Abstract
Background and Objective
Human β-defensins have been identified in the oral cavity and are predicted to play a role in the defense against pathogenic bacteria. Homologous rat β-defensins (RBDs) have been identified, but their expression in the oral cavity has not been examined. Therefore, the aim of this study was to investigate the expression of innate immune mediators in the rat gingival epithelium.
Material and Methods
Rats were pretreated with antibiotics to depress the normal oral flora, followed by the introduction of Actinobacillus actinomycetemcomitans in their food to allow colonization and the development of periodontal disease. At various time points, animals were killed and the gingival epithelium was extracted. Semiquantitative reverse transcription–polymerase chain reaction was performed to measure RBD and Toll-like receptor (TLR) mRNA levels.
Results
Three β-defensins (RBD-1, -2 and -5) and two TLRs (TLR-3 and -4) are expressed in normal rat gingival epithelium. After the introduction of A. actinomycetemcomitans, RBD-1 and RBD-2 mRNA levels increased for the first week followed by a return to basal levels. No change in TLR mRNA levels was observed.
Conclusion
The rat model provides a good system for experimental analysis of the innate immune response to periopathogenic bacteria in the oral cavity, as well as the potential role of β-defensins in the host response to colonization.
Keywords: innate immunity, animal model, periodontitis
The innate immune response provides the mucosal epithelium with its first line of defense against invading pathogens. The cells that comprise the innate immune response are primarily phagocytes (including neutrophils and macrophages) and the cells lining the epithelial mucosa. As part of this defense, these cells express the genes that encode antimicrobial peptides. These small peptides are characterized by a cationic charge and a broad spectrum of antimicrobial activity against gram-positive and -negative bacteria, fungi and enveloped viruses (1). Among these peptides are the defensins – cationic peptides 20–40 amino acids in length, containing six cysteines which form three intramolecular disulfide bonds. The two major families of defensins (the α- and β-defensins) differ based upon the position of their disulfide bonds (2). The most well studied human β-defensins (HBDs) are HBD-1, HBD-2 and HBD-3, which have been isolated from the kidney, genitourinary tract, small intestine, lung, gingival epithelium and skin (3). While HBD-1 is constitutively expressed in the epithelium, HBD-2 mRNA and protein levels can be induced in airway epithelium by live gram-negative bacteria through a Toll-like receptor (TLR)-4-mediated pathway (4), and by lipopeptide through a TLR-2 pathway (5). Furthermore, both HBD-2 and -3 can be induced by lipopolysaccharide, interleukin-1β and tumor necrosis factor-α (TNF-α) in vitro (6), and by bacterial infection and inflammation in vivo (7).
In the oral cavity, the magnitude of microbial organisms necessitates a protective mechanism to prevent the onset of disease, but which has no detrimental effect on commensal organisms. In human gingival tissue, HBD-1 and HBD-2 are both expressed in normal, uninflamed tissue, at the highest levels at the gingival margin near the site of plaque formation, and within the sulcular epithelium during states of inflammation (8). In vitro studies have demonstrated that Porphyromonas gingivalis induces HBD-1 expression in cultured human gingival epithelial cells, while Fusobacterium nucleatum and Actinobacillus actinomycetemcomitans stimulate the production of HBD-2 and -3 (9,10). In neither case, however, is the induction mediated by lipopolysaccharide, suggesting that regulation of the induction of these mediators of innate immunity in the oral mucosa is controlled by more complex mechanisms of innate immunity.
To elucidate the in vivo response of β-defensin genes to oral pathogens in the gingival epithelium we used a rat model of A. actinomycetemcomitans-induced periodontitis (11). A. actinomycetemcomitans is a gram-negative, capnophilic coccobacillus, which is associated both with chronic periodontitis as well as localized aggressive periodontitis (12). This affects mainly prepubescent adolescents of African–American descent (13), and is defined by destruction of the periodontal ligament and alveolar bone of the first molar and central incisors (14). Feeding A. actinomycetemcomitans to rats in their food results in stable colonization by A. actinomycetemcomitans (15) and consequently leads to gingival detachment and alveolar bone loss (11). Studies in the rat initially demonstrated the expression of homologues to HBD-1 and -2 in kidney and lung (16), respectively, and later in several other tissues (17). Subsequently, numerous new rat β-defensin (RBD) genes were identified, with the expression of at least three new molecules (Defb 3, 5 and 36) in the intestinal epithelium (18). Here, we describe the identification of β-defensins in the rat gingival epithelium and examine the response of these genes in A. actinomycetemcomitans-induced periodontal disease.
Material and methods
Inoculation of rats
Rats were infected with A. actinomycetemcomitans, as described in Schreiner et al. (11), with the following modifications. Pathogen-free, Sprague-Dawley male rats, 6–8-wk of age and weighing 150–250 g (Taconic Laboratories, Germantown, NY, USA) were fed powdered Laboratory Rodent Meal Diet 5001 (Purina Mills Feeds, St Louis, MO, USA) for 2 wk. To depress the normal oral flora, antibiotics (kanamycin and ampicillin, 20 mg/d) were included in the rats' drinking water, daily, for 4 d. On the last 2 d of antibiotic treatment, their oral cavities were swabbed with chlorhexidine gluconate (0.12%, Peridex; Proctor and Gamble, Cincinnati, OH, USA). After 3 d without antibiotic treatment, rats were divided into two groups of 26. Group 1 was fed with A. actinomycetemcomitans and group 2 served as the uninoculated control. A. actinomycetemcomitans strain Aa1005Rif was grown, as described previously (11), on AAGM solid plates supplemented with BV (bacitracin at a final concentration of 75 μg/ml and vancomycin at a final concentration of 5 μg/ml). Liquid cultures of 125 ml of AAGM-BV of A. actinomycetemcomitans were inoculated and grown in T-175 flasks for 2 d in an incubator containing 10% CO2/90% air atmosphere and at a temperature of 37°C. Four flasks containing 125 ml in each flask were required for each day of feeding to yield an approximate inoculum of 108 cells. Cells were harvested by washing three times with 25 ml of phosphate-buffered saline and then scraped into a solution of phosphate buffered saline containing 3% sucrose. Feeding was performed, according to previously published methods (11), for 8 d. After the feeding/inoculation regimen, rats were returned to a regular powdered food diet. Rats were killed, after stopping feeding, at 3, 7, 21, 42 and 84 d. At each time point, rats from each group were killed by CO2 asphyxiation for 5 min. Gingiva was dissected from three rats from each group. The mandibular buccal and lingual surfaces and maxillary buccal surfaces were extracted using a spoon excavator. Tissue was immediately snap frozen in liquid nitrogen and stored at −80°C.
RNA preparation and reverse transcription–polymerase chain reaction
Tissue was placed on dry ice and ground with a mortar and pestle. RNA was isolated by the following protocol using the Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA, USA) for animal tissue, according to the manufacturer's instructions. Reverse transcription–polymerase chain reaction (RT–PCR) was performed using the SuperScript II Reverse Transcriptase and PCR guidelines (Invitrogen, Carlsbad, CA, USA). PCR primers for the amplification of RBDs and TLR are listed in Table 1. PCR thermal cycler conditions are initial denaturation (94°C for 3 min), followed by 30 cycles of denaturation (94°C for 1 min), annealing (temperature in Table 2 for 30 s) and elongation (72°C for 1 min), with a final elongation at 72°C for 5 min. Products were electrophoresed on agarose gels and stained with ethidium bromide to visualize. Semiquantitative DNA analysis was performed on 2% agarose gels using fluorimage analysis (Typhoon; Molecular Dynamics, GE Healthcare, Piscataway, NJ, USA) and calculated for comparative intensities relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels.
Table 1.
Primer sequences and their predicted product sizes
| Primers | Forward (5′ to 3′) | Reverse (5′ to 3′) | Product size (bp) | Reference |
|---|---|---|---|---|
| RBD-1 | GGACGCAGAACAGATCAATACCGA | TCTTCAAACCACTGTCAACTCCTG | 225 | (16) |
| RBD-2 | TTAATTTGGTTTGTTTTGTGCAT | CATGCCTGACCAAAGGAGGCGTA | 132 | (16) |
| Defb3 | CTCTTCTCATTTCTCCTGGTGCT | GTGCCACAACTGCCAATCTCT | 150 | (18) |
| Defb5 | CTCTTCTCATTTCTCCTGGTGCT | TCTCACTTTACCCAGTCCACAA | 164 | (18) |
| Defb36 | AGGCCCATCATGAAGCTCCTGC | TTCCGGCTTCCGGGCAGTTAG | 227 | (18) |
| TLR-2 | GCACTTGAGCGAGTCTGCTTTC | GAACAAATAGAACTGGGGGATGTG | 361 | |
| TLR-3 | GCTTCTCACCCCAACATTGACG | ACCCTCCAACAAGTCCTCGTTC | 564 | |
| TLR-4 | GGCTGTGGAGACAAAAATGACCTC | AGGCTTGGGCTTGAATGGAGTC | 272 | |
| TLR-5-1 | TCAACACGACTGAGAGGCTCCTAC | CCCTGAAAAGCGTCTGGATTC | 217 | |
| TLR-5-2 | TCACCCTATTCGGTTCTCCTGC | CGTTCTGTGCCCATTCAAAGTC | 333 | |
| TLR-6-1 | GAGAACCCATCGTGGAGAGTTTC | GGATTTTGTGCTTGGTGACAGG | 155 | |
| TLR-6-2 | GCATCAAGAGAAGTGGTGGAGG | ACTGCCATAGCATCCTGAGATACC | 223 | |
| TLR-7-1 | CCAAGCATCTCTCCAGACTCCTTC | TGGCAAAATGGTGGGGACAG | 441 | |
| TLR-7-2 | CGACTCTCTCCTTAGCCAAAAATGG | AACCCACCAGACAAACCACACAGC | 362 | |
| TLR-9-1 | GGACGGGAACTGCTACTACAAGAAC | AGGTGGCTCAGGTGATGGAAAG | 336 | |
| TLR-9-2 | AGCCTGATTCATCTGGACCTGTC | ACTGCCACACTTCACACCGTTAG | 492 | |
| GAPDH | AGACAGCCGCATCTTCTTGT | CTTGCCGTGGGTAGAGTCAT | 207 |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RBD, rat β-defensin; TLR, Toll-like receptor.
Table 2.
Primer annealing temperatures
| RBD-1 | 60°C | TLR-5-1 | 56°C |
| RBD-2 | 60°C | TLR-5-2 | 56°C |
| Defb3 | 53.6°C | TLR-6-1 | 53.4°C |
| Defb5 | 54.1°C | TLR-6-2 | 56°C |
| Defb36 | 53.6°C | TLR-7-1 | 55°C |
| TLR-2 | 55°C | TLR-7-2 | 55°C |
| TLR-3 | 55°C | TLR-9-1 | 58.6°C |
| TLR-4 | 55°C | TLR-9-2 | 58.3°C |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RBD, rat β-defensin; TLR, Toll-like receptor.
Results
In order to examine the innate immune response of the oral cavity in the rat, we first defined the expression of β-defensins in gingival epithelium. Primers for RBD-1 and -2 (16), and for Defb3, -5 and -36 (18), were synthesized according to published sequences (see the noted references). These latter β-defensins were chosen because of their high homology to RBD-2 and expression in intestinal epithelium. RT–PCR of mRNA from rat gingival epithelium shows the expression of RBD-1 and -2 and of Defb5 (referred to hereon as RBD-5) (Fig. 1). Kidney and lung are shown as positive controls of rat β-defensins, while GAPDH is a housekeeping gene. We did not detect the expression of Defb3 or -36 in this tissue (data not shown).
Fig. 1.
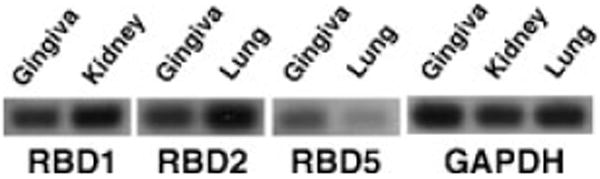
β-defensin expression in rat gingival epithelium. mRNA was isolated from rat gingiva and amplified by reverse transcription–polymerase chain reaction. The amplification products were subjected to agarose-gel electrophoresis then examined for the presence of rat β-defensin (RBD)-1, RBD-2 and Defb5. Kidney and lung were included as positive controls. Glyceraldehyde-3-phosphate dehydrogenase (GAP-DH) was included as a housekeeping gene.
To elucidate the potential transcriptional control of these β-defensin genes, we examined the putative promoter regions by computer analysis (MATINSPECTOR, http://www.genomatix.de). Examination of this region demonstrated the presence of potential transcription factor-binding sites for signal transducers and activators of transcription (STAT5) and for Interferon regulatory factor (IRF-3) and -7 upstream from RBD-1, and of nuclear factor-κB (NF-κB) and CCAAT/enhancer binding protein/β (CEBP/β) transcription factor-binding sites upstream from RBD-2. The RBD5 gene appears to have both STAT and IRF sites, as well as NF-κB and CEBP/β sites (Fig. 2). This suggested that these genes may participate in an innate immune response to both viral (IRF and STAT regulation) and bacterial (NF-κB and CEBP/β) pathogens. To determine if this type of response occurred in the gingival epithelium, we examined this tissue for the presence of TLRs. We designed primers to rat TLR-2, -3, -4, -5, -6, -7 and -9 based on published cDNA sequences. The results indicate the presence of mRNA for only TLR-3 and -4 in rat gingiva (Fig. 3). Bone marrow is shown as a positive control for TLRs, and GAPDH is a housekeeping gene.
Fig. 2.
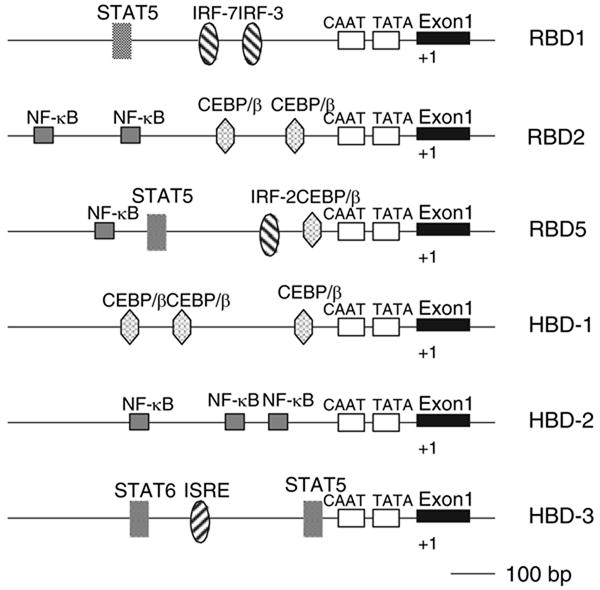
Map of the promoter regions in rat β-defensins (RBDs). The 5′ flanking regions (to–1000 bp) for RBD-1, -2 and -5 were analyzed for the presence of putative transcription factor binding sequences. Flanking regions for human β-defensin (HBD)-1, -2 and -3 are shown for comparison. CEBP, CCAAT/enhancer binding protein; IRF, Interferon regulatory factor; ISRE, Interferon-stimulated response element; NF-κB, nuclear factor-κB; STAT, signal transducers and activators of transcription.
Fig. 3.
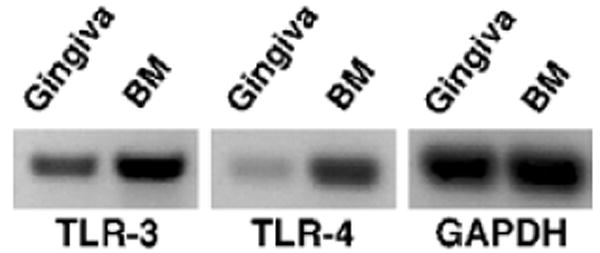
Toll-like receptor (TLR) expression in rat gingival epithelium. mRNA from rat gingiva was analyzed for the expression of TLR-2, -3, -4, -5, -6, -7 and -9. The only positive signals obtained by gel electrophoresis were for TLR-3 and -4 (shown). Bone marrow (BM) is included as the positive control for TLRs (positive signals were observed for all TLRs; data not shown), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was included as a housekeeping gene.
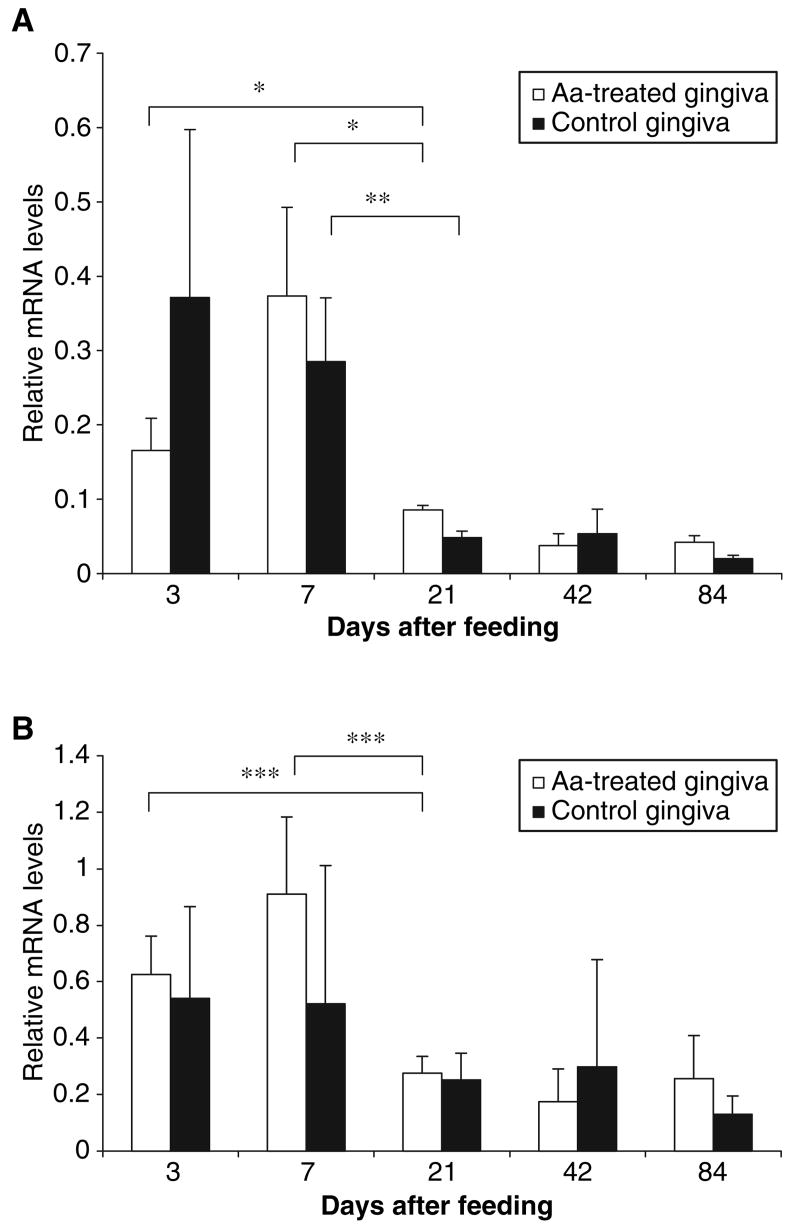
These findings suggested that the oral tissue could be responsive to pathogenic microorganisms by the induction of β-defensins, as seen in other tissues. To examine this potential response, we introduced A. actinomycetemcomitans into the rat by oral inoculation, and killed the animals at 3, 7, 21, 42 and 84 d. Bacteria (both A. actinomycetemcomitans and normal flora) could be cultured from the experimental animals, while only normal flora could be cultured from control animals (data not shown). The expression of RBD-1, -2 and -5 was measured by RT–PCR of extracted gingival epithelium at each time point, and the relative levels were quantified. Our results indicate that there was a higher level of mRNAs for RBD-1 and -2 in both infected animals in the first two time points (i.e. 3 and 7 d after the end of the feeding period, Fig. 4), in comparison with the later time points. A statistically significant increase was also observed in RBD-1 levels at 7 d in comparison with the later time points in the control animals. No difference was observed in levels of RBD-5 throughout the experiment (data not shown). In addition, we also examined TLRs and discovered no change in the levels of TLR expression at each time point (data not shown).
Fig. 4.
Expression of rat β-defensins in the gingival epithelium in response to bacterial challenge. Rats were treated with antibiotic for 4 d prior to feeding with either Actinobacillus actinomycetemcomitans (Aa)-incorporated food or normal food, as described in the Material and methods. Rats (n = 3) were killed on the day indicated, and mRNA was isolated from the gingival epithelium and amplified by the semiquantitative reverse transcription-polymerase chain reaction. Levels of mRNA for rat β-defensin 1 (RBD-1) (A) and RBD-2 (B) are shown relative to the mRNA levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Error bars + standard error of the mean are shown. Asterisks represent statistical significance, determined by the Student's t-test (*p < 0.04; **p < 0.03; ***p < 0.01).
Discussion
The initial site for interaction between the host and periodontal pathogens is the gingival epithelium. Therefore, examination of the response mechanism of this tissue to pathogens will provide a useful tool for using to understand the host–pathogen interaction in the development of periodontal disease. β-defensins have been proposed to play an important role in this first line of defense against colonization by periodontal pathogens (19). HBD-1, -2 and -3 have been identified in the gingival epithelium and are expressed at different levels in health and periodontitis (20,21). However, in contrast to other epithelia, the inducible β-defensins (HBD-2 and -3) are expressed in normal, uninflamed gingiva. It has been hypothesized that commensal bacteria can stimulate these defensins in order to maintain a level of defense against potential pathogens (22). Our goal was to examine the potential of the rat as an experimental model for studying the innate immune response in the oral epithelium, using β-defensins as a model response mediator. Earlier studies have suggested that the rat is a good model for the defensin-mediated host defense, because, in contrast to mice, which lack α-defensins in their neutrophils (23), the rat expresses four in their neutrophils (24), as does the human (25). Our study provides the basis of a model for using to examine the expression of these and other innate immune mediators in response to colonization by bacteria and the development of periodontal inflammation and infection. We showed that, similarly to humans, three β-defensins (RBD-1, -2 and -5) are expressed in the gingival epithelium. The expression of RBD-1 and -2 was initially described primarily in the kidney and lung, respectively. The gene encoding RBD-5 (defB5) has been identified through bioinformatics screening (18); however, this is the first description of its expression in any tissue.
Promoter analysis of the two inducible human β-defensin genes (those encoding HBD-2 and -3) describe different response elements. Active NF-κB transcription factor-binding sites, which are utilized upon induction with bacteria, have been described upstream from HBD-2 (2). These sites lack the HBD-3 promoter, which instead has STAT and interferon response elements. This suggests that HBD-2 can be regulated as part of a standard TLR-2 or -4-mediated pathway (which has been shown for several other epithelial tissues), while HBD-3 may be controlled by other stimuli, including virally induced, interferon-based pathways. Indeed, Chung & Dale have reported both NF-κB-dependent and -independent pathways leading to the induction of HBD-2 in oral epithelial cells (26). Our analysis of the RBD genes suggests that the rat also has the capability to up-regulate genes through different response pathways. Whether these pathways are functional in the gingival epithelium, where a large number of resident pathogens exist commensally, remains to be determined. The fact that the gingival epithelium expresses TLR-3 and -4 (Fig. 3), which recognizes double-stranded RNA and bacterial lipopolysaccharide, respectively, suggests that it may be a responsive tissue. Higher-order control mechanisms, including adapter molecules, such as MyD88, MD-2 and others, may play a regulatory role in this complex environment. Based on our results, the rat represents a good model for the further experimental examination of this response.
In a first attempt to elucidate this response, we challenged rats with a periopathogenic bacterium, A. actinomycetemcomitans, by incorporating live microorganisms in their food. Prior to feeding, however, the rats were treated with antibiotics (ampicillin and kanamycin, 20 mg/d for 4 d) to reduce the normal flora. Our results demonstrated that the levels of β-defensin mRNAs rose for the first week after normal feeding was resumed, when the oral cavities were recolonized with commensal microorganisms as well as A. actinomycetemcomitans (11). As a homeostasis of the oral flora was reached, the levels of β-defensin mRNA dropped to a basal level. This may reflect the natural state, whereby the gingival epithelium can respond to an initial recognition of microorganisms. Persistent colonization may lead to a tolerance phenotype, whereby the presence of commensal bacteria is not sufficient to induce further expression. It is surprising that the levels of RBD-1 increased in response to bacteria, as our promoter analysis suggested that this gene would be responsive to viral stimulation. This may be explained by induction through a secondary pathway, utilizing other transcription factor binding sites not shown in Fig. 2. As pro-inflammatory cytokines, such as interleukin-1β, can induce HBDs both in oral tissues (10) (G. D., unpublished data) as well as in other epithelia (27), it may be that a secondary induction requires an inflammatory response. Thus, β-defensins, while present at basal levels in normal conditions, may play a role in the response and defense against periodontal disease during an infection and inflammation phase.
Acknowledgments
We wish to thank David Furgang for helpful advice and Maribel Vega for technical assistance. This work was supported by the US Public Health Service grants R01DE14897 (to G.D.) and R01DE14897S1 (to A.R.K.), and a grant from the UMDNJ Foundation (to H.S.).
References
- 1.Hancock REW, Diamond G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000;8:402–410. doi: 10.1016/s0966-842x(00)01823-0. [DOI] [PubMed] [Google Scholar]
- 2.Kaiser V, Diamond G. Expression of mammalian defensin genes. J Leukoc Biol. 2000;68:779–784. [PubMed] [Google Scholar]
- 3.Diamond G, Laube D, Klein-Patel ME. Mammalian β-defensins in mucosal defences. In: Hancock REW, Devine DA, editors. Mammalian Host Defence Peptides. Cambridge, UK: Cambridge University Press; 2004. pp. 111–138. [Google Scholar]
- 4.Legarda D, Klein-Patel ME, Yim S, Yuk MH, Diamond G. Suppression of NF-κB-mediated beta-defensin gene expression in the mammalian airway by the Bordetella type III secretion system. Cell Microbiol. 2005;7:489–497. doi: 10.1111/j.1462-5822.2004.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hertz CJ, Wu Q, Porter EM, et al. Activation of Toll-like receptor 2 on human tracheobronchial epithelial cells induces the antimicrobial peptide human beta defensin-2. J Immunol. 2003;171:6820–6826. doi: 10.4049/jimmunol.171.12.6820. [DOI] [PubMed] [Google Scholar]
- 6.Froy O. Regulation of mammalian defensin expression by Toll-like receptor-dependent and independent signalling pathways. Cell Microbiol. 2005;7:1387–1397. doi: 10.1111/j.1462-5822.2005.00590.x. [DOI] [PubMed] [Google Scholar]
- 7.Komatsuzawa H, Ouhara K, Yamada S, et al. Innate defences against methicillin-resistant Staphylococcus aureus (MRSA) infection. J Pathol. 2006;208:249–260. doi: 10.1002/path.1898. [DOI] [PubMed] [Google Scholar]
- 8.Dale BA, Kimball JR, Krisanaprakornkit S, et al. Localized antimicrobial peptide expression in human gingiva. J Periodont Res. 2001;36:285–294. doi: 10.1034/j.1600-0765.2001.360503.x. [DOI] [PubMed] [Google Scholar]
- 9.Feucht EC, DeSanti CL, Weinberg A. Selective induction of human beta-defensin mRNAs by Actinobacillus actinomycetemcomitans in primary and immortalized oral epithelial cells. Oral Microbiol Immunol. 2003;18:359–363. doi: 10.1046/j.0902-0055.2002.00097.x. [DOI] [PubMed] [Google Scholar]
- 10.Vankeerberghen A, Nuytten H, Dierickx K, Quirynen M, Cassiman JJ, Cuppens H. Differential induction of human beta-defensin expression by periodontal commensals and pathogens in periodontal pocket epithelial cells. J Periodontol. 2005;76:1293–1303. doi: 10.1902/jop.2005.76.8.1293. [DOI] [PubMed] [Google Scholar]
- 11.Schreiner HC, Sinatra K, Kaplan JB, et al. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc Natl Acad Sci USA. 2003;100:7295–7300. doi: 10.1073/pnas.1237223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slots J, Ting M. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease: occurrence and treatment. Periodontol 2000. 1999;20:82–121. doi: 10.1111/j.1600-0757.1999.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 13.Loe H, Brown LJ. Early onset periodontitis in the United States of America. J Periodontol. 1991;62:608–616. doi: 10.1902/jop.1991.62.10.608. [DOI] [PubMed] [Google Scholar]
- 14.Henderson B, Nair SP, Ward JM, Wilson M. Molecular pathogenicity of the oral opportunistic pathogen Actinobacillus actinomycetemcomitans. Annu Rev Microbiol. 2003;57:29–55. doi: 10.1146/annurev.micro.57.030502.090908. [DOI] [PubMed] [Google Scholar]
- 15.Fine DH, Goncharoff P, Schreiner H, Chang KM, Furgang D, Figurski D. Colonization and persistence of rough and smooth colony variants of Actinobacillus actinomycetemcomitans in the mouths of rats. Arch Oral Biol. 2001;46:1065–1078. doi: 10.1016/s0003-9969(01)00067-x. [DOI] [PubMed] [Google Scholar]
- 16.Jia HP, Mills JN, Barahmand-Pour F, et al. Molecular cloning and characterization of rat genes encoding homologues of human beta-defensins. Infect Immun. 1999;67:4827–4833. doi: 10.1128/iai.67.9.4827-4833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Froy O, Hananel A, Chapnik N, Madar Z. Differential expression of rat beta-defensins. IUBMB Life. 2005;57:41–43. doi: 10.1080/15216540500088912. [DOI] [PubMed] [Google Scholar]
- 18.Patil AA, Cai Y, Sang Y, Blecha F, Zhang G. Cross-species analysis of the mammalian beta-defensin gene family: presence of syntenic gene clusters and preferential expression in the male reproductive tract. Physiol Genomics. 2005;23:5–17. doi: 10.1152/physiolgenomics.00104.2005. [DOI] [PubMed] [Google Scholar]
- 19.Weinberg A, Krisanaprakornkit S, Dale BA. Epithelial antimicrobial peptides: review and significance for oral applications. Crit Rev Oral Biol Med. 1998;9:399–414. doi: 10.1177/10454411980090040201. [DOI] [PubMed] [Google Scholar]
- 20.Bissell J, Joly S, Johnson GK, et al. Expression of beta-defensins in gingival health and in periodontal disease. J Oral Pathol Med. 2004;33:278–285. doi: 10.1111/j.0904-2512.2004.00143.x. [DOI] [PubMed] [Google Scholar]
- 21.Dommisch H, Acil Y, Dunsche A, Winter J, Jepsen S. Differential gene expression of human beta-defensins (hBD-1-2-3) in inflammatory gingival diseases. Oral Microbiol Immunol. 2005;20:186–190. doi: 10.1111/j.1399-302X.2005.00211.x. [DOI] [PubMed] [Google Scholar]
- 22.Dale BA, Fredericks LP. Antimicrobial peptides in the oral environment: expression and function in health and disease. Curr Issues Mol Biol. 2005;7:119–133. doi: 10.1093/jac/dki103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisenhauer PB, Lehrer RI. Mouse neutrophils lack defensins. Infect Immun. 1992;60:3446–3447. doi: 10.1128/iai.60.8.3446-3447.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisenhauer P, Harwig S, Szlarek D, Ganz T, Lehrer RI. Polymorphic expression of defensins in neutrophils from outbred rats. Infect Immun. 1990;58:3899–3902. doi: 10.1128/iai.58.12.3899-3902.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehrer RI, Ganz T. Antimicrobial polypeptides of human neutrophils. Blood. 1990;76:2169–2181. [PubMed] [Google Scholar]
- 26.Chung WO, Dale BA. Innate immune response of oral and foreskin keratinocytes: utilization of different signaling pathways by various bacterial species. Infect Immun. 2004;72:352–358. doi: 10.1128/IAI.72.1.352-358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsutsumi-Ishii Y, Nagaoka I. Modulation of human beta-defensin-2 transcription in pulmonary epithelial cells by lipopolysaccharide-stimulated mononuclear phagocytes via proinflammatory cytokine production. J Immunol. 2003;170:4226–4236. doi: 10.4049/jimmunol.170.8.4226. [DOI] [PubMed] [Google Scholar]