Abstract
Rice bacterial artificial chromosome clones containing centromeric DNA were isolated by using a DNA sequence (pSau3A9) that is present in the centromeres of Gramineae species. Seven distinct repetitive DNA elements were isolated from a 75-kilobase rice bacterial artificial chromosome clone. All seven DNA elements are present in every rice centromere as demonstrated by fluorescence in situ hybridization. Six of the elements are middle repetitive, and their copy numbers range from ≈50 to ≈300 in the rice genome. Five of these six middle repetitive DNA elements are present in all of the Gramineae species, and the other element is detected only in species within the Bambusoideae subfamily of Gramineae. All six middle repetitive DNA elements are dispersed in the centromeric regions. The seventh element, the RCS2 family, is a tandem repeat of a 168-bp sequence that is represented ≈6,000 times in the rice genome and is detected only in Oryza species. Fiber-fluorescence in situ hybridization analysis revealed that the RCS2 family is organized into long uninterrupted arrays and resembles previously reported tandem repeats located in the centromeres of human and Arabidopsis thaliana chromosomes. We characterized a large DNA fragment derived from a plant centromere and demonstrated that rice centromeres consist of complex DNA, including both highly and middle repetitive DNA sequences.
Centromeres are one of the most characteristic landmarks of eukaryotic chromosomes. The centromeric region is the site for mitotic and meiotic spindle fiber attachment and is responsible for sister chromatid association. Thus centromeres play a central role in the process of chromosomal segregation and transmission in cell divisions. The molecular organization of centromeres has been studied extensively in yeast, Drosophila melanogaster, and humans. Whereas the centromeres of budding yeast (Saccharomyces cerevisiae) chromosomes are structurally simple, specified by only a 125-bp, single-copy DNA sequence (1–2), the centromeres from higher eukaryotic species, such as D. melanogaster and humans, encompass several hundred kilobases (kb) or even megabases of DNA and contain repetitive DNA sequences (3–7).
Thus far, only limited information is available for the organization of plant centromeres. Peacock et al. (8) first isolated a repetitive DNA element from the maize knobs that can act as neocentromeres in certain genetic backgrounds. A repetitive DNA element also was cloned from the centromeres of the supernumerary B chromosomes of maize (9–10). Part of this B-specific DNA element shows strong homology to the maize knob sequences. A 180-bp tandem repeat (pAL1 family) is the major component of the centromeric regions of Arabidopsis thaliana chromosomes. The genomic organization of this repeat family shares similarities to the alpha satellite DNA at the human centromeres (11–14). Recently, two repetitive DNA elements, pSau3A9 and CCS1, were isolated from sorghum (Sorghum bicolor) (15) and Brachypodium sylvaticum (16), respectively. These two repeats were detected in the centromeres of various grass species. The conservation of these sequences across distantly related plant species may imply a role in centromere function.
By screening a rice bacterial artificial chromosome (BAC) library using the pSau3A9 sequence as a probe, we identified a number of BAC clones derived from the centromeres of rice chromosomes. Seven different repetitive DNA families were cloned from a 75-kb rice BAC. The sequences and molecular organization of these repeats are presented in this paper.
MATERIALS AND METHODS
Materials.
The rice BAC library used in the present study was constructed from an indica rice (Oryza sativa ssp. Indica) line IR-BB21 and consists of 11,000 clones (17). The cereal centromeric DNA element pSau3A9 (15) was used to isolate the rice centromere-specific BAC clones. Rice lines used in the present study include a javanica rice (O. sativa ssp. Javanica) line DV85, a japonica rice (O. sativa ssp. Japonica) line Norin 28, an indica rice line IR72, and four other Oryza species (O. glaberrima, O. rufipogon, O. officinalis, and O. alta). Gramineae species used in conservation studies include two species from the Bambusoideae subfamily [bamboo (Bambusa vulgaris), Pharus sp.], three species from the Panicoideae subfamily [sorghum, maize (Zea mays), and sugar cane (Saccharum officinarum)], six species form the Pooideae subfamily [Agropyron intermedium, barley (Hordeum vulgare), oats (Avena sativa), rye (Secale cereale), wheat (Triticum aestivum), and Aegilops squarrosa]. Three non-Gramineae species, Juncus effusus, Cyperus alternifolius, and A. thaliana, and rye and maize lines containing B chromosomes also were included.
BAC Library Screening.
BAC filter preparations and BAC library screening were as described (17, 18). BAC clones were isolated by using pSau3A9 as a probe, and their cytological locations were confirmed by fluorescence in situ hybridization (FISH).
Subcloning and Sequencing.
DNA fragments recovered from agarose gels were subcloned into pUC18 plasmids as previously described (15). Cycle sequencing reactions were performed by using Applied Biosystems AmpliTaq DNA polymerase, FS Dye Terminator Ready Reactions kit, and a Perkin–Elmer Thermocycler (model 2400). Reaction products were analyzed on an Applied Biosystems DNA sequencer (model 373).
Southern Blot Hybridization.
Plant genomic DNA was isolated as described (19). BAC DNA was prepared by using an alkaline lysis method (20) and purified by CsCl ultracentrifugation. Gel transfers, prehybridizations, hybridizations, and posthybridization washing were all as previously described (15).
Slot Blot Hybridization.
Copy number of each subclone in rice genome was determined by slot blot hybridization (21). Band intensities were measured on the autoradiographs by IPLab Spectrum v3.1 software.
FISH.
Detailed protocols for FISH and Fiber-FISH were described previously (22, 23). The formamide in the hybridization mixture was 50% and 30% in regular and low stringency hybridizations, respectively. Washing was conducted at either low [2× saline sodium citrate (SSC) at 42°C for 15 min], medium (50% formamide at 45°C for 15 min), or high stringency (70% formamide at 50°C for 15 min).
RESULTS
Isolation of BAC Clones Derived from Rice Centromeric Regions.
We demonstrated previously that DNA sequences homologous to the sorghum centromeric DNA family Sau3A9 are present in the centromeric regions of various grass species, including rice (15). A rice BAC library (17) was screened by using pSau3A9 as a probe. Twenty-two clones showed unambiguous positive hybridizations. Ten of the 22 clones were analyzed cytologically by FISH. Eight clones hybridized to the centromeric or/and paracentromeric regions of all rice chromosomes (data not shown). Clone 17p22 showed bright and sharp signals specific to the centromeric regions. At a low hybridization stringency, this clone also hybridized exclusively to the centromeric regions of chromosomes from sorghum, maize, wheat, barley, oats, and rye (data not shown).
DNA from clone 17p22 was digested with nine restriction enzymes (BamHI, DraI, EcoRI, HaeIII, HindIII, MspI, PstI, Sau3AI, and SalI) and blotted onto nylon membrane. Small DNA fragments ranging from 0.5 to 3 kb were subcloned, and their distinctiveness was confirmed by Southern hybridization using blots containing 17p22 DNA digested with the nine restriction enzymes. Seven different DNA families, including two Sau3AI fragments (subclones pRCS1 and pRCS2), three HindIII fragments (subclones pRCH1, pRCH2, and pRCH3), and two EcoRI fragments (subclones pRCE1 and pRCE2), were identified (Table 1). These seven families hybridized to all of the fragments generated by the nine enzymes. FISH and Southern hybridization analysis indicated that all seven elements are repetitive in the rice genome (see below).
Table 1.
Summary of the seven rice centromeric repetitive DNA families
| Family | GenBank accession no. | Size, bp | GC content, % | Organization pattern | Copy number* | Conservation |
|---|---|---|---|---|---|---|
| RCS1 | AF058901 | 877 | 40 | Dispersed | 130 | Gramineae |
| RCS2 | AF058902 | 639 | 41 | Tandem | 6,200† | Oryza |
| RCH1 | AF058903 | 827 | 45 | Dispersed | 53 | Gramineae |
| RCH2 | AF058904 | 1,201 | 46 | Dispersed | 99 | Gramineae |
| RCH3 | AF058905 | 1,341 | 48 | Dispersed | 67 | Gramineae |
| RCE1 | AF058906 | 701 | 39 | Dispersed | 287 | Bambusoideae |
| RCE2 | — | 2,700 | — | Dispersed | 305 | Gramineae |
Based on the haploid genome of rice as 424 Mb (24).
The copy number of the 168-bp monomer in the rice genome.
The RCS1 Family.
Clone pRCS1 contains a 877-bp Sau3AI fragment that hybridizes to the pSau3A9 sequence. Sequencing analysis revealed that the 259 bp at the 3′ end of pRCS1 had 80% sequence identity to the central part (bases 338–602) of the pSau3A9 sequence (15). The first 95 bp in pRCS1 had 76% sequence identity to a Ty3/gypsy class of retrotransposon sequence reported in maize (AF030633). Nucleotides 171–228 of pRCS1 had 70% sequence identity to a Ty3/gypsy class of retrotransposon sequence reported in Lilium henryi (X13886). We also found that the pSau3A9 sequence in sorghum has similar sequence identities to the Ty3/gypsy class of retrotransposons. These results indicated that both pSau3A9 and pRCS1 probably were derived from retrotransposon-related DNA sequences.
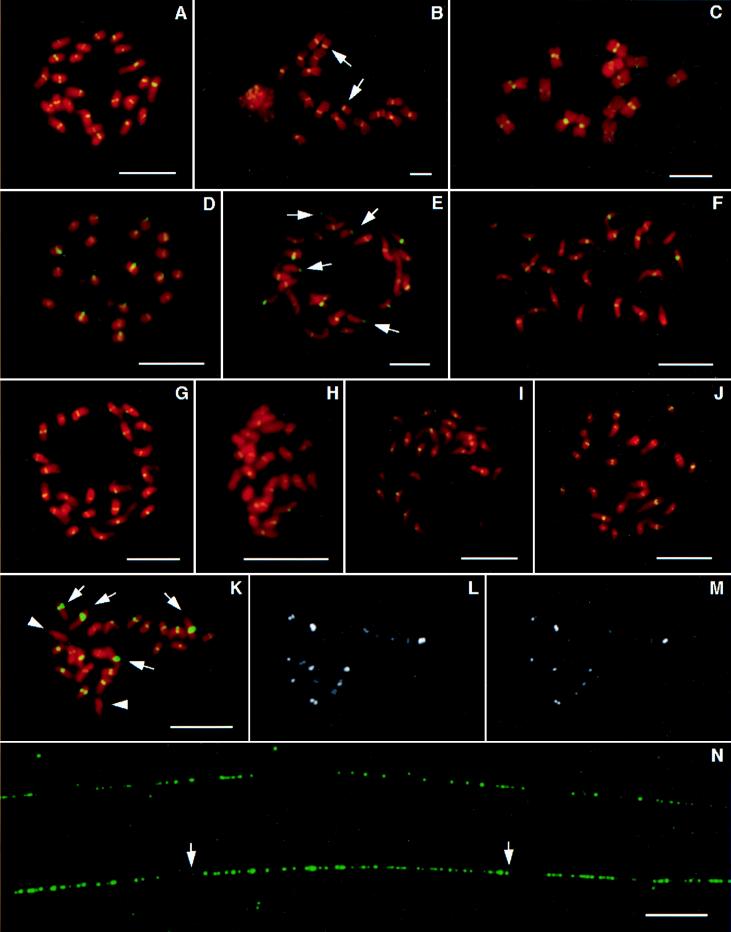
The RCS1 sequence was located in the centromeric regions of all 24 rice chromosomes by FISH (Fig. 1A). The sizes and intensities of the FISH signals were uniform on different chromosomes, suggesting that all rice chromosomes contain a similar number of copies of this element. Slot blot analysis suggested that there are about 130 copies of RCS1 present in the haploid genome of japonica rice DV85 (Table 1).
Figure 1.
FISH analysis of the rice centromeric DNA elements. The probes were biotinylated and hybridized in situ to rice chromosomes or DNA fibers. The probes were detected by fluorescein isothiocyanate-conjugated antibodies (green color) and the chromosomes were stained with propidium iodide (red color). Probe pRCS1 hybridized exclusively to the centromeric regions of the chromosomes from rice (A), rye (B), barley (C), sorghum (D), and maize (E). FISH signals also were detected in the centromeric regions of the acrocentric B chromosomes (arrows) from rye (B) and maize (E). Similarly, rice centromeric DNA families RCH2 (F), RCH1 (G), RCH3 (H), RCE1 (I), RCE2 (J), and RCS2 (K) all were located in the centromere of every rice chromosome. RCS2 showed major difference in size and intensity of FISH signals on different chromosomes (K). Two pairs of chromosomes with the strongest signals are indicated by arrows and the third pair with the weakest signals by arrowheads (K). The same metaphase cell (K) was washed under medium (L) and high (M) strigencies, and most signals are still discernible. (N) Fiber-FISH analysis using pRCS2 as a probe. The RCS2 family is organized into various sizes of uninterrupted arrays. The marked array between two arrows is 51 μm long and represents approximately 151-kb DNA. All bars are 10 μm.
Rice genomic DNA was digested with several restriction enzymes and probed with the 259-bp fragment conserved between rice and sorghum. One or few major bands and several minor bands were detected in most of the lanes (Fig. 2). Fiber-FISH using pRCS1 as a probe did not generate clustered signals (data not shown). These results suggested that the RCS1 sequence is dispersed in the centromeric regions of rice chromosomes.
Figure 2.
