Abstract
Both genotoxic and oncogenic stress activates the nuclear factor-kappa B (NF-κB) and p53 proteins; however, the p53 activity is antagonized by NF-κB signaling. Dmp1 is a Myb-like transcription factor that activates the Arf-p53 pathway. The Dmp1 promoter was activated by a classical NF-κB activator tumor necrosis factor α, but repressed by treatment of cells with non-classical NF-κB activators, anthracyclins and UV-C. p65 and other subsets of NF-κB proteins were bound to the Dmp1 promoter following anthracyclin/UV-C treatment of rodent fibroblasts. This resulted in the downregulation of Dmp1 mRNA and protein. Repression of the Dmp1 transcription by anthracyclins depended on the unique NF-κB site on the promoter. Downregulation of p65 significantly attenuated the repression of the Dmp1 promoter by anthracyclins/UV-C. The amount of Dmp1 bound to the Arf promoter decreased significantly upon anthracyclin treatment; this, in turn, downregulated the Arf levels. Repression of the Arf promoter by p65 or anthracyclins depended on Dmp1, which was significantly attenuated in Dmp1−/− cells. Both Dmp1−/−and Arf−/−cells showed resistance to anthracyclin-induced cell death compared to wild-type cells; non-immortalized p65-knockdown cells were much more sensitive. Thus, the Dmp1-Arf pathway is repressed by p65 in response to genotoxic stress, which implicates a novel mechanism of p53 inactivation by NF-κB.
Keywords: Dmp1, Arf, p53, NF-κB, anthracyclin, UV-C
Introduction
The nuclear factor-kappa B (NF-κB) transcription factors were originally isolated and characterized for controlling gene expression and function in the immune system (Hayden and Ghosh, 2004). However, the importance of NF-κB in modulating stress signaling, cellular cycles, apoptosis and carcinogenesis has also been well documented (Karin et al., 2002; Hayden and Ghosh, 2004; Karin, 2006). In unstimulated cells, the NF-κB dimers are bound by inhibitory IκB molecules and stay in the cytoplasm. Since transcriptional activation by NF-κB requires its nuclear translocation, signal-induced degradation of IκB molecules is considered to be critical in NF-κB activation (Hayden and Ghosh, 2004). There are five distinct subunits of NF-κB, namely p65 (relA), relB, c-rel, p50/p105 (NF-κB1) and p52/p100 (NF-κB2). Among these, p50:p65 heterodimers are most abundantly expressed subunits in mammalian cells while p52 and c-rel subunits are mainly detected in hemato-poietic/lymphoid cells (Hayden and Ghosh, 2004, Karin et al., 2002). Owing to the presence of a strong transactivation domain, p65 is responsible for most NF-κB transcriptional activity.
Generally, NF-κB is a well-documented anti-apoptotic factor, which is simultaneously activated with p53 in response to chemotherapeutic agents’ DNA damage, oncogenes and by tumor necrosis factor α (TNFα) (for review see, Perkins, 2004). These transcription factors modulate each other’s activities. For instance, the p53 promoter is activated by NF-κB in response to TNFα and to anticancer drugs. NF-κB plays essential roles in p53-mediated programmed cell death (Ryan et al., 2000). On the other hand, p53 stabilization is decreased upon NF-κB activation by chemotherapeutic reagents through the upregulation of Mdm2 (Tergaonkar et al., 2002). Competition for limiting pools of transcriptional coactivators p300 and CBP has also been reported to mediate a mutual repression between NF-κB and p53 (Webster and Perkins, 1999). The third mechanism of antagonism involves the activation of anti-apoptotic genes by NF-κB, which competes with proapoptotic pathways activated by p53 (Tergaonkar et al., 2002; Perkins, 2004). Indeed, many human cancers have constitutive activation of NF-κB and inhibition of NF-κB has been documented to lead to increased efficacy of anti-cancer drugs (Karin et al., 2002; Karin, 2006).
The activity of p53 is positively regulated by p19Arf (p14ARF in humans) in response to oncogenic stress. p19Arf is an alternative reading frame gene product generated from the Ink4a/Arf locus (Sherr, 2001; Lowe and Sherr, 2003). Since the single genetic locus encodes two independent tumor suppressor proteins that regulate the p53 and the Rb pathways, this locus is often disrupted in human cancer (Ruas and Peters, 1998). Arf is induced by potentially harmful growth-promoting signals stemming from overexpression of various oncoproteins (Sherr, 2001; Lowe and Sherr, 2003). This drives incipient cancer cells to undergo p53-dependent and p53-independent cell cycle arrest or apoptosis, providing a powerful mode of tumor suppression (Sherr 2001, 2006). Among known Arf activators, the Dmp1 transcription factor (cyclin D binding myb-like protein-1, also called Dmtf1) is a unique tumor suppressor (Inoue et al., 2000, 2001, 2007). Dmp1 was originally isolated in a yeast two-hybrid screen of a murine T-lymphocyte library with cyclin D2 as bait. Importantly, Dmp1 directly binds to the Arf promoter to activate its expression, thereby inducing p53-dependent cell cycle arrest (Inoue et al., 1999). Dmp1-null mice are prone to spontaneous tumor development, which was accelerated when the animals were neonatally treated with ionizing radiation or dimethylbenzanthracene (Inoue et al., 2000, 2001). Dmp1 is haplo-insufficient for tumor suppression and is a physiological regulator of the Arf-p53 pathway in vivo (Inoue et al., 2001). The Dmp1 promoter is activated by the oncogenic Ras-Raf-MEK-ERK pathway, and the induction of Arf by Ras is Dmp1-dependent (Sreeramaneni et al., 2005). On the other hand, the Dmp1 promoter is repressed by overexpression of E2Fs and physiological mitogenic signaling. Thus, Dmp1 is a novel marker of cells that have exited from the cell cycle (Mallakin et al., 2006).
In the present study, we examined the roles of NF-κB in the regulation of the Dmp1-Arf tumor suppressor pathway, especially through the non-classical NF-κB pathways activated by anthracyclins and UV-C. Here we demonstrate that NF-κB directly binds and inhibits the Dmp1 promoter and decreases its expression in response to genotoxic stress.
Results
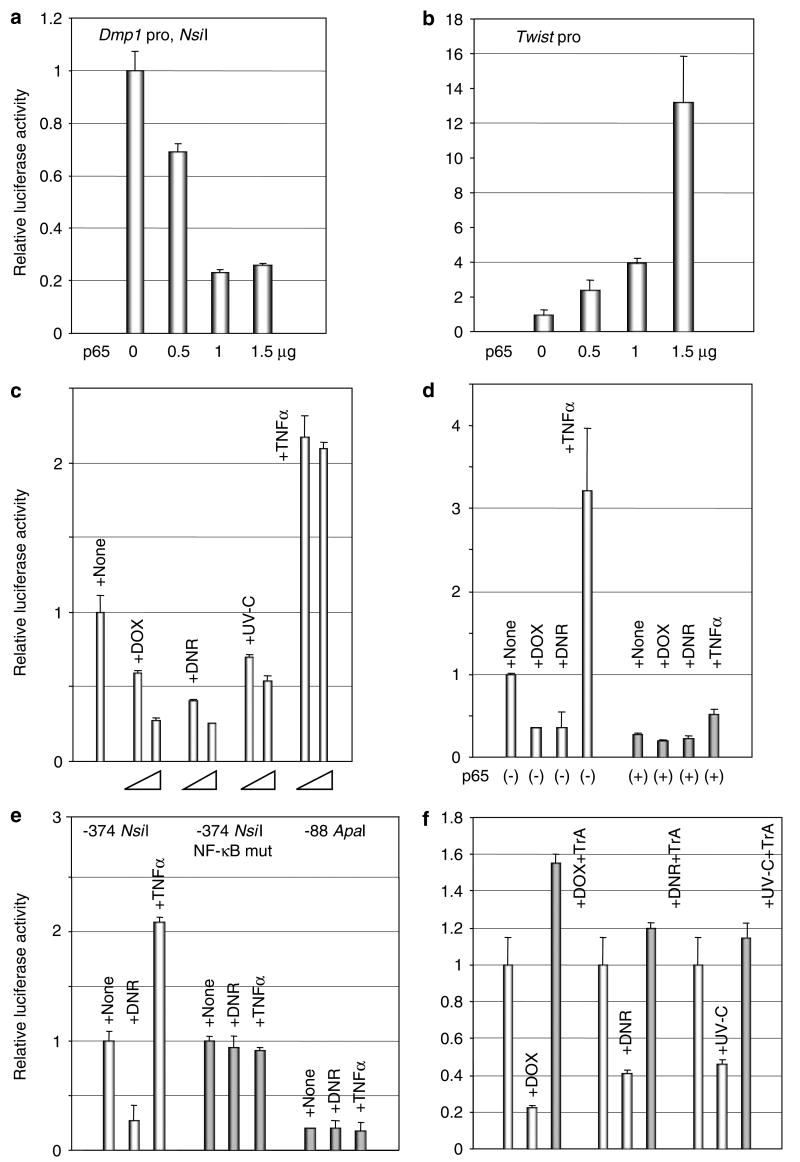
Responsiveness of the Dmp1 promoter to p65 and atypical NF-κB activators
We studied the responsiveness of the Dmp1 promoter to p50 and p65, the most ubiquitously expressed subunits of NF-κB in mammalian cells. Although p50 had little effect on the Dmp1 promoter (data not shown), p65 efficiently inhibited its transcription (~fivefold) (Figure 1a). Conversely, the Twist promoter was activated considerably by p65, demonstrating that repression of the Dmp1 promoter by p65 was very specific (Figure 1b). The Dmp1 promoter was efficiently repressed by all of the non-classical NF-κB activators (doxorubicin (DOX), daunorubicin (DNR)) or by UV-C, although the effect was more striking with anthracyclins (~75% repression) than with UV-C (~50% repression) (Figure 1c). Interestingly, the Dmp1 promoter was activated by stimulation of murine embryonic fibroblasts (MEFs) with TNFα (Figure 1c). To study the degree to which repression of the Dmp1 promoter by anthracyclins depended on NF-κB, a reporter assay was performed by transfecting the cells with a p65 expression vector (Figure 1d). The basal Dmp1 promoter levels decreased in cells transfected with p65 and the promoter became less responsive to DOX, DNR or TNFα (Figure 1d). The responsiveness of the Dmp1 promoter to TNFα or genotoxic stress was completely lost by mutation of the unique NF-κB site of the promoter (−374 NsiI NF-κB mut) or in the deletion construct that lacks this unique NF-κB site (−88 ApaI) (Figure 1e, Supplementary Figure 1). Furthermore, the repression of the Dmp1 promoter was completely (DOX, DNR) or partially (UV-C) reversed by the addition of Trichostatin A to the tissue culture media, implicating the involvement of histone deacetylases in Dmp1 promoter repression (Figure 1f). These results suggest that transcriptional repression of the Dmp1 promoter by DOX, DNR or UV-C depends on the unique NF-κB binding site on the Dmp1 promoter and involves histone deacetylases.
Figure 1.
Responsiveness of the murine Dmp1 promoter to anthracyclins and UV-C. (a) The Dmp1 promoter (−374 NsiI fragment; Supplementary Figure 1) is repressed by p65 overexpression in 3T3 cells. (b) The classical nuclear factor-kappa B (NF-κB) target, the Twist promoter is strongly activated by p65. (c) Responsiveness of the Dmp1 promoter to doxorubicin (DOX), daunorubicin (DNR), UV-C or to tumor necrosis factorα (TNFα). NIH 3T3 cells were transfected with the −374 NsiI construct and were treated with DOX (2–5 μM), DNR (1–2 μM), UV-C (20–40 J/m2) or TNFα (10–20 ng/ml). Reporter assays were performed 14 h after the treatment of cells with DOX, DNR or UV-C and 2 h for TNFα. (d) The responsiveness of the Dmp1 promoter to anthracyclins and UV-C is subverted by overexpressing p65. (e) Repression and activation of the Dmp1 promoter by DNR and TNFα are dependent on the unique NF-κB site on the promoter. (f) Repression of the Dmp1 promoter by DOX, DNR or UV-C is histone deacetylase-dependent. The Dmp1 promoter assay was conducted in the presence (black bars) or absence (white bars) of the HDAC inhibitor, Trichostatin A (TrA).
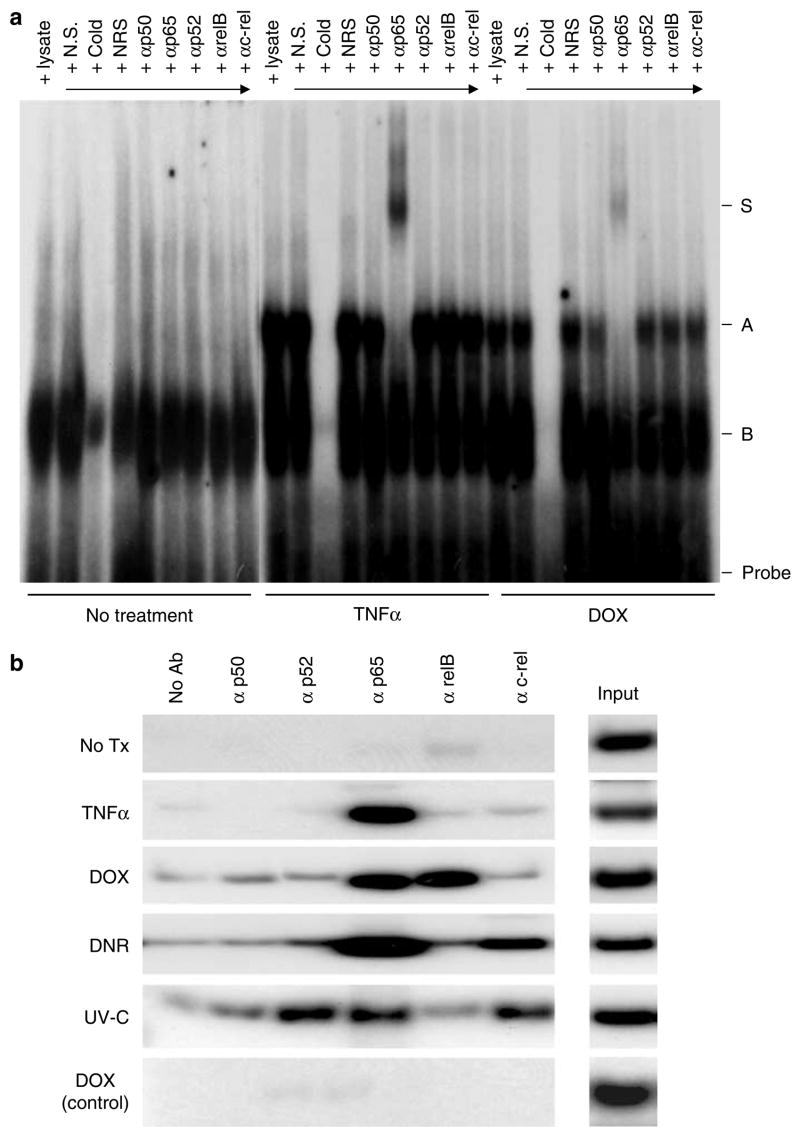
NF-κB binds to the Dmp1 promoter upon TNFα and anthracyclin treatment
To confirm the binding of NF-κB proteins to the Dmp1 promoter in response to TNFα and anthracyclins, an electrophoretic mobility shift assay (EMSA) was performed with a probe covering the NF-κB consensus sequence on the Dmp1 promoter. When 3T3 cells were treated with TNFα, a specific complex A was formed and was supershifted with antibody to p65 (complex S, Figure 2a, middle panel). Other NF-κB proteins (p50, p52, relB and c-rel) were not found to associate with the Dmp1 promoter upon TNFα treatment (Figure 2a, middle panel). When the cells were treated with 5 μM DOX for 90 min, complex A was generated, which was supershifted by incubating the complex with antibody to p65 (Figure 2a, right panel). This supershift was not observed with antibodies to p50, p52, relB or c-rel (Figure 2a, right panel). The pattern of EMSA and supershift assay with 1 μM DNR treatment was essentially identical with that of DOX, that is, p65 was the major component directly bound to the Dmp1 promoter (data not shown). NF-κB binding to the Dmp1 promoter was also studied by chromatin immunoprecipitation (ChIP) in MEFs (Figure 2b). p65 was the only subunit detected on the Dmp1 promoter upon TNFα stimulation, as predicted from the EMSA (Figure 2b, second panel). p65 and relB were bound to the Dmp1 promoter on DOX treatment of the cells, whereas p65 and c-rel were bound upon DNR treatment (Figure 2b, third and fourth panels). On the other hand, p52, p65 and c-rel were found on the Dmp1 promoter upon UV-C treatment (Figure 2b, fifth panel). The specificity of NF-κB binding to the proximal region of the Dmp1 promoter was confirmed by PCR amplification of the genomic DNA fragment located 2 kb upstream from the transcription initiation site as a control (Figure 2b, bottom panel). Taken together, our data suggest that (i) homodimers of p65 activate the Dmp1 promoter in response to TNFα and (ii) subsets of NF-κB proteins, including p65, repress the Dmp1 promoter upon anthracyclin and UV-C treatment of rodent fibroblasts.
Figure 2.
Binding of nuclear factor-kappa B (NF-κB) proteins to the Dmp1 promoter. (a) Left and middle panels: 3T3 cells were treated with or without 20 ng/ml of tumor necrosis factorα (TNFα) for 30 min and electrophoretic mobility shift assay (EMSA) was performed with oligonucleotides that covered the NF-κB consensus sequence (−229 to −238; Supplementary Figure 1a). NS: non-specific cold oligonucleotides; Cold: non-labeled oligonucleotide probe; NRS: normal rabbit serum. Right panel: 3T3 cells were treated with 5 μM doxorubicin for 90 min and EMSA was performed with the same probe. (b) Detection of the NF-κB proteins on the Dmp1 promoter by chromatin immunoprecipitation (ChIP) assay. Wild-type murine embryonic fibroblasts before crisis were treated with TNFα, DOX, DNR or with UV-C and ChIP assays were performed with specific antibodies to NF-κB proteins with primers covering the possible NF-κB sequence on the Dmp1 promoter. No significant signals were obtained with control primers that amplify the Dmp1 promoter sequence 2 kb upstream from the NF-κB site (DOX, control). Input indicates the relative intensity of signals from the standardized aliquot of the total input chromatin (0.2%).
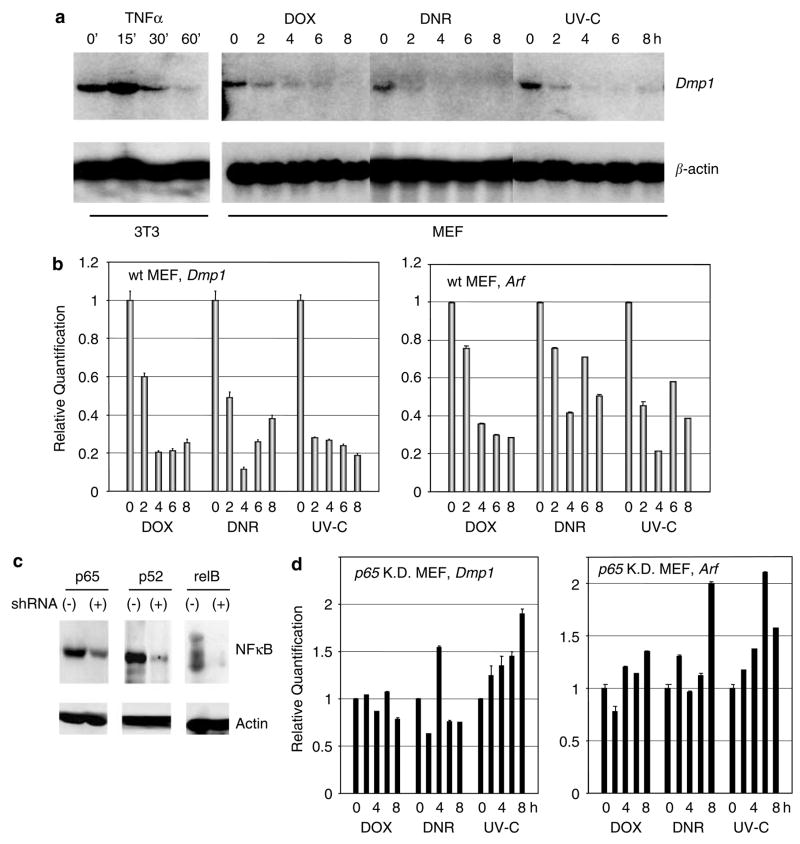
Anthracyclin and UV-C treatment downregulate Dmp1 and Arf in a p65-dependent fashion
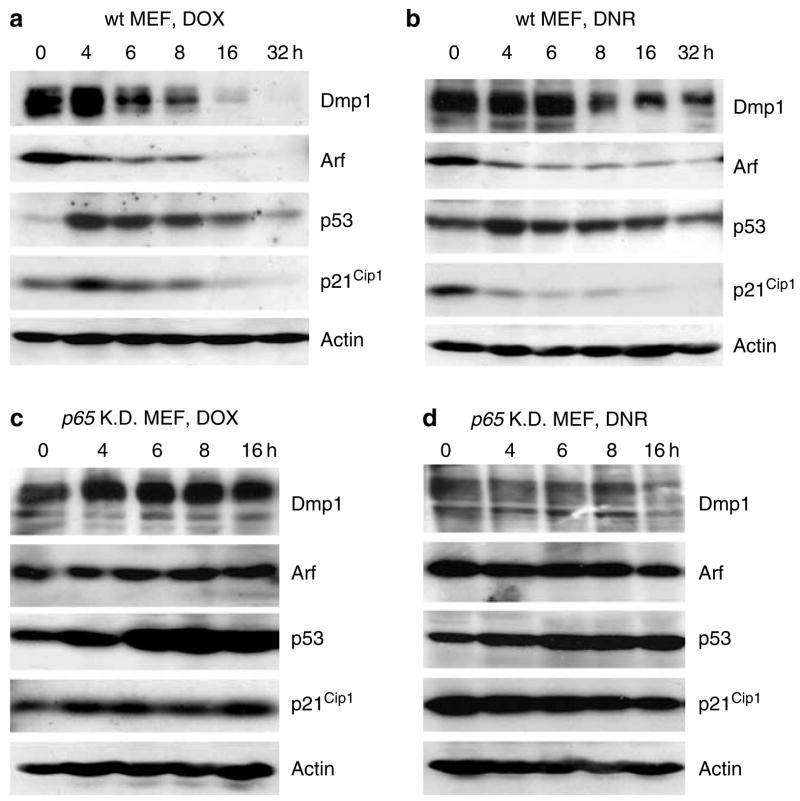
Whether classical and non-classical NF-κB activators would affect Dmp1 expression levels was then investigated. When 3T3 cells were treated with TNFα, Dmp1 mRNA increased rapidly, peaking 15 min after treatment (Figure 3a, left panel). Conversely, treatment of wild-type MEFs with 5 μM DOX resulted in marked reduction of Dmp1 mRNA by 4 h and was sustained through 8 h (Figure 3a, right panel). DNR (1 μM) and UV-C (40 J/m2) treatment of MEFs affected Dmp1 mRNA levels in a manner similar to DOX (Figure 3a). Real-time PCR demonstrated that Dmp1 mRNA decreased to 20% with DOX, 10% with DNR and 18% with UV-C as compared to untreated cells (Figure 3b). These results were confirmed by conventional semi-quantitative RT–PCR analysis (Supplementary Figure 2). The classical Dmp1 target Arf mRNA was also downregulated by DOX/DNR/UV-C treatment of MEFs at similar kinetics (Figure 3b). Next we examined whether the Dmp1 promoter repression was dependent on p65 or on other NF-κB proteins by downregulating their expression with shRNA (Figure 3c). The level of Dmp1 mRNA did not change significantly in non-immortalized p65-knockdown cells with any of the genotoxic treatments (Figure 3d, left panel). The level of Arf mRNA either remained unchanged or was paradoxically increased, in p65-knockdown cells with genotoxic stress (Figure 3d, right panel). Conversely, Dmp1 was downregulated in p52- or relB-knockdown cells treated with DOX or DNR (Supplementary Figure 2d). These data suggest that downregulation of Dmp1 or Arf mRNA by anthracyclins/UV-C is dependent on p65. We then examined the kinetics of Dmp1 and Arf protein expression following genotoxic stress (Figure 4). Dmp1 protein was significantly downregulated (80–90% decrease) at 6–32 h with DOX and 8–32 h with DNR treatment (Figures 4a and b, top panels). p19Arf was also downregulated with DOX/DNR at 6–32 h after exposure (second panels). p53 transiently increased at 4 h with DOX or DNR treatment and then declined when both Dmp1 and Arf were downregulated (third panels). A striking decline in the expression of the p53 target, p21Cip1 was observed 6–32 h after anthracyclin treatment (fourth panels). In contrast, Dmp1 and p19Arf protein levels did not change significantly with DOX or DNR treatment in p65-knockdown cells (Figures 4c and d, top and second panels). Although there was significant accumulation of p53 in p65-knockdown cells, there was no decrease of p53 and its target p21Cip1 8–16 h after DOX or DNR exposure (third and fourth panels). Collectively, these results indicate that both Dmp1 and Arf protein expression are downregulated by anthracyclins, which are dependent on p65.
Figure 3.
Downregulation of the Dmp1 and Arf by doxorubicin (DOX), daunorubicin (DNR) or UV-C depends on the p65 subunit of NF-κB. (a) Detection of the Dmp1 mRNA by northern blotting. NIH 3T3 or wild-type murine embryonic fibroblasts MEFs were treated with 20 ng/ml TNFα, 5 μM DOX, 1 μM DNR or with 40 J/m2 UV-C and RNA was extracted at each time point. (b) Relative quantification of the Dmp1 and Arf mRNA by real-time PCR. (c) Downregulation of NF-κB proteins by shRNA. Wild-type MEFs were infected with retroviruses that express shRNA for NF-κB genes, selected with puromycin and the NF-κB proteins were detected by western blotting. (d) Relative quantitation of the Dmp1 and Arf mRNA levels in p65-knockdown cells (p65 KD) by real-time PCR.
Figure 4.
Regulation of the Dmp1 and p19Arf protein by nuclear factor-kappa B activators. (a, b) Both Dmp1 and p19Arf proteins are downregulated in murine embryonic fibroblasts (MEFs) treated with 5 μM doxorubicin (DOX) (a) or with 1 μM daunorubicin (DNR) (b). Both p53 and its target protein p21Cip1 were downregulated when Dmp1 and p19Arf decreased. (c, d) Non-immortal p65-knockdown MEFs treated with 5 μM DOX (c) or with 1 μM DNR (d). p53 and p21Cip1 were not downregulated after DOX/DNR treatment, reflecting the sustained activation of the Arf-p53 pathway.
Dmp1 plays critical roles in transcriptional repression of the Arf promoter by p65
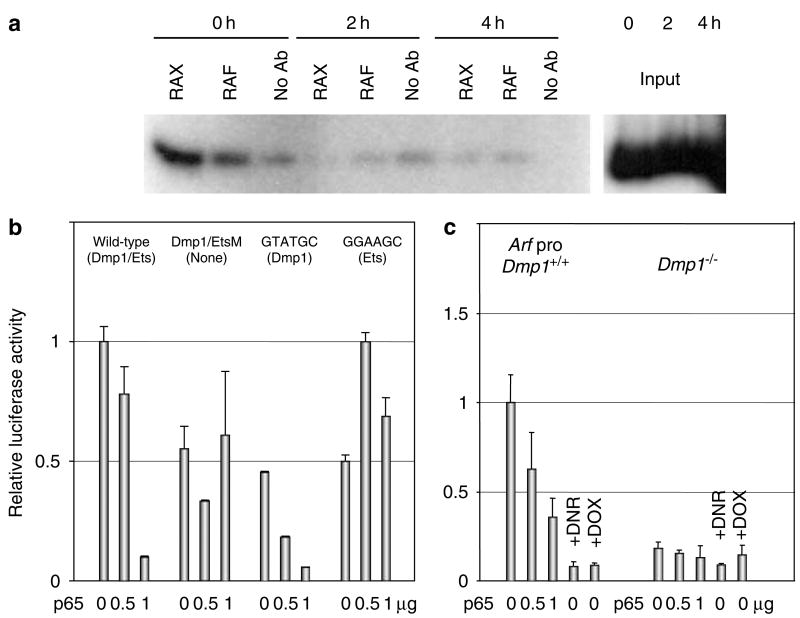
Although, we observed downregulation of p19Arf in response to DOX, DNR or UV-C, the Arf promoter lacks typical NF-κB binding sequences (Inoue et al., 1999). Therefore, we hypothesized that Dmp1 has essential roles in the downregulation of p19Arf with these treatments. A significant level of endogenous Dmp1 was bound to the Arf promoter before treatment of wild-type MEFs (Figure 5a). Interestingly, Dmp1 became undetectable on the Arf promoter 2–4 h after DOX treatment. Accordingly, the wild-type Arf promoter was significantly repressed by overexpression of p65 in 3T3 cells (Figure 5b, wild-type). Disruption of the Dmp1/Ets locus on the Arf promoter abolished its responsiveness to p65 (Figure 5b, Dmp1/Ets M). We then mutated the Arf promoter to the sequence that is recognized by Dmp1 (CCCGTATGC) or by Ets family proteins (CCCGGAAGC). The GTATGC mutant was repressed by p65, while the GGAAGC mutant was slightly activated by p65 (Figure 5b). These results suggest that Dmp1 is crucial for Arf promoter repression by p65. These data were confirmed by an independent experiment employing Dmp1+/+ and Dmp1−/− MEFs. Overexpression of p65 as well as treatment of cells with DNR or DOX repressed the Arf promoter in wild-type cells (Figure 5c, left panel). The Arf promoter was unresponsive or significantly less responsive to p65 or DNR/DOX treatment in Dmp1−/− MEFs (Figure 5c, right panel). Taken together, our data suggest that anthracyclins downregulate the Arf levels through downregulation of Dmp1.
Figure 5.
The Arf promoter repression by anthracyclins depends on Dmp1. (a) Chromatin immunoprecipitation assay demonstrating the Dmp1 binding to the proximal promoter region of the Arf promoter. RAX and RAF designate the two different antibodies to Dmp1. Input indicates the relative intensity of signals obtained from 1% of the total chromatin. (b) The repression of the Arf promoter by p65 depends on the Dmp1/Ets site. The Arf promoter reporter assay was performed with −281 BamHI fragment with increasing amount of the p65 expression vectors. (c) Repression of the Arf promoter by p65 or by anthracyclins largely depends on Dmp1. The Arf promoter reporter assay was conducted in both wild-type and Dmp1−/− MEFs with increasing amount of the p65 expression vector or by treating the cells with doxorubicin or daunorubicin.
Both Dmp1-null and Arf-null cells are resistant to anthracyclin-induced cell death
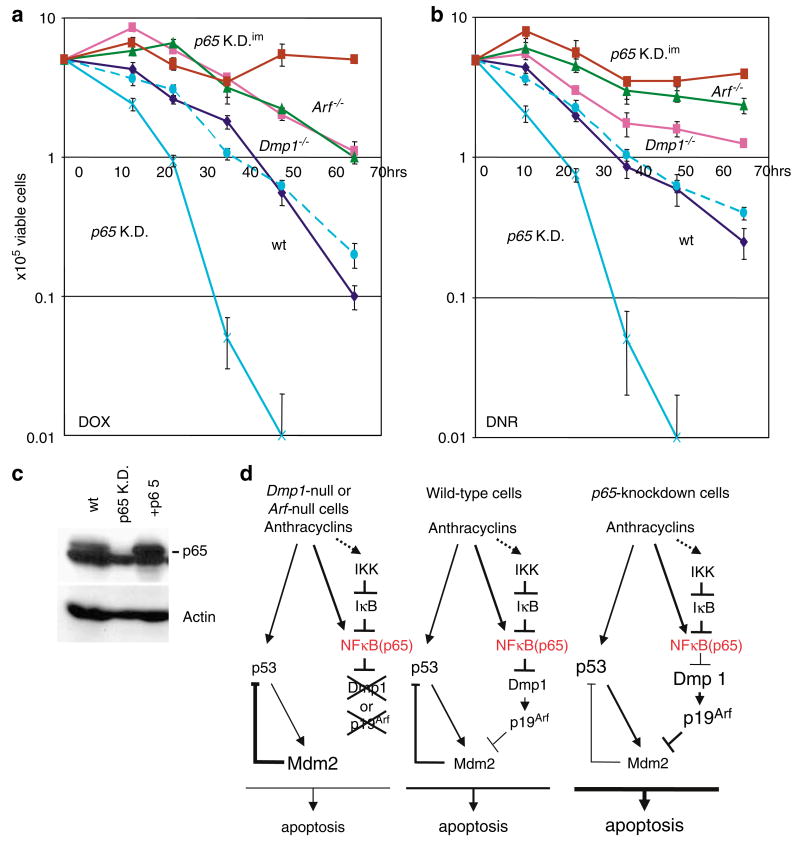
To study the biological significance of the downregulation of the Dmp1-Arf pathway in response to genotoxic stress, wild-type, Dmp1-null, Arf-null and p65-knockdown cells were treated with DOX and DNR to study their survival (Figures 6a and b). Ninety-eight % and 95 percent of wild-type cells died after 64 h with 5 μM DOX or 1 μM DNR (dark blue lines) treatment, respectively. In Dmp1−/− and Arf−/− cells, the number of cells initially increased with both DOX and DNR treatment and then began to undergo apoptotic cell death; however, >20% of the cells still survived after 64 hours of exposure (Figure 6, pink lines for Dmp1−/− and green lines for Arf−/−). A genomic DNA fragmentation assay confirmed that the cells were undergoing apoptosis (data not shown). No significant difference in the survival of Dmp1−/− cells versus the survival of Arf−/− cells with 5 μM DOX treatment was observed. Arf−/− cells showed more drug resistance than Dmp1−/− cells with 1 μM DNR treatment (Figure 6, green and pink lines), consistent with the results of the reporter assay shown in Figure 5c. Importantly, the freshly established p65-knockdown (p65KD) cells were very sensitive to DOX or DNR; all cells were dead within 36–48 h of drug treatment (Figure 6, light blue). The sensitivity of the p65 KD cells to DOX/DNR reverted to normal MEF levels after expression of p65 by retroviral infection (Figure 6, broken light blue lines; Figure 6c), suggesting critical roles for p65 in preventing anthracyclin-induced apoptotic cell death. On the other hand, the immortalized p65-knockdown cells (p65 KDim) were extremely resistant to DOX or DNR treatment (Figure 6, brown lines) and the sensitivity of the cells to DOX/DNR did not change significantly after p65 restoration (survival curves not shown). These results suggest that the absence of Dmp1 or p19Arf is associated with relative resistance of the cells to anthracyclins and thus, normal cells escape from anthracyclin-induced cell death by downregulating the activity of the Dmp1-Arf-p53 pathway.
Figure 6.
Both Dmp1−/− and Arf−/− murine embryonic fibroblasts are resistant to anthracyclin-induced cell death. Wild-type (dark blue), Dmp1-null (pink), Arf-null (green) and p65-knockdown (light blue) cells were treated with either 5 μM doxorubicin (DOX) (a) or 1 μM daunorubicin (DNR) (b) to study their survival. Broken light blue lines: non-immortalized p65-knockdown cells restored with p65; brown lines: immortalized p65-knockdown cells. (c) Western blotting analysis shows restoration of p65 in p65-knockdown cells after retroviral infection. (d) Our current hypothesis regarding the downregulation of the Dmp1-Arf pathway by atypical NF-κB activators.
Our current hypothesis regarding the downregulation of the Dmp1-Arf pathway by atypical NF-κB activators is summarized in Figure 6d. In wild-type cells, the p65 subunit of NF-κB is directly or indirectly activated by anthracyclins or by UV-C, which, in turn binds to the Dmp1 promoter and decreases its expression. This results in the downregulation of p19Arf, which will attenuate the apoptotic cell death mediated by p53. On the other hand, the non-immortalized p65-knockdown cells are more sensitive to anthracyclins/UV-C-induced cell death due to the increased activity of the Arf-p53 pathway. The p53 activity is also regulated by genotoxic stimuli through phosphorylation by ATM (Bakkenist and Kastan, 2004; Kurz et al., 2004).
Discussion
The classical NF-κB activators, such as inflammatory cytokines and some classes of cytotoxic agents, induce a form of NF-κB to promote anti-apoptotic gene expression (Karin et al., 2002; Karin, 2006). On the other hand, the second class of activators, which include anthracyclins and UV-C, induce NF-κB to act as repressors of anti-apoptotic gene expression through p65 phosphorylation and association with HDACs (Campbell et al., 2004). Thus, NF-κB is an important mediator of apoptotic cell death as well. Our data show that both Dmp1−/− and Arf−/− MEFs are more resistant to DOX or DNR-induced cell death than wild-type cells, suggesting the proapoptotic roles of the Dmp1-Arf pathway in programmed cell death. Consistently, it was reported that p14ARF expression sensitizes normal and cancer cells to apoptosis in the presence of DOX, although p14ARF did not induce cell death by itself (Gallagher et al., 2005). Therefore, atypical NF-κB activators downregulate both proapoptotic (Dmp1, Arf) and anti-apoptotic (Bcl-xL, XIAP and A20) genes and thus, the roles of NF-κB in response to genotoxic stress are more intricate than previously believed.
Our results indicated that the response of immortalized p65-knockdown cells to genotoxic drugs was completely different from that of non-immortalized cells. It has been reported that immortalized p65-null MEFs are more resistant to UV-C-induced cell death than wild-type cells (Campbell et al., 2004). In agreement, our experiments also showed that spontaneously immortalized p65-knockdown MEFs were very resistant to DOX/DNR-induced cell death. However, non-immortalized p65-knockdown cells were much more sensitive to DOX/DNR-induced apoptosis (Figure 6). Therefore, p65 appears to behave as an anti-apoptotic protein in non-immortalized cells. Our results are consistent with the previous report that showed increased cell death and p53 induction with DOX treatment in IKK1/2-null cells that lacked detectable NF-κB activity (Tergaonkar et al., 2002). Since immortalization of p65-knockout cells always involves inactivation of p19Arf or p53 (Gapuzan et al., 2005), the responsiveness of p65-knockdown cells to anthracyclins/UV-C is completely different depending on the integrity of the Arf-p53 pathway.
Although different NF-κB family proteins bind to the Dmp1 promoter in response to DOX, DNR or UV-C signaling, downregulation of p65 was sufficient to abolish the inhibitory effects of these geno-toxic stimuli on the Dmp1-Arf pathway. On the other hand, downregulation of other NF-κB proteins, such as relB or p52, had little effect on the Dmp1 promoter repression although they were clearly detected on the Dmp1 promoter by ChIP. Since p65 was the only NF-κB subunit directly bound to the Dmp1 promoter by EMSA in response to DOX/DNR, we speculate that other NF-κB proteins bind to the Dmp1 promoter indirectly through their interaction with p65. The only exception to this hypothesis would be relB binding to the Dmp1 promoter, since it has been reported that relB does not make heterodimers with p65 (Hayden and Ghosh, 2004). It is possible that relB binds to the Dmp1 promoter indirectly as an intertwined homodimer (Huang et al, 2005) or by making heterodimers with residual levels of p50 or p52 (Figure 2b).
DOX treatment of cells stimulates nuclear accumulation and phosphorylation of p53, a process of which is mediated by ATM (Kurz et al., 2004). Our data show that Dmp1-Arf and p53 are differentially regulated by anthracyclins, within at least 4 h after drug treatment; however, both p53 and p21Cip1 decreased when Dmp1 and p19Arf were downregulated. On the other hand, p53 or p21Cip1 was not downregulated in p65-knockdown cells, where both Dmp1 and p19Arf remain elevated and the cells underwent extensive apoptosis 24–36 h after the genotoxic drug treatment when approximately 20% of wild-type cells survived. Thus, downregulation of the Dmp1–Arf pathway by p65 appears to mediate protection of normal cells from the extensive cell death induced by genotoxic drugs. This mechanism will be especially important with respect to the side effects of chemotherapeutic agents in normal tissues. The major mechanisms of action of anthracyclins have been considered to be stabilization of topoisomerase IIα cleavage complexes and generation of reactive oxygen intermediates (DeVita et al., 2005). The former causes protein-linked double- and single-stranded DNA breaks, which are converted into cytotoxic DNA damage and cell death. Thus, cancer cells are generally much more sensitive to anthracyclins than normal tissues even when they have ARF deletions or p53 mutations simply because they divide more frequently than normal cells.
Both the TNFα-responsive element and the anthra-cyclin/UV-C-responsive elements were mapped to the unique NF-κB site on the Dmp1 promoter. p65 was the major player in stimulation of the Dmp1 promoter by TNFα, although other subsets of NF-κB were also detected on the promoter in response to DOX/DNR/UV-C. Our data agree with previous reports that suggested the involvement of subunits of NF-κB other than the classical p50:p65 heterodimer following geno-toxic and other stresses (Rocha et al., 2003; Saccani et al., 2003). Thus, fine differences in the NF-κB subunits that are bound to the Dmp1 promoter could explain differences in the responsiveness of the Dmp1 promoter to TNFα and genotoxic stress. It has been reported that TNF stimulation of cells activates the JNK/SAPK, p38MAPK and Jak-STAT pathways in addition to the classical IKK-IκB-NF-κB pathway (Watts, 2005; Yoshimura, 2006). Since the Dmp1 promoter was repressed solely by overexpressing p65, p65 might interact with transcription factors that act downstream of these signaling pathways to activate Dmp1 transcription in response to TNFα. Although it is not clear which other factors are involved in the Dmp1 promoter activation by TNFα, apparently p65 plays a major role in this process since the responsive element was clearly mapped to the unique NF-κB site on the Dmp1 promoter and direct binding of p65 was confirmed by EMSA.
In conclusion, our present study identified a novel pathway that directly links NF-κB activation and Dmp1–Arf signaling. Since Arf is a positive regulator of p53, our study implicates a novel mechanism of p53 inactivation by NF-κB.
Materials and methods
Electrophoretic mobility-shift assay
To detect proteins that bind to the NF-κB consensus sequence on the murine Dmp1 promoter, lysates were prepared from 3T3 cells treated with 20 ng/ml TNFα for 30 min, 1 μM DNR for 90 min or with 5 μM DOX for 90 min. The nuclear lysate was incubated with [α-32P]-labeled oligonucleotide probe covering the NF-κB consensus sequence of the murine Dmp1 promoter obtained by annealing sense oligonucleotide 5′-ACGCCTGCCAAGGGAAGACCCTCCTTTACGTCT-3′ and its reverse complementary sequence (the NF-κB consensus sequence is underlined). For competition assays, a 100-fold excess of unlabeled oligonucleotides was added to reaction mixtures before probe incubation. To verify the identity of the proteins in shifted complexes, reaction mixtures were incubated with control non-immune rabbit serum or with specific antibodies to p50 (sc-7178x), p52 (sc-7386x), p65 (sc-7151x and sc-372x), relB (sc-226x) or c-rel (sc-71x) proteins.
Western blotting
For analysis of Dmp1, p19Arf, p53, p21Cip1 and actin, affinity–purified polyclonal antibodies to the mouse Dmp1 (RAX, Mallakin et al., 2006), p19Arf (sc-32748, Santa Cruz Biotechnology, Santa Cruz, CA, USA), p53 (sc-6243G), p21Cip1 (sc-6246) or Actin (sc-1615), were used, followed by incubation of the filters with HRP-conjugated second antibodies, reaction with the enhanced ECL detection kit (PerkinElmer, Boston, MA, USA).
Supplementary Material
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Acknowledgments
We thank Dr George Kulik and Ms Karen Klein for critical reading of the manuscript and Drs Neil Perkins, Alexander Hoffmann, Bruce Torbett and Mario Tschan for helpful discussions and sharing unpublished data. We are grateful to Drs Gioacchino Natoli, Andrew Thorburn, Michael Ostrowski, Charles Sherr and Martine Roussel for plasmid DNAs. We also thank Ramesh Sreeramaneni and Scott Barton for technical assistance. This work was supported by NIH/NCI 5R01CA106314 (K Inoue) and by a Wake Forest University Comprehensive Cancer Center ‘Push’ grant (Center grant CA12197-31) to K Inoue.
References
- Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Campbell JK, Rocha S, Perkins ND. Active repression of antiapoptotic gene expression by RelA(p65) NF-κB. Mol Cell. 2004;13:853–865. doi: 10.1016/s1097-2765(04)00131-5. [DOI] [PubMed] [Google Scholar]
- DeVita VT, Hellman S, Rosenberg SA. Cancer: Principles & Practice of Oncology. 7. Lippincott Williams & Willkins; Hagerstown, MD, USA: 2005. [Google Scholar]
- Gallagher S, Kefford RF, Rizos H. Enforced expression of p14ARF induces p53-dependent cell cycle arrest but not apoptosis. Cell Cycle. 2005;4:465–472. doi: 10.4161/cc.4.3.1526. [DOI] [PubMed] [Google Scholar]
- Gapuzan ME, Schmah O, Pollock AD, Hoffmann A, Gilmore TD. Immortalized fibroblasts from NF-kappaB RelA knockout mice show phenotypic heterogeneity and maintain increased sensitivity to tumor necrosis factor alpha after transformation by v-Ras. Oncogene. 2005;24:6574–6583. doi: 10.1038/sj.onc.1208809. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Huang DB, Vu D, Ghosh G. NF-kappaB RelB forms an intertwined homodimer. Structure. 2005;13:1365–1373. doi: 10.1016/j.str.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Inoue K, Mallakin A, Frazier DP. Dmp1 and tumor suppression (review) Oncogene. 2007 doi: 10.1038/sj.onc.1210226. Epub ahead of print Jan 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Roussel MF, Sherr CJ. Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc Natl Acad Sci USA. 1999;96:3993–3998. doi: 10.1073/pnas.96.7.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Wen R, Rehg JE, Adachi M, Cleveland JL, Roussel MF, et al. Disruption of the ARF transcriptional activator DMP1 facilitates cell immortalization, Ras transformation and tumorigenesis. Genes Dev. 2000;14:1797–1809. [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Zindy F, Randle DH, Rehg JE, Sherr CJ. Dmp1 is haplo-insufficient for tumor suppression and modifies the frequencies of Arf and p53 mutations in Myc-induced lymphomas. Genes Dev. 2001;15:2934–2939. doi: 10.1101/gad.929901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- Karin M, Cao Y, Greten FR, Li Z-W. NF-κB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- Kurz EU, Douglas P, Lees-Miller SP. Doxorubicin activates ATM-dependent phosphorylation of multiple downstream targets in part through the generation of reactive oxygen species. J Biol Chem. 2004;279:53272–53281. doi: 10.1074/jbc.M406879200. [DOI] [PubMed] [Google Scholar]
- Lowe S, Sherr CJ. Tumor suppression by Ink4a-Arf: Progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Mallakin A, Taneja P, Matise LA, Willingham MC, Inoue K. Expression of Dmp1 in specific differentiated, nonproliferating cells and its repression by E2Fs. Oncogene. 2006;25:7703–7713. doi: 10.1038/sj.onc.1209750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND. NF-κB: tumor promoter or suppressor? Trends Cell Biol. 2004;14:64–69. doi: 10.1016/j.tcb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Rocha S, Martin AM, Meek DW, Perkins ND. p53 represses cyclin D1 transcription through down regulation of Bcl-3 and inducing increased association of the p52 NF-kappaB subunit with histone deacetylase 1. Mol Cell Biol. 2003;23:4713–4727. doi: 10.1128/MCB.23.13.4713-4727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochem Biophys Acta Rev Cancer. 1998;1378:F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- Ryan KM, Ernst MK, Rice NR, Vousden KH. Role of NF-κB in p53-mediated programmed cell death. Nature. 2000;404:892–897. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G. Modulation of NF-κB activity by exchange of dimers. Mol Cell. 2003;11:1563–1574. doi: 10.1016/s1097-2765(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. The INK4a/ARF network in tumor suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- Sreeramaneni R, Chaudhry A, McMahon M, Sherr CJ, Inoue K. Ras-Raf-Arf signaling critically depends on Dmp1 transcription factor. Mol Cell Biol. 2005;25:220–232. doi: 10.1128/MCB.25.1.220-232.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tergaonkar V, Pando M, Vafa O, Wahl G, Verma I. p53 stabilization is decreased upon NFκB activation: a role for NFκB in acquisition of resistance to chemotherapy. Cancer Cell. 2002;1:493–503. doi: 10.1016/s1535-6108(02)00068-5. [DOI] [PubMed] [Google Scholar]
- Watts TH. TNF/TNFR family members in costimulation of T cell responses. Ann Rev Immunol. 2005;23:23–68. doi: 10.1146/annurev.immunol.23.021704.115839. [DOI] [PubMed] [Google Scholar]
- Webster GA, Perkins ND. Transcriptional cross talk between NF-κB and p53. Mol Cell Biol. 1999;19:3485–3495. doi: 10.1128/mcb.19.5.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A. Signal transduction of inflammatory cytokines and tumor development. Cancer Sci. 2006;97:439–447. doi: 10.1111/j.1349-7006.2006.00197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).