Abstract
We have investigated the role of two polymorphic sites (R190W and N192K) on the binding and activation of the formyl peptide receptor (FPR) by viral and formyl peptides. WEDWVGWI, a peptide with antiviral activity derived from the membrane proximal region of feline immunodeficiency virus, binds with high affinity to FPR. The three tryptophans in the peptide are all essential for FPR binding, just as they were essential for antiviral activity [S. Giannecchini, A. Di Fenza, A.M. D'Ursi, D. Matteucci, P. Rovero, M. Bendinelli, Antiviral activity and conformational features of an octapeptide derived from the membrane-proximal ectodomain of the feline immunodeficiency virus transmembrane glycoprotein, J. Virol. 77 (2003) 3724]. Formyl-NleWEDWVGWI behaved as a weak partial agonist with FPR W190/N192 but a stronger partial agonist with FPR R190/K192 and FPR R190/N192. Formyl-NleNleWEDWVGWI behaved as a full agonist toward all three FPRs but exhibited a much higher EC50 with W190/N192 FPR (300 ± 45 nM) than for R190/K192 FPR (40 ± 3 nM) or R190/N192 (60 ± 8 nM). Formyl-MYKWPWYVWL preferentially activated R190/K192 and R190/N192 FPRs by > 5 fold compared to W190/N192 FPR. Formyl-MFEDAVAWF, a peptide derived from a protein in Mycobacterium avium subsp. paratuberculosis and formyl-MFTFEPFPTN, a peptide derived from the N-terminus of chemotaxis inhibitory protein of Staphylococcus aureus with an added N-terminal formyl-methionine exhibited the greatest selectivity for R190/K192 and R190/N192 FPRs with ∼ 10 fold lower EC50s than that observed with FPR W190/N192. Thus, individuals with the W190 polymorphism may display a reduced ability to detect certain formyl peptides.
Abbreviations: FPR, formyl peptide receptor; CHIPS, chemotaxis inhibitory protein of Staphylococcus aureus; CHO S, Chinese hamster ovary cells designed for suspension culture; HRSV, human respiratory syncytial virus; FIV, feline immunodeficiency virus; fMLF, N-formyl-methionyl-leucyl-phenylalanine; FLIPr, FPRL1 inhibitory protein; AIDS, Acquired Immunodeficiency Syndrome; SIV, Simian Immunodeficiency Virus; HIV, human immunodeficiency virus; SARS, severe acute respiratory syndrome; GP-41, 41 kilodalton glycoprotein; GP-36, 36 kilodalton glycoprotein; HR, Heptade Repeat; FITC, Fluorescein isothiocyanate; formyl-Nle-Leu-Phe-Nle-Tyr-Lys-FITC, formyl-Nle-Leu-Phe-Nle-Tyr-Lys labeled at the Lys residue with Fluorescein isothiocyanate; formyl-Nle-Leu-Phe-Nle-Tyr-Lys, labeled at the Lys residue with Alexa Fluor N-hydroxy-succinimide; fMLF, formyl-Met-Leu-Phe; TMH, transmembrane helix; FPRL1, formyl peptide receptor like 1; GTPγS, guanosine 5′-3-O-(thio)triphosphate
Keywords: Formyl peptides, Signal Transduction, G protein-coupled receptor, polymorphism, Feline immunodeficiency virus, Chemotaxis inhibitory protein of Staphylococcus aureus
1. Introduction
Neutrophils play an essential role in innate immunity. In addition to activating phagocytosis and secreting superoxides and hydrolytic enzymes, the neutrophil must be able to chemotax, or migrate toward the source of a chemoattractant [1], [2]. The formyl peptide receptor (FPR) is a chemoattractant G protein-coupled receptor found on the surface of phagocytes and it is thought to play an important role in allowing phagocytic cells to recognize the presence of bacteria [3] and damaged cells, since only eubacteria, mitochondria, and chloroplasts initiate their protein synthesis with formyl-methionine [4]. FPR expressing cells also exhibit chemotaxis toward peptides derived from the GP-41 envelope protein of HIV-1 [5], [6]. A peptide derived from herpes simplex virus type 2 elicited chemotaxis and superoxide production in neutrophils in a process that appeared to involve FPR [7]. In addition, we recently observed that peptides from the proximal membrane region of the fusion proteins of human immunodeficiency viruses 1 and 2, severe acute respiratory syndrome coronavirus, coronaviruses 229E and HKU, and Ebola virus were potent inhibitors of FPR [8]. Thus FPR appears to respond to the presence of virally derived peptides in addition to bacterially derived ones (for a review of ligands affecting FPR see [9]). Two proteins secreted by Staphylococcus aureus are inhibitors of FPR ([10], [11], [12]). Chemotaxis inhibitory factor of S. aureus (CHIPS) inhibits FPR and the C5a receptor and a homologue of CHIPS, FPRL1 inhibitory protein (FLIPr) inhibits FPRL1 and to a lesser extent FPR, but not C5a. The N-terminus of CHIPS is essential for binding to FPR since removal of the n-terminal phenylalanine reduces affinity for FPR ∼ 1000 fold. Peptides derived from the N-terminus are also inhibitors of FPR but with ∼ 10,000 fold lower affinity than CHIPS.
Polymorphisms in FPR are very common. Sahagun-Ruiz et al. [13] did an extensive haplotype investigation of FPR and reported finding at least 23 haplotypes for FPR. No polymorphisms were found in the closely related receptor FPRL1 [13]. At present there is no explanation for the wide sequence diversity of FPR. Numerous studies have indicated that patients with aggressive periodontitis exhibit a ∼ 2 fold reduction in chemotaxis toward fMLF [14], [15], [16], [17], [18], [19] indicating that a reduced ability to exhibit chemotaxis toward formyl peptides may be associated with this disease. Several studies have attempted to correlate FPR polymorphisms with aggressive periodontitis but have produced conflicting results [20], [21], [22].
Lentiviruses are associated with immunological impairment in their respective hosts, and both human immunodeficiency and feline immunodeficiency viral infections increase the likelihood of secondary bacterial infections [23], [24]. Recently, Kubes et al. [25] demonstrated that feline neutrophils exhibited a marked (> 90%) reduction of neutrophil chemotaxis toward fMLF following infection with FIV. Ueda et al. noted that the HIV-1 envelope GP-41 was able to downregulate the expression of FPR and several chemokine receptors at low nanomolar concentrations, and that the downregulation of FPR and the chemokine receptors was dependent upon expression of CD4 [26].
Peptides derived from the HR2 and proximal membrane regions of HIV-1, SIV, HRSV, human parainfluenza virus type 3, measles virus, and a coronavirus have all been shown to be able to block virus infection [27], [28], [29], [30]. An eight mer peptide derived from the proximal membrane region of FIV GP-36, WEDWVGWI, was a potent (low nanomolar) inhibitor of FIV infection [31]. The three W residues were all essential for activity whereas the other residues were unimportant for activity.
We previously observed that FPR W190/N192 exhibited an enhanced affinity to bind some viral peptides but had a reduced affinity for the formyl peptide formyl-NleLeuPhe when compared with FPR R190/N192 or FPR R190/K192 [8]. Here we assessed whether the three W residues in WEDWVGWI essential to viral inhibition [31] are also are important in FPR binding. We also undertook a more extensive analysis of three FPR polymorphisms using a variety of formyl peptides of varying sequences in order to identify formyl peptides which might behave markedly different toward the different polymorphisms. We identified several formyl peptides that exhibit high selectivity for activation of R190/K192 and R190/N192 compared with W190/N192. We also identified possible secondary structure changes which might result from amino acid changes by secondary structure prediction [32] and correlateld these structural changes with altered binding and activation of FPR.
2. Materials and methods
2.1. FPR expressing cells
CHO S cells expressing W190/N192, R190/K192, and R190/N192 were prepared as described previously [8].
2.2. Materials
Peptides were obtained from Genscript and were > 75% pure. Mass spectral data was provided with each peptide and the major mass peak matched the expected mass of the peptide in all cases. WEDWVGWI, AEDWVGVWI, WEDAVGWI, and WEDWVGAI were N acetylated and amidated at the C-terminus. All formyl peptides and had free C-termini. FTFEPFPTN had a free N-terminus and a free C-terminus. Formyl-Nle-Leu-Phe was obtained from Sigma and used without further purification. CHIPS was purchased from Cell Sciences.
2.3. FACScan analysis of ligand binding to FPR W190/N192, R190/K192 or R190/N192 expressed in CHO-s cells
CHO S cells (100 μl in Gibco CHO-S-SFMII) were added to 3 mM KCl, 100 mM NaCl, 10 mM sodium phosphate, pH 7.4, 1 mM Mg++, 1 mM Ca++ containing 5% fetal bovine serum and incubated at 4 °C for 60 min with varying concentrations peptide and 0.38 nM formyl-Nle-Leu-Phe-Nle-Tyr-Lys-FITC. The mean fluorescence of the cells was determined, and the mean fluorescence observed in non-expressing CHO S cells (non-specific binding) was subtracted. The K i was then determined by nonlinear least squares analysis using the known K d for formyl-Nle-Leu-Phe-Nle-Tyr-Lys-FITC for each FPR variant [8] and the observed EC50 of each peptide. The K i was determined from the equation K i = EC50/(1 + (ligand/K d) using the graphics/statistics program Graphpad Prizm.
2.4. Downregulation of surface FPR
CHO S cells expressing FPR were incubated in Gibco CHO-S-SFMII at 37 °C for 1 h (or the indicated time for Fig. 2A) with the indicted concentration of peptide. DMSO was 0.4% in all cases. The cells were washed 2× with 10 ml 10 mM phosphate, pH 7.4, 3 mM KCl, 100 mM NaCl at 4 °C. The cells were resuspended in 100 mM NaCl, 3 mM KCl, 10 mM sodium phosphate, pH 7.4, 1 mM Mg++, 1 mM Ca++, 5% fetal bovine serum and 50 nM formyl-Nle-Leu-Phe-Nle-Tyr-Lys-FITC and incubated for 1 h at 4 °C and analyzed by FAScan as describe above. For the receptor recovery experiment, the cells were washed 2× with 10 ml 10 mM phosphate, pH 7.4, 3 mM KCl, 100 mM NaCl at 4 °C after incubation with peptide, resuspended in Gibco CHO-S-SFMII at 37 °C for the indicated time. Then the cells were washed with 10 ml 10 mM phosphate, pH 7.4, 3 mM KCl, 100 mM NaCl at 4 °C and then analyzed by FACScan as above. The mean fluorescence of the cells was determined, and the mean fluorescence observed in non-expressing CHO S cells (non-specific binding) was subtracted. The data were then analyzed by nonlinear least squares analysis and the EC50 and D max determined. For partial agonist activity, D max was the % downregulation of FPRs W190/N192, R190/K192, or R190/N192 compared with 20 μM formyl-Nle-Leu-Phe carried out on the same day.
Fig. 2.
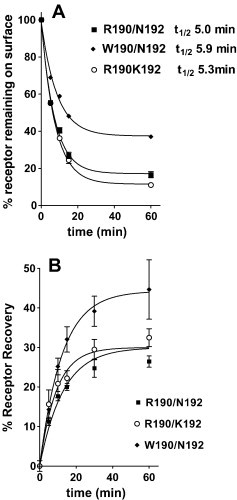
(A) Downregulation of FPR surface expression in response to preincubation at 37 °C for the indicated times with 3 μM formyl-Nle-Leu-Phe for FPR R190/N192 (▪), R190/K192 (○) or W190/N192 (♦). (B) Recovery from downregulation with 3 μM formyl-Nle-Leu-Phe for 60 min at 37 °C after its removal for FPR R190/N192 (▪), R190/K192 (○) or W190/N192.
2.5. Statistical analysis
Experimental results are expressed as the mean ± standard error of the mean. The significance was evaluated by unpaired t test (2 tailed) using the computer program Graphpad Prizm 2.0.
3. Results and discussion
FIV peptides (N-acetylated and C-amidated) derived from the proximal membrane region of GP-36 have been assessed for antiviral activity [31]. Therefore N-acetylated and C-amidated, WEDWVGWI, AEDWVGVWI, WEDAVGWI, and WEDWVGAI were synthesized and evaluated for their ability to inhibit formyl-Nle-Leu-Phe-Nle-Tyr-Lys-FITC binding to FPR. Fig. 1 shows the effect of these peptides on binding to W190/N192, R190/K192, and R190/N192. Only WEDWVGWI exhibited appreciable inhibition similar to what Giannecchini et al. [31] observed for inhibition of viral infection by FIV inhibition. The dose dependence of WEDWVGWI, formyl-NleWEDWVGWI, and formyl-NleNleWEDWVGWI inhibition of formyl-Nle-Leu-Phe-Nle-Tyr-Lys-FITC binding to W190/N192, R190/K192, and R190/N192 was determined. The results are shown in Table 1 . Thus the addition of a formyl-Nle had no affect on affinity and the addition of formyl-NleNle increased the affinity to FPR ∼ 2-fold. W190/N192 exhibited a ∼ 2 fold lower affinity for all three peptides.
Fig. 1.
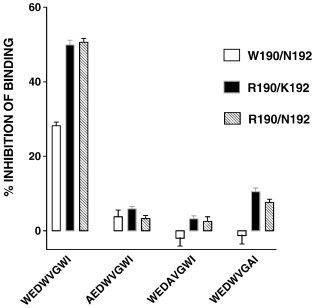
Inhibition of formyl-Nle-Leu-Phe-Nle-Tyr-Lys-FITC binding by 5 μM WEDWVGWI, AEDWVGVWI, WEDAVGWI, and WEDWVGWI in FPRs R190/N192, R190/K192, or W190/N192.
Table 1.
Ki (nM ± SEM) for binding of peptides to R190/N192, R190/K192, or W190/N192 FPR
| R190/N192 | R190/K192 | W190/N192 | |
|---|---|---|---|
| WEDWVGWI | 1300 ± 200 | 1300 ± 300 | 2600 ± 400 |
| Formyl-NleWEDWVGWI | 1600 ± 100 | 1700 ± 100 | 2800 ± 200⁎ |
| Formyl-NleNleWEDWVGWI | 480 ± 30 | 480 ± 20 | 1000 ± 100⁎ |
| Formyl-MFEDAVAWF | 7700 ± 500 | 7000 ± 400 | 30,000 ± 2000⁎⁎ |
| FTFEPFPTN | > 50,000 | > 50,000 | > 50,000 |
| formyl-MFTFEPFPTN | 16.0 ± 0.7⁎⁎⁎ | 9.5 ± 0.2⁎⁎⁎ | 71 ± 4⁎⁎⁎ |
P < 0.001 W190/N192 vs. either R190/N192 or R190/K192.
P < 0.0001 W190/N192 vs. either R190/N192 or R190/K192.
P < 0.0001 vs. both other variants.
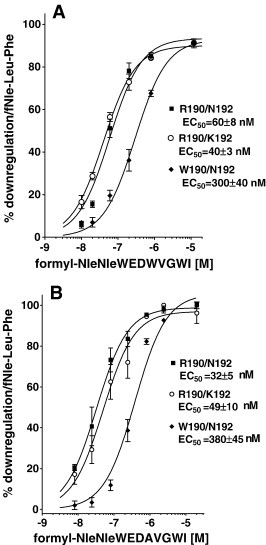
Since downregulation is a hallmark of receptor activation [33] and the trafficking of FPR expressed in CHO cells to endocytotic vesicles following treatment with fMLF has been thoroughly investigated [34], we chose this assay as a highly precise way to evaluate ligand activation of receptor. Fig. 2A shows a time course of loss of surface receptor for the three FPR variants following treatment with 3 μM formyl-Nle-Leu-Phe. All exhibited t1/2 of ∼ 5–6 min but W190/N192 downregulated to a lesser maximum extent. Fig. 2B shows recovery of surface receptor after treatment with formyl-Nle-Leu-Phe for 1 h. All three variants exhibited partial recovery of surface receptor after downregulation with slightly higher recovery observed with W190/N192. We then determined the peptides concentration dependent downregulation surface expression of the receptor to assess whether the peptides exhibited agonist, partial agonist, or antagonist activity. As we observed for other viral peptides [8], WEDWVGWI did not cause any downregulation at concentrations up to 25 μM, but 25 μM WEDWVGWI inhibited the downregulation with 300 nM formyl-Nle-Leu-Phe by ∼ 50% in all three FPRs (data not shown). However, formyl-NleWEDWVGWI behaved as a partial agonist (Fig. 2B) when compared with formyl-Nle-Leu-Phe (Fig. 3A). Only weak partial agonist activity was observed with W190/N192 (34 ± 3%, P < 0.0001 vs. either R190/K192 or R190/N192), and greater partial agonist activity was observed with R190/K192 (55 ± 2%) and R190/N192 (58 ± 4%). W190/N192 also exhibited a higher EC50 for downregulation than R190/K192 or R190/N192 but the difference was not significant. Formyl-NleNleWEDWVGWI behaved as a full agonist for all three FPRs (Fig. 4A), but the EC50 for W190/N192 (300 ± 40 nM, P < 0.0001 vs. either R190/K192 or R190/N192) was 5–7 fold higher than either R190/K192 (40 ± 3 nM) or R190/N192 (60 ± 8 nM) despite the observation that the K is differed by only two fold. The observed EC50s were more than 10 fold below the respective K is for R190/K192 and R190/N192 and 3 fold below the K i of W190/R190, implying that formyl-NleNleWEDWVGWI was 3 times more effective in activating R190/K192 and R190/N192 than W190/N192. This large difference in EC50 vs. K i is in marked contrast to what we observe with formyl-Nle-Leu-Phe (Fig. 3A) where the EC50s are similar to the K i s we previously reported [8]; EC50 = 175 ± 20 vs. K i = 160 ± 25 for W190/N192; EC50 = 75 ± 5 vs. K i = 62 ± 8 for R190/K192; EC50 = 50 ± 7 vs. K i = 67 ± 16 for R190/N192. Thus, formyl-NleNleWEDWVGWI appears to behave as a “super agonist” with R190/K192 and R190/N192, so that occupation of only 5% of the receptors is able to bring about the internalization of 50% of them. We also tested formyl-NleNleWEDAVGWI (Fig. 4B) for its ability to cause downregulation of the receptor. Its behavior was essentially identical to that seen with formyl-NleNleWEDWVGWI. This is in contrast to WEDAVGWI which was > 10 fold weaker than WEDWVGWI in binding to the three FPRs. We also tested formyl-NleNleWED. This peptide exhibited very little downregulation at concentrations up to 50 μM with all three FPRs. Thus the VGWI portion of the peptide, formyl-NleNleWEDWVGWI, appears to be essential for FPR activation.
Fig. 3.
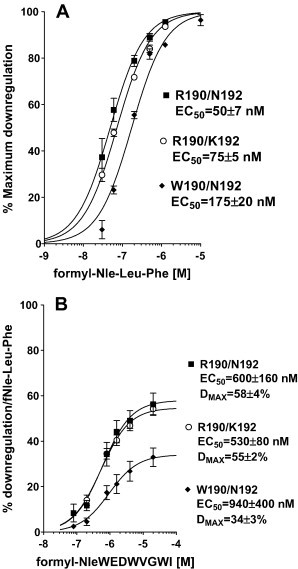
Downregulation of FPR surface expression in response to preincubation with varying concentrations of formyl-Nle-Leu-Phe (A) or formyl-NleWEDWVGWI (B) with FPR R190/N192 (▪), R190/K192 (○) or W190/N192 (♦). Maximum downregulation with formyl-Nle-Leu-Phe was 83 ± 2%, 74 ± 1%, and 60 ± 2% for R190/N192, R190/K192, and W190/N192, respectively.
Fig. 4.
Downregulation of FPR surface expression in response to preincubation with varying concentrations of formyl-NleNleWEDWVGWI (A) or formyl-NleNleWEDAVGWI (B) with FPR R190/N192 (▪), R190/K192 (♦) or W190/N192 (♦).
We tested formyl-MYVKWPWYVWL, which we had previously shown to have essentially identical K is for W190/N192, R190/K192 and R190/N190 [8] of 130 nM, for its ability to downregulate surface receptor (Fig. 5 A). R190/K192 exhibited the lowest EC50 of 44 ± 4 nM about 3 fold below the observed K i and a maximum downregulation very similar (89%) to that seen with formyl-Nle-Leu-Phe (Fig. 3A). R190/N192 had a 2 fold higher EC50 (P = 0.005) than R190/K192 and maximum downregulation similar to that observed with formyl-Nle-Leu-Phe. W190/N192 had a EC50 14 fold greater than K192/N192 (P = 0.0002) and 6 fold greater than R190/N192 (P = 0.0006) and a maximum downregulation slightly reduced (74%) compared to that seen with formyl-Nle-Leu-Phe. This reduction is consistent with the reduced (65%) stimulation of GTPγS binding of W190/N192 FPR by 300 nM formyl-MYVKWPWYVWL compared to R190/K192 or R190/N192 FPRs reported previously [8].
Fig. 5.
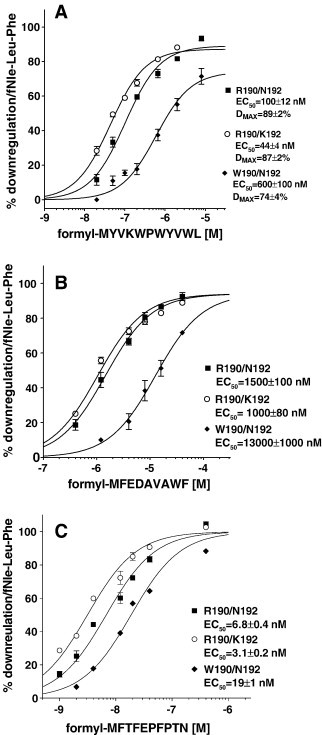
Downregulation of FPR surface expression in response to preincubation with varying concentrations of formyl-MYKWPWYVWL (A) or formyl-MFEDAVAWF (B) or formyl-M MFTFEPFPTN (C) with FPR R190/N192 (▪), R190/K192 (○) or W190/N192 (♦).
We carried out a protein blastp search (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi?PAGE=Proteins&PROGRAM=blastp&BLAST_PROGRAMS=blastp&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on) with the sequence MMWEDAVGWI (Nle is a oxidation resistant analog of M) to identify sequences which were most similar in sequence to formyl-NleNleWEDAVGWI. Bacteria (taxid:2) was used as the search organism. Two sequences of reasonable similarity to MMWEDAVGWI at their N-terminus were identified (Only N-termini would be formylated). One was protein MAP 4176 from Mycobacterium avium subsp. paratuberculosis and the other was protein JNB 01080 from Janibacter. Only the MAP 4176 sequence was investigated and it had MFEDAVAWF as its N-terminal sequence. Formyl-MFEDAVAWF was synthesized and evaluated for binding and downregulation. Formyl-MFEDAVAWF's K i was 4 fold higher for W190/N192 than for R190/K192 or R190/N192 (Table 1). We also determined the EC50 for downregulation. Formyl-MFEDAVAWF behaved as a full agonist with all three FPRs (Fig. 5B) and this peptide activates W190/N190 ∼ 10 fold more weakly than either R190/K192 or R190/N192. EC50s for W190/N192, R190/K192 and R190/N192 were 13,000 ± 1000 nM, 1000 ± 80 nM, and 1500 ± 100 nM, respectively (P < 0.0001, W190/N192 vs. either R190/K192 or R190/N192).
Formyl-MFEDAVAWF exhibited some similarity to the N-terminus of CHIPS so we synthesized the peptide FTFEPFPTN, which corresponds to the N-terminus of CHIPS, and also formyl-MFTFEPFPTN. We tested commercially available CHIPS for its ability to inhibit formyl-Nle-Leu-Phe-Nle-Tyr-Lys-FITC binding but could not detect any inhibition at 50 nM with any of the FPR variants. FTFEPFPTN produced very little inhibition of formyl-Nle-Leu-Phe-Nle-Tyr-Lys-FITC binding also with at most 30% inhibition at 100 μM with the three FPR variants. Formyl-MFTFEPFPTN was very effective at inhibiting all three variants (Table 1) and exhibited significantly different K is for W190/N192, R190/K192 and R190/N192 with K is of 71 ± 4 nM, 16 ± 0.7 nM, and 9.5 ± 0.2 nM, respectively (P < 0.0001 for each variant vs. each of the other two). We also tested formyl-MFTFEPFPTN for its ability to downregulate W190/N192, R190/K192 and R190/N192 (Fig. 5C). The EC50s were 19 ± 1 nM, 3.1 ± 0.2 nM, and 6.8 ± 100 nM, respectively (P < 0.0001 for each variant vs. each of the other two) and the maximum downregulation was very similar to that see with formyl-Nle-Leu-Phe. The addition of formyl-methionine to the N-terminus of FTFEPFPTN enhanced the affinity ∼ 10,000 fold, and formyl-MFTFEPFPTN exhibited an affinity similar to that seen with CHIPS (K d ∼ 35 nM) [11]. It should be noted that while CHIPS was originally isolated as a secreted protein from S. aureus [35], many of the experiments were done using CHIPS expressed in Escherichia coli [10], [11], [36], and the properties of FLIPr were determined only on the E. coli expressed protein [12]. E. coli expressed proteins would be expected to have an N terminal formyl-methionine. However, the authors noted no difference with the E. coli expressed CHIPS [11].
3.1. Effects of variants W190 and K192 on predicted secondary structure
We analyzed the possible effects on secondary structure that might occur in response to changes at positions 190 or 192 using the program PSIPRED which gives a quantitative estimation of secondary structure (http://bioinf.cs.ucl.ac.uk/psipred/) [32]. The results are shown in Table 2 . The sequence prediction of rhodopsin is also given since its crystal structure is known [37]. TMH V in rhodopsin begins at E202 and is the first residue predicted to have a non-zero value for helix propensity. Underlined residues in the rhodopsin sequence are identical in FPR. W190/N192 exhibits a reduced helix propensity of 113 vs. 140 for R190/N192 or 137 for R190/K192 (summation of helix propensity for residues N-terminal to FPR P213 or rhodopsin P215; underlined in Table 2). PRISPED analysis of all three FPR sequences suggests that TMH V is longer than that observed with rhodopsin as we previously suggested [8] based on Kyte and Doolittle [38] analysis, but the predicted beginning of helix V varies depending upon the FPR sequence. Most interesting is the predicted effect W190/N192 has on the helix propensity of the residues N-terminal to FPR P213. P213 was previously shown to be important in FPR structure, since P213A did not fold properly and was retained in the endoplasmic reticulum [39]. Since proline is secondary amine, