membrane protein that is specifically involved in HSPG biosynthesis in vivo (12). In Drosophila, Ttv is required for diffusion of the segment-polarity protein hedgehog (Hh). It has been proposed that Ttv either facilitates endocytosis of the secreted Hh protein by target cells or permits Hh diffusion between responding cells in the developing wing (12). Hh, along with its vertebrate homologues, Indian hedgehog (Ihh), Desert hedgehog, and Sonic hedgehog, forms a conserved family of secreted proteins involved in short- and long-range intercellular signaling, necessary for the developmental patterning of tissues, including bone, foregut, and muscle. Hh proteins undergo autoproteolysis to give the active signaling form (Hh-N), an 18 kDa N-terminal fragment that is covalently coupled to a cholesterol moiety (for a detailed review of Hh biology see ref. 29). Total ablation of ttv is fatal at the pupal stage (30). Analysis of somatic clones that lack Ttv function indicates that this gene product is needed to allow short-range diffusion of Hh and therefore for the induction of downstream genes required for normal development (12). Interestingly, while biochemical studies have confirmed that ttv mutants exhibit a marked reduction in HS biosynthesis in vivo, it appears that not all HSPG-dependent signaling pathways are equally impaired (31). This raises the possibility that Ttv may not be the only HS-Pol in Drosophila, a hypothesis that is supported by the existence of a Drosophila EXT2 homologue. This would imply that Ttv may be a selective HS-Pol, only decorating Hh-specific PGs with HS. Alternatively, Hh diffusion may be extremely restrictive, requiring HSPGs, while other GAGs such as chondroitin sulfate may be able to replace HS for other signaling molecules (31).
An EXT1 knockout mouse model has shown that EXT1 is essential for HS biosynthesis in early embryonic development and that ablation of EXT1 results in embryonic lethality due to a failure to gastrulate (32). Interestingly, despite a 50% reduction in HS expression, heterozygous EXT1+/– mice do not develop exostoses, although their bone length is diminished by 10%. Recent studies of diseased chondrocytes from HME patients have revealed several abnormalities in cell structure, including a stellate appearance and an atypical cytoskeleton characterized by a very rigid structure made up of huge bundles of α-actin striated with α-actinin (33). Thus, disruptions in HS biosynthesis may lead to abnormal maturation of human chondrocytes.
Investigating the molecular etiology of HME
Extensive genetic analysis of genomic DNA from HME patients has led to the identification of a number of mutations in both EXT1 and EXT2 that appear to be disease-related. The majority of the mutations are splice-site, frameshift, or nonsense mutations that result in premature termination. However, a small number of single amino acid (missense) mutations have been identified, repeatedly in some cases, mostly affecting amino acids located in the N-terminal half of the EXT1 and EXT2 proteins (shown schematically in Figure 1). The sog9 cell HSV infectivity assay has proven to be particularly useful for studying the function of these etiological mutant EXT1 proteins, as it measures in vivo HS biosynthesis and transduction to the cell surface, unlike in vitro enzyme assays, which only measure the addition of a single sugar to an artificial substrate. Indeed, McCormick et al. (19) found that neither of two previously reported etiological EXT1 alleles, bearing missense mutations G339D and R340C, can rescue HS biosynthesis in this assay. By contrast, recent results obtained by extensive analysis of all 12 putative etiological mutations identified to date in the EXT1 gene have revealed that four of the reported mutations do not abrogate HS biosynthesis in vivo (34). Moreover, neither substitution nor deletion of each of these four residues was found to eliminate HS biosynthesis, indicating that several of the putative etiological mutants retain the ability to synthesize and express HS on the cell surface. It is possible that the corresponding missense mutations represent rare genetic polymorphisms in the EXT1 gene or that they interfere with as-yet undefined functions of EXT1 that are involved in HME pathogenesis. Analysis of each of the nontruncating mutations in EXT2 may prove to be useful in pinpointing the different functional domains of the second EXT protein, thereby helping to further elucidate the molecular mechanisms responsible for the pathology of HME.
A working model for the formation of exostoses
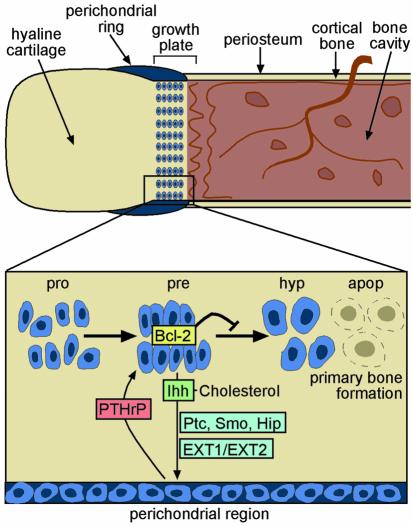
Recent advances in the understanding of the molecular biology of endochondral ossification have also proved valuable to the understanding of HME. It is well established that proper bone development depends upon the tight regulation of the cartilage progenitor cells, the growth plate chondrocytes, which go through subsequent stages of proliferation, prehypertrophy, hypertrophy, and finally apoptosis. Elegant in vivo studies have shown that two signaling molecules, the mammalian Hh homologue Ihh and parathyroid hormone-related peptide (PTHrP), negatively regulate chondrocyte progression from proliferation to hypertrophy in a coordinated way. According to the current signaling model (illustrated in Figure 2), the intermediate prehypertrophic chondrocytes localized within the growth plate, also known as borderline chondrocytes, produce Ihh, which binds to its receptor, Patched (Ptc), on the osteogenic cells of the periarticular perichondral region. This signal stimulates chondrocyte proliferation by upregulating the second signaling molecule, PTHrP. PTHrP binds to the PTH/PTHrP receptor on a subpopulation of proliferating and prehypertrophic chondrocytes, postponing differentiation and eventual cell death by inducing production of Bcl-2, a well known antiapoptotic protein (reviewed in ref. 35). This feedback loop favors continued longitudinal cartilage growth until a shift in the expression of Ihh or PTHrP disrupts the equilibrium, leading to chondrocyte apoptosis and subsequent ossification. Consistent with this model, knockout mice missing either PTHrP or its receptor are small, with excessive and unmodulated bone formation and prematurely ossified growth plates.
Figure 2.
A working model of mammalian bone development. Hypothetical representation of the growth plate during endochondral bone formation (adapted from ref. 44). Prehypertrophic chondrocytes (pre) localized within the growth plate produce a cell signaling molecule, Ihh, which diffuses to the receiving cells via HS proteoglycans (HSPGs) that are glycosylated by EXT1 and EXT2 HS-Pol activity. By analogy with what is known about the Hh signaling cascade in Drosophila, it is likely that signaling is initiated when Ihh binds to a two-component receptor/signal transducer complex on the surface of a responding cell, which is composed of both Patched (Ptc) and Smoothened (Smo). When Ihh binds to Ptc, Smo is de-repressed and additional intracellular components of the signaling pathway are activated (reviewed in ref. 29). Recent work suggests that there may be an additional member in this cascade, Hedgehog-interacting protein (Hip), which could act to attenuate Ihh signaling. In bone, Ptc is expressed on the osteogenic cells of the perichondral region, and Ihh binding induces chondrocyte proliferation by upregulating a second signaling molecule, PTHrP. PTHrP binds to the PTH/PTHrP receptor on a subpopulation of proliferating (pro) and prehypertrophic chondrocytes, thereby inducing production of Bcl-2, a well known antiapoptotic protein. In the absence of negative feedback, chondrocytes differentiate into hypertrophic chondrocytes (hyp), which undergo apoptosis and are then replaced by bone-forming osteoblasts. According to this model, hereditary exostosis formation would occur when a chondrocyte bearing a germline mutation in either EXT1 or EXT2 develops an inactivating “hit” in the second copy of the same gene, thereby abrogating HS expression. Clonal expansion of this mutant chondrocyte would then lead to a local perturbation in Ihh diffusion with, as a result, release from negative feedback control. The neighboring population of proliferating chondrocytes would therefore undergo premature chondrocyte differentiation, apoptosis, and subsequent ossification.
Insights into the role played by the EXT proteins in normal bone development have come from an unexpected source, the invertebrate D. melanogaster. Since the Drosophila EXT1 homologue Ttv is responsible for the synthesis of HSPGs that specifically enhance Hh diffusion during development (12), mammalian EXT proteins probably synthesize HSPGs that are required for the diffusion and/or efficient signaling by Ihh in the growth plate of developing bone. Consistent with this model (illustrated in Figure 2), in situ hybridization studies in wild-type mice have shown that EXT1 and EXT2 are expressed in the proliferative and prehypertrophic chondrocytes, but not in the hypertrophic zone, and that their expression pattern overlaps with that of Ihh (36). Furthermore, Lin et al. (32) showed recently that Ihh is incapable of associating with the cell surface of target cells in murine EXT1–/– embryos, indicating that HS expression is essential for Ihh binding.
Such a role for EXT in normal bone formation would help to explain the etiology of HME; a defect in HS biosynthesis due to a second, somatic mutation in EXT1 or EXT2 would cause a localized disruption in the negative feedback loop that regulates chondrocyte proliferation and maturation, permitting premature differentiation, and thus aberrant bone growth in the immediate region. Furthermore, taking into account the wide range of ligands bound by HS, it has been proposed that HS, and thus the EXT proteins, may also negatively regulate bone growth by mediating the binding of additional factors such as bone morphogenic proteins and FGFs (32, 36). In agreement with this, it has been proposed that a general function of both transmembrane and glycosylphosphatidylinositol-linked HSPGs could be to deliver ligand, such as growth factors, to neighboring cells (37).
Other PG-related bone disorders
Considering the key roles that PGs, and their GAG moieties in particular, play in morphogenesis and growth regulation in both vertebrates and invertebrates (for recent reviews on PGs see refs. 37–41), it is not surprising that a gene involved in the biosynthesis of HS is at the root of a developmental disorder of bone. HS, along with several other GAGs such as chondroitin sulfate, dermatan sulfate, and keratan sulfate, decorate PGs such as betaglycan, perlecan, and members of the syndecan and glypican families, which are all present in bone and cartilage. Studies of PGs and their ligands indicate that GAG-ligand interactions are critical for the coordination of proper bone development. For example, perturbations in the levels of the HS-ligand FGF-2 accelerate ossification of growth plate cartilage in vivo and in vitro, while targeted disruption in mice of glypican-3, perlecan, or biglycan leads to various skeletal defects, indicating that each of these PGs is important for normal cartilage and endochondral ossification. Furthermore, syndecan-3 has been found to regulate chondrocyte proliferation and maturation (reviewed in ref. 42), while syndecan-1 (particularly its HS chains) appears to enhance differentiation of bone-forming osteoblasts and to reduce differentiation of bone-resorbing osteoclasts. Consistent with these observations, a number of human skeletal dysplasias are known to be caused by changes in molecular interactions that rely on HS or other GAGs. For instance, achondroplasia, the most common form of dwarfism, hypochondroplasia, and thanatophoric dysplasia are all caused by mutations in the gene encoding the FGF receptor 3, a cell surface protein that binds FGF only in the presence of HS. Diastrophic dysplasia and multiple epiphyseal dysplasia have both been linked to mutations in a sulfate transporter gene, which probably results in undersulfated glycoproteins and GAGs. When all of these observations are considered together, it is evident that bone development is a process exquisitely sensitive to changes in PGs, particularly in GAG expression and structure.
EXT and HME: the unanswered questions
A wealth of information has been generated in the last few years concerning the EXT family of putative tumor suppressors, due to important contributions coming from many diverse disciplines including genetics, cell and developmental biology, and biochemistry. Each study has helped to give us a clearer picture of the link between the glycosyltransferase enzymes of the HS biosynthesis pathway and the process of endochondral bone formation. Not only have these advances contributed to our understanding of glycobiology in vertebrates and invertebrates, but they have, in some cases, suggested practical applications including a greater ease of diagnosis and treatment in the clinical setting for individuals with hereditary bone tumors. Future research will continue to focus on the molecular pathogenesis of HME, with a detailed analysis of the known etiological mutant EXT proteins and of the corresponding diseased chondrocytes isolated from HME patients. In addition, researchers will begin to address several important unresolved issues, including the preoccupying question of why a dominantly inherited syndrome, caused by a germline defect in either of two ubiquitously expressed proteins, EXT1 or EXT2, only manifests itself in bone tissue. From a clinical perspective, the underlying basis of pathogenesis for patients that do not harbor detectable defects in EXT1 or EXT2 remains to be determined. Despite these questions, there has been tremendous progress in understanding the molecular players involved in HME. It is likely that rapid progress will be made toward understanding the connections between HS biosynthesis and HME, and perhaps more importantly, whether EXT proteins harbor additional functions not yet identified.
Acknowledgments
This work was supported by grants to F. Tufaro from the Medical Research Council of Canada and the Canadian Genetic Diseases Network. G. Duncan is supported by the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Craig McCormick’s present address is: Department of Microbiology and Immunology, University of California, San Francisco, San Francisco, California, USA.
References
- 1.Solomon L. Hereditary multiple exostosis. J Bone J Surg. 1963;45:292–304. [Google Scholar]
- 2.Porter DE, Emerton ME, Villanueva-Lopez F, Simpson AH. Clinical and radiographic analysis of osteochondromas and growth disturbance in hereditary multiple exostoses. J Ped Ortho. 2000;20:246–250. [PubMed] [Google Scholar]
- 3.Porter DE, Simpson AH. The neoplastic pathogenesis of solitary and multiple osteochondromas. J Pathol. 1999;188:119–125. doi: 10.1002/(SICI)1096-9896(199906)188:2<119::AID-PATH321>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 4.Lüdecke HJ, et al. Molecular dissection of a contiguous gene syndrome: localization of the genes involved in the Langer-Giedion syndrome. Hum Mol Genet. 1995;4:31–36. doi: 10.1093/hmg/4.1.31. [DOI] [PubMed] [Google Scholar]
- 5.Wuyts W, et al. Refinement of the multiple exostoses locus (EXT2) to a 3-cM interval on chromosome 11. Am J Hum Genet. 1995;57:382–387. [PMC free article] [PubMed] [Google Scholar]
- 6.Le Merrer M, et al. A gene for hereditary multiple exostoses maps to chromosome 19p. Hum Mol Genet. 1994;3:717–722. doi: 10.1093/hmg/3.5.717. [DOI] [PubMed] [Google Scholar]
- 7.Wuyts W, et al. Positional cloning of a gene involved in hereditary multiple exostoses. Hum Mol Genet. 1996;5:1547–1557. doi: 10.1093/hmg/5.10.1547. [DOI] [PubMed] [Google Scholar]
- 8.Ahn J, et al. Cloning of the putative tumour suppressor gene for hereditary multiple exostoses (EXT1) Nat Genet. 1995;11:137–143. doi: 10.1038/ng1095-137. [DOI] [PubMed] [Google Scholar]
- 9.Clines GA, Ashley JA, Shah S, Lovett M. The structure of the human multiple exostoses 2 gene and characterization of homologs in mouse and Caenorhabditis elegans. Gen Res. 1997;7:359–367. doi: 10.1101/gr.7.4.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin X, Wells D. Isolation of the mouse cDNA homologous to the human EXT1 gene responsible for hereditary multiple exostoses. DNA Seq. 1997;7:199–202. doi: 10.3109/10425179709034035. [DOI] [PubMed] [Google Scholar]
- 11.Stickens D, Evans GA. Isolation and characterization of the murine homolog of the human EXT2 multiple exostoses gene. Biochem Mol Med. 1997;61:16–21. doi: 10.1006/bmme.1997.2588. [DOI] [PubMed] [Google Scholar]
- 12.Bellaiche Y, The I, Perrimon N. Tout-velu is a Drosophila homologue of the putative tumour suppressor Ext-1 and is needed for Hh diffusion. Nature. 1998;394:85–88. doi: 10.1038/27932. [DOI] [PubMed] [Google Scholar]
- 13.Wise CA, Clines GA, Massa H, Trask BJ, Lovett M. Identification and localization of the gene for EXTL, a third member of the multiple exostoses gene family. Genome Res. 1997;7:10–16. doi: 10.1101/gr.7.1.10. [DOI] [PubMed] [Google Scholar]
- 14.Wuyts W, et al. Identification and characterization of a novel member of the EXT gene family, EXTL2. Eur J Hum Genet. 1997;5:382–389. [PubMed] [Google Scholar]
- 15.Van Hul W, et al. Identification of a third EXT-like gene (EXTL3) belonging to the EXT gene family. Genomics. 1998;47:230–237. doi: 10.1006/geno.1997.5101. [DOI] [PubMed] [Google Scholar]
- 16.Saito T, et al. Structure, chromosomal location, and expression profile of EXTR1 and EXTR2, new members of the multiple exostoses gene family. Biochem Biophys Res Comm. 1998;243:61–66. doi: 10.1006/bbrc.1997.8062. [DOI] [PubMed] [Google Scholar]
- 17.McCormick C, et al. The putative tumour suppressor EXT1 alters the expression of cell-surface heparan sulfate. Nat Genet. 1998;19:158–161. doi: 10.1038/514. [DOI] [PubMed] [Google Scholar]
- 18.Lind T, Tufaro F, McCormick C, Lindahl U, Lidholt K. The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J Biol Chem. 1998;273:26265–26268. doi: 10.1074/jbc.273.41.26265. [DOI] [PubMed] [Google Scholar]
- 19.McCormick C, Duncan G, Goutsos KT, Tufaro F. The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc Natl Acad Sci USA. 2000;97:668–673. doi: 10.1073/pnas.97.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salmivirta M, Lidholt K, Lindahl U. Heparan sulfate: a piece of information. FASEB J. 1996;10:1270–1279. doi: 10.1096/fasebj.10.11.8836040. [DOI] [PubMed] [Google Scholar]
- 21.Shukla D, Spear PG. Herpesvirus and heparan sulfate: an intimate relationship in aid of viral entry. J Clin Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitagawa H, Shimakawa H, Sugahara K. The tumor suppressor EXT-like gene EXTL2 encodes an alpha 1, 4-N-acetylhexosaminyltransferase that transfers N-acetylgalactosamine and N-acetylglucosamine to the common glycosaminoglycan-protein linkage region. The key enzyme for the chain initiation of heparan sulfate. J Biol Chem. 1999;274:13933–13937. doi: 10.1074/jbc.274.20.13933. [DOI] [PubMed] [Google Scholar]
- 23.Kitagawa H, et al. rib-2, a Caenorhabditis elegans homolog of the human tumor suppressor EXT genes encodes a novel alpha1,4-N-acetylglucosaminyltransferase involved in the biosynthetic initiation and elongation of heparan sulfate. J Biol Chem. 2001;276:4834–4838. doi: 10.1074/jbc.C000835200. [DOI] [PubMed] [Google Scholar]
- 24.Kim BT, et al. Human tumor suppressor EXT gene family members EXTL1 and EXTL3 encode alpha 1,4- N-acetylglucosaminyltransferases that likely are involved in heparan sulfate/ heparin biosynthesis. Proc Natl Acad Sci USA. 2001;98:7176–7181. doi: 10.1073/pnas.131188498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi S, et al. Identification of a receptor for reg (regenerating gene) protein, a pancreatic beta-cell regeneration factor. J Biol Chem. 2000;275:10723–10726. doi: 10.1074/jbc.275.15.10723. [DOI] [PubMed] [Google Scholar]
- 26.Senay C, et al. The EXT1/EXT2 tumor suppressors: catalytic activities and role in heparan sulfate biosynthesis. EMBO Rep. 2000;1:282–286. doi: 10.1093/embo-reports/kvd045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei G, et al. Location of the glucuronosyltransferase domain in the heparan sulfate copolymerase EXT1 by analysis of Chinese hamster ovary cell mutants. J Biol Chem. 2000;275:27733–27740. doi: 10.1074/jbc.M002990200. [DOI] [PubMed] [Google Scholar]
- 28.Simmons AD, et al. A direct interaction between EXT proteins and glycosyltransferases is defective in hereditary multiple exostoses. Hum Mol Genet. 1999;8:2155–2164. doi: 10.1093/hmg/8.12.2155. [DOI] [PubMed] [Google Scholar]
- 29.Ingham PW. Transducing Hedgehog: the story so far. EMBO J. 1998;17:3505–3511. doi: 10.1093/emboj/17.13.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The I, Bellaiche Y, Perrimon N. Hedgehog movement is regulated through Tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol Cell. 1999;4:633–639. doi: 10.1016/s1097-2765(00)80214-2. [DOI] [PubMed] [Google Scholar]
- 31.Toyoda H, Kinoshita-Toyoda A, Fox B, Selleck SB. Structural analysis of glycosaminoglycans in animals bearing mutations in sugarless, sulfateless, and tout-velu. Drosophila homologues of vertebrate genes encoding glycosaminoglycan biosynthetic enzymes. J Biol Chem. 2000;275:21856–21861. doi: 10.1074/jbc.M003540200. [DOI] [PubMed] [Google Scholar]
- 32.Lin X, et al. Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice. Dev Biol. 2000;224:299–311. doi: 10.1006/dbio.2000.9798. [DOI] [PubMed] [Google Scholar]
- 33.Bernard MA, et al. Cytoskeletal abnormalities in chondrocytes with EXT1 and EXT2 mutations. J Bone Miner Res. 2000;15:442–450. doi: 10.1359/jbmr.2000.15.3.442. [DOI] [PubMed] [Google Scholar]
- 34.Cheung PK, et al. Etiological point mutations in the hereditary multiple exostoses gene Ext1: a functional analysis of heparan sulfate polymerase activity. Am J Hum Genet. 2001;69:55–66. doi: 10.1086/321278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horton WE, Jr, Feng L, Adams C. Chondrocyte apoptosis in development, aging and disease. Matrix Biol. 1998;17:107–115. doi: 10.1016/s0945-053x(98)90024-5. [DOI] [PubMed] [Google Scholar]
- 36.Stickens D, Brown D, Evans GA. EXT genes are differentially expressed in bone and cartilage during mouse embryogenesis. Dev Dyn. 2000;218:452–464. doi: 10.1002/1097-0177(200007)218:3<452::AID-DVDY1000>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 37.Lander AD, Selleck SB. The elusive functions of proteoglycans: in vivo veritas. J Cell Biol. 2000;148:227–232. doi: 10.1083/jcb.148.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perrimon N, Bernfield M. Specificities of heparan sulphate proteoglycans in developmental processes. Nature. 2000;404:725–728. doi: 10.1038/35008000. [DOI] [PubMed] [Google Scholar]
- 39.Bernfield M, et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 40.Selleck SB. Proteoglycans and pattern formation: sugar biochemistry meets developmental genetics. Trends Genet. 2000;16:206–212. doi: 10.1016/s0168-9525(00)01997-1. [DOI] [PubMed] [Google Scholar]
- 41.Iozzo RV. Matrix proteoglycans: from molecular design to cellular function. Annu Rev Biochem. 1998;67:609–652. doi: 10.1146/annurev.biochem.67.1.609. [DOI] [PubMed] [Google Scholar]
- 42.Kosher RA. Syndecan-3 in limb skeletal development. Microsc Res Tech. 1998;43:123–130. doi: 10.1002/(SICI)1097-0029(19981015)43:2<123::AID-JEMT5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 43.Sugahara K, Kitagawa H. Recent advances in the study of the biosynthesis and functions of sulfated glycosaminoglycans. Curr Opin Struct Biol. 2000;10:518–527. doi: 10.1016/s0959-440x(00)00125-1. [DOI] [PubMed] [Google Scholar]
- 44.Stickens D, Evans GA. A sugar fix for bone tumors? Nat Genet. 1998;19:110–111. doi: 10.1038/458. [DOI] [PubMed] [Google Scholar]