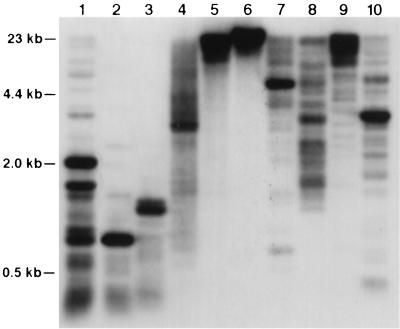
Genomic organization of the RCS1 family. Rice genomic DNA was digested with Sau3AI (lane 1), DpnII (lane 2), HaeIII (lane 3), MspI (lane 4), HpaII (lane 5), SalI (lane 6), BamHI (lane 7), DraI (lane 8), EcoRI (lane 9), and HindIII (lane 10), and probed with pRCS1. One or few major and a number of minor hybridization bands were detected in most lanes.
FISH analysis revealed that pRCS1 also hybridized exclusively to centromeric regions of chromosomes from other Gramineae species (Fig. 1 B–E). The FISH results on rye (Fig. 1B) and barley (Fig. 1C) chromosomes showed that hybridization was exclusive to the primary constrictions. FISH signals also were detected in the centromeres of the supernumerary B chromosomes from both rye and maize (Fig. 1 B and E). Positive Southern hybridization signals were detected in all other Gramineae species analyzed, including bamboo, Pharus sp., oats, wheat, sugar cane, Ae. squarrosa, and Ag. intermedium (data not shown). However, we could not detect homologous sequences by Southern hybridization analysis in dicot species and any monocot species outside of Gramineae, indicating that the RCS1 family is sufficiently conserved only in the grass family Gramineae.
The RCS2 Family.
Clone pRCS2 contains a 639-bp Sau3AI fragment consisting of four copies of a tandemly arranged repeat with a consensus sequence of 168 bp (Fig. 3). The four copies were 84–91% identical with one another. The third copy of the repeat contains a 6-bp insertion (TTGGCC) at base 147. A search of the GenBank database found a highly significant match to a repetitive DNA element isolated from O. sativa (U63977). No publication or description is available for this previously sequenced rice DNA fragment.
Figure 3.
Nucleotide sequence of element pRCS2. The 639-bp insert of clone pRCS2 contains four copies of a tandemly arranged repeat. The four members (A–D) range from 155 to 165 bp and share 84–91% sequence identity with one another. F represents the consensus sequence of the four members.
Southern hybridization analysis of rice genomic DNA using probe pRCS2 revealed ladder patterns using several restriction enzymes, including DpnII, Sau3AI, MspI, HpaII, and HaeIII, indicating that the RCS2 family is tandemly arranged in the rice genome (Fig. 4). Several enzymes produced digestion profiles comprised of monomer and multiples (dimer, trimer, tetramer, etc.) of the 168-bp basic repeat.
Figure 4.
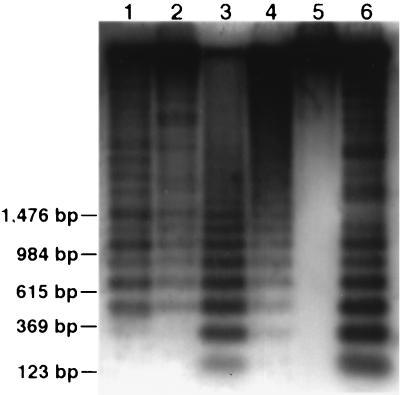
Genomic organization of the RCS2 family. Rice genomic DNA was digested with DpnII (lane 1), Sau3AI (lane 2), MspI (lane 3), HpaII (lane 4), SalI (lane 5), and HaeIII (lane 6), and probed with pRCS2. Five lanes showed ladder patterns, indicating a tandem repeat nature of this element in the rice genome.
Probe pRCS2 hybridized only to the centromeric regions on all rice chromosomes (Fig. 1K). Significant variation in the size and intensity of the FISH signals was detected in different centromeres. Two pairs of chromosomes had strong signals, and a third pair had very faint signals (Fig. 1K). All of the signals became weaker as the posthybridization washing stringency was increased (Fig. 1L). However, even after washing in 70% formamide at 50°C for 15 min most signals were still discernible (Fig. 1M), suggesting that the signal disparity reflects difference in copy numbers rather than sequence divergence of the RCS2 family in different rice centromeres. Though the longest chromosome (chromosome 1) had the strongest signals, it was not possible to relate the copy numbers to the chromosome sizes. It was evident that the weakest signals were not on the smallest chromosomes (Fig. 1K).
Three subspecies of O. sativa (AA genome), together with O. glaberrima (AA), O. rufipogon (AA), O. alta (CCDD), and O. officinalis (CC) were included for FISH analysis. FISH signals were detected in the centromeric regions from all of the chromosomes of these species (data not shown). Southern hybridization analysis revealed that the RCS2 family is present only in the species within genus Oryza (data not shown). We could not detect homologous sequences even at a low stringency in any plant species outside of genus Oryza.
RCS2 is the most abundant element isolated from BAC 17p22 and has about 1,550 copies, corresponding to 6,200 monomers, in the haploid genome of DV85 (Table 1). BAC 17p22 contains about 46 copies of this element, corresponding to approximately 39% of the BAC insert. Fiber-FISH analysis demonstrated that the RCS2 family is organized into various sizes of uninterrupted arrays in the rice genome. Fiber-FISH signals with interspersed gaps smaller than 2 μm can be considered to be derived from continuous DNA sequences (S.A.J. and J.J., unpublished data). The longest observed block with small interspersed gaps (<2 μm) was 51 μm (Fig. 1N). Based on a 2.96-kb/μm resolution of the Fiber-FISH technique (S.A.J. and J.J., unpublished data), this block represents approximately 151 kb of uninterrupted RCS2 sequences. The longest observed single Fiber-FISH signal with interspersed gaps larger than 2 μm was 188 μm, representing approximately 556 kb of centromeric DNA sequences.
Other Rice Centromeric DNA Families.
The other five centromeric DNA elements isolated from rice BAC clone 17p22 were analyzed by FISH, and all of them hybridized exclusively to the centromeric regions of all rice chromosomes (Fig . 1 F–J). One or two pairs of rice metaphase chromosomes showed weak hybridization when pRCH2, pRCH3, and pRCE2 were used as probes. No relationship can be confirmed between signal intensities and the sizes of the chromosomes. pRCH1 also was used as a restriction fragment length polymorphism probe for genetic linkage mapping, and two polymorphic bands were mapped to the centromeres of rice chromosomes 2 and 12, respectively (S. McCouch, personal communication).
The sequence information for these families is listed in Table 1. Searching in the GenBank database did not find any significant matches to these sequences except for pRCH2. Bases 39–102 and 204–232 in pRCH2 had sequence identities to the centromeric CCS1 sequence isolated from B. sylvaticum (16, 25). Interestingly, about 120 bp (bases 8–130) of this element had 80% sequence identity to the spacer sequence that separates the rice 5S rRNA genes. The possibility that this element associates with the 5S rDNA locus was excluded because the FISH signals from pRCH2 was located proximal to those from the 5S rDNA locus (data not shown).
In Southern hybridization analysis, all five elements produced one or few major bands and several minor bands under several restriction enzymes (data not shown), similar to the RCS1 family (Fig. 2), suggesting that they all are dispersed in the rice centromeric regions. The copy numbers of these elements ranged from 53 to 305 copies per haploid rice genome (Table 1).
All five elements were hybridized to various plant species by Southern hybridization. The RCE1 family was present only in the species from the Bambusoideae subfamily, including rice, bamboo, and Pharus sp. (Fig. 5B), whereas RCH1, RCH2, RCH3, and RCE2 all were conserved across the Gramineae species (Fig. 5A for RCH1, data not shown for the others). Species from subfamily Panicoideae and Bambusoideae generally had stronger hybridization signals than those from subfamily Pooideae (Fig. 5A).
Figure 5.
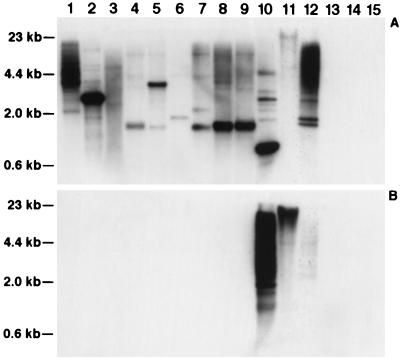
Conservation of the RCH1 and RCE1 families in Gramineae species. Genomic DNA from sorghum (lane 1), maize (lane 2), sugar cane (lane 3), Ag. intermedium (lane 4), barley (lane 5), oats (lane 6), rye (lane 7), wheat (lane 8), Ae. squarrosa (lane 9), rice (lane 10), bamboo (lane 11), Pharus sp. (lane 12), J. effusus (lane 13), C. alternifolius (lane 14), and A. thaliana (lane 15) was digested with HindIII and probed with pRCH1 (A) and pRCE1 (B). The pRCH1 sequence is conserved across the species within the Gramineae family, whereas the pRCE1 sequence is present only in species within the Bambusoideae subfamily.
Methylation of the Rice Centromeric DNA Sequences.
The cytosine nucleotides, especially those in dinucleotide sequence 5′CpG3′, are the most common sites for methylation in plant genomes. Methylation occurs at lower frequencies when the C and G are separated by 1–2 A/T nucleotides (26). Enzymes MspI and HpaII are isoschizomers that recognize the 5′CCGG3′ sequence. Neither enzyme can cut when the 5′C is methylated, and only MspI can cleave when the internal cytosine is methylated. Though both enzymes produced similar digestion profiles of rice genomic DNA (data not shown), MspI generated much smaller-sized hybridization bands from all of the rice centromeric DNA probes than HpaII did (Fig. 2 for RCS1, Fig. 4 for RCS2, data not shown for the others). For the RCS2 element, monomers of the 168-bp basic repeat could be found in MspI lane, and most of the hybridization was in the fragments smaller than 2 kb, whereas the majority of hybridization in the HpaII lane was larger than 2 kb (Fig. 4). For the other centromeric elements, DNA fragments smaller than 5 kb were not detected in HpaII lanes (Fig. 2 for RCS1). These results suggest that the cytosine of the CpG dinucleotides are heavily methylated in the rice centromeric DNA sequences. Restriction enzyme SalI recognizes 5′GTCGAC3′ and is sensitive to the methylation of CpG dinucleotides. Small fragments (<10 kb) that hybridized to the centromeric elements were not detected in the SalI lanes (Figs. 2 and 4).
DISCUSSION
The well characterized eukaryotic centromeres can be grouped into three major types. The first major type represented by the centromeres of S. cerevisiae chromosomes contains only about 125 bp of unique sequence (1–2). The second major type of eukaryotic centromeres consists of a single class of repetitive DNA. For example, the repetitive alpha satellite DNA is the major component of human centromeres (5–7). The third major type of eukaryotic centromeres, such as those from fission yeast (Schizosaccharomyces pombe) and D. melanogaster chromosomes, consists of complex DNA including various classes of repetitive DNA elements. The centromeric regions of S. pombe chromosomes contain several classes of repetitive DNA elements, including K, L, B, J, and M, and these elements are organized into patterns that are peculiar for each centromere (27–32). Unique sequences and repetitive elements specific to a particular centromere also were identified (33). It has been demonstrated that the centromere-specific repeats are essential for stable mitotic and meiotic segregation and maintaining sister chromatid attachment in meiosis I (2, 33). Repetitive DNA element K is absolutely required for centromere function (34–35). The essential core of the centromere in a D. melanogaster minichromosome Dp1187 is contained within a 220-kb region called the Bora Bora island (4). This essential core contains significant amounts of complex DNA, consisting of single copy and middle repetitive sequences (3). Normal chromosome stability also requires about 200 kb of DNA on either side of the essential core. The flanking DNA predominantly contains the AATAT satellite family in Drosophila (4).
Like the fission yeast and Drosophila centromeres, the present study suggests that the rice centromeres also consist of complex DNA, including highly repetitive and middle repetitive DNA sequences. FISH results showed every rice centromere contains members of all seven repetitive DNA elements. DNA sequences specific to one or few rice centromeres were not identified. However, each rice centromere may encompass several megabases of DNA. BAC clone 17p22 contains approximately 75 kb of DNA and represents only part of a rice centromere. Thus, DNA sequences specific to one or few centromeres and/or more centromeric repetitive DNA elements cannot be excluded at present. The most abundant DNA element in the A. thaliana centromeric regions is a 180-bp tandem repeat that is organized as the RCS2 family in rice (see below). A number of middle repetitive DNA elements also were isolated from the centromeric regions of A. thaliana chromosomes (36–37). Like the rice centromeric DNA elements, all of the centromeric DNA elements so far isolated from A. thaliana are also present in every centromere (37, 38). These results suggest that rice and A. thaliana centromeres have similar types of DNA components, including both highly and middle repetitive DNA sequences.
Highly repetitive tandem repeats or satellite DNA have been reported as putative parts of functional centromeres in a number of eukaryotic species, including humans (5–6), mouse (39), D. melanogaster (4), A. thaliana (14), sorghum (40), and presently in rice. The genomic organization of the 180-bp repeat in A. thaliana and the 168-bp RCS2 element in rice share many similarities. Both of them are highly abundant in the genome. In fact, the 180-bp repeat is the most abundant repetitive DNA family, except for the 5.8S-18S-26S rRNA gene clusters, identified so far in A. thaliana. The 180-bp repeat is conserved only in closely related Arabidopsis species (41). Among the seven repetitive DNA elements isolated from BAC 17p22, the tandem repeat family RCS2 is present only in closely related species within Oryza. Other six elements are conserved at different extents in other Gramineae species (Table 1). This result indicates that the RCS2 family has diverged faster than other rice centromeric DNA sequences. Both the 180-repeat (14) and the RCS2 family are organized as long uninterrupted arrays in the centromeres. This organization pattern resembles the alpha satellite DNA in human centromeres.
Evolutionary conservation of DNA sequences is a likely indication of functional importance. For example, the human telomeric DNA sequence was identified and cloned by screening for evolutionary conserved repetitive DNA sequences among the mammalian species (42). The Gramineae species such as rice, maize, and wheat diverged about 60–100 million years ago (43–44). No repetitive DNA element, except the telomeric DNA sequences and a number of multicopy gene families such as the ribosomal RNA gene families, has been reported to be conserved among these species. The conservation of the five rice centromeric sequences across the entire Gramineae family suggests that they might be part of the functional centromeres of rice chromosomes. Direct evidence will come only from functional analysis of these elements. The recent success of Agrobacterium-mediated genetic transformation of large genomic DNA fragments (45) opens the door for future functional analysis of these centromeric elements.
Acknowledgments
We are grateful to Dr. R. D. Vierstra for critical reading of the manuscript. This research is supported by Hatch Funds (142-3935 and 142-D395) and Funds 135-0534 and 135-0528 from the Graduate School of the University of Wisconsin-Madison to J.J.
ABBREVIATIONS
- FISH
fluorescence in situ hybridization
- BAC
bacterial artificial chromosome
- kb
kilobases
Footnotes
References
- 1.Clarke L, Carbon J. Nature (London) 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- 2.Clarke L. Trends Genet. 1990;6:150–154. doi: 10.1016/0168-9525(90)90149-z. [DOI] [PubMed] [Google Scholar]
- 3.Le M-H, Duricka D, Karpen G H. Genetics. 1995;141:283–303. doi: 10.1093/genetics/141.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy T D, Karpen G H. Cell. 1995;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willard H F, Waye J S. Trends Genet. 1987;3:192–198. [Google Scholar]
- 6.Tyler-Smith C, Oakey R J, Larin Z, Fisher R B, Crocker M, Affara N A, Ferguson-Smith M A, Muenke M, Zuffardi O, Jobling M A. Nat Genet. 1993;5:368–375. doi: 10.1038/ng1293-368. [DOI] [PubMed] [Google Scholar]
- 7.Harrington J J, Bokkelen G V, Mays R W, Gustashaw K, Willard H F. Nat Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 8.Peacock W J, Dennis E S, Rhoades M M, Pryor A J. Proc Natl Acad Sci USA. 1981;78:4490–4494. doi: 10.1073/pnas.78.7.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alfenito M R, Birchler J A. Genetics. 1993;135:589–597. doi: 10.1093/genetics/135.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaszas E, Birchler J A. EMBO J. 1996;15:5246–5255. [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Zapater J, Estelle M A, Somerville C R. Mol Gen Genet. 1986;204:417–423. [Google Scholar]
- 12.Simoens C R, Gielen J, Montagu M V, Inze D. Nucleic Acids Res. 1988;16:6753–6766. doi: 10.1093/nar/16.14.6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maluszynska J, Heslop-Harrison J S. Plant J. 1991;1:159–166. [Google Scholar]
- 14.Round E K, Flowers S K, Richards E J. Genome Res. 1997;7:1045–1053. doi: 10.1101/gr.7.11.1045. [DOI] [PubMed] [Google Scholar]
- 15.Jiang J, Nasuda S, Dong F, Scherrer C W, Woo S-S, Wing R A, Gill B S, Ward D C. Proc Natl Acad Sci USA. 1996;93:14210–14213. doi: 10.1073/pnas.93.24.14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aragón-Alcaide L, Miller T, Schwarzacher T, Reader S, Graham M. Chromosoma. 1996;105:261–268. doi: 10.1007/BF02524643. [DOI] [PubMed] [Google Scholar]
- 17.Wang G-L, Holsten T E, Song W-Y, Wang H-P, Ronald P C. Plant J. 1995;7:525–533. doi: 10.1046/j.1365-313x.1995.7030525.x. [DOI] [PubMed] [Google Scholar]
- 18.Hoheisel J D, Maier E, Mott R, McCathy L, Grigoriev A V, Schalkwyk L C, Nizetic D, Francis F, Lehrach H. Cell. 1993;73:109–120. doi: 10.1016/0092-8674(93)90164-l. [DOI] [PubMed] [Google Scholar]
- 19.Gill K S, Lubbers E L, Gill B S, Raupp W J, Cox T S. Genome. 1991;34:362–374. [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 1.25–1.26. [Google Scholar]
- 21.Zhao X, Wu T, Xie Y, Wu R. Theor Appl Genet. 1989;78:201–209. doi: 10.1007/BF00288800. [DOI] [PubMed] [Google Scholar]
- 22.Jiang J, Hulbert S H, Gill B S, Ward D C. Mol Gen Genet. 1996;252:497–502. doi: 10.1007/BF02172395. [DOI] [PubMed] [Google Scholar]
- 23.Fransz P F, Alonso-Blanco C, Liharka T B, Peeters A J M, Zabel P, de Jong J H. Plant J. 1996;9:421–430. doi: 10.1046/j.1365-313x.1996.09030421.x. [DOI] [PubMed] [Google Scholar]
- 24.Arumuganathan K, Earle E D. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- 25.Abbo S, Dunford R P, Foote T, Reader S M, Flavell R B, Moore G. Chromosome Res. 1995;3:5–15. doi: 10.1007/BF00711156. [DOI] [PubMed] [Google Scholar]
- 26.Gruenbaum Y, Naveh-Many T, Cedar H, Razin A. Nature (London) 1981;292:860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- 27.Clarke L, Amstutz H, Fishel B, Carbon J. Proc Natl Acad Sci USA. 1986;83:8253–8257. doi: 10.1073/pnas.83.21.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakaseko Y, Adachi Y, Funahashi S, Niwa O, Yanagida M. EMBO J. 1986;5:1011–1021. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakaseko Y, Kinoshita N, Yanagida M. Nucleic Acids Res. 1987;15:4705–4715. doi: 10.1093/nar/15.12.4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fishel B, Amstutz H, Baum M, Carbon J, Clarke L. Mol Cell Biol. 1988;8:754–763. doi: 10.1128/mcb.8.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M. Cell. 1989;57:739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- 32.Clarke L, Baum M, Marschall L G, Ngan V K, Steiner N C. Cold Spring Harbor Symp Quant Biol. 1993;58:687–695. doi: 10.1101/sqb.1993.058.01.076. [DOI] [PubMed] [Google Scholar]
- 33.Clarke L, Baum M P. Mol Cell Biol. 1990;10:1863–1872. doi: 10.1128/mcb.10.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hahnenberger K M, Carbon J, Clarke L. Mol Cell Biol. 1991;11:2206–2215. doi: 10.1128/mcb.11.4.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baum M, Ngan V, Clarke L. Mol Biol Cell. 1994;5:747–761. doi: 10.1091/mbc.5.7.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson H L, Schmidt R, Dean C. Nucleic Acid Res. 1996;24:3017–3022. doi: 10.1093/nar/24.15.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson H L, Schmidt R, Brandes A, Heslop-Harrison J S, Dean C. Mol Gen Genet. 1996;253:247–252. doi: 10.1007/s004380050319. [DOI] [PubMed] [Google Scholar]
- 38.Brandes A, Thompson H, Dean C, Heslop-Harrison J S. Chromosome Res. 1997;5:238–246. doi: 10.1023/a:1018415502795. [DOI] [PubMed] [Google Scholar]
- 39.Kipling D, Mitchell A R, Masumoto H, Wilson H E, Nicol L, Cooke H J. Mol Cell Biol. 1995;15:4009–4020. doi: 10.1128/mcb.15.8.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, J. T., Jackson, S. A., Nasuda, S., Gill, B. S., Wing, R. A. & Jiang, J. (1998) Theor. Appl. Genet., in press.
- 41.Kamm A, Galasso I, Schmidt T, Heslop-Harrison J S. Plant Mol Biol. 1995;27:853–862. doi: 10.1007/BF00037014. [DOI] [PubMed] [Google Scholar]
- 42.Moyzis R K, Buckingham J M, Cram L S, Dani M, Deaven L L, Jones M D, Meyne J, Ratliff R L, Wu J-R. Proc Natl Acad Sci USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin W, Gierl A, Saedler H. Nature (London) 1989;339:46–48. [Google Scholar]
- 44.Wolfe K H, Gouy M, Yang Y-W, Sharp P M, Li W-H. Proc Natl Acad Sci USA. 1989;86:6201–6205. doi: 10.1073/pnas.86.16.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamilton C M, Frary A, Lewis C, Tanksley S D. Proc Natl Acad Sci USA. 1996;93:9975–9979. doi: 10.1073/pnas.93.18.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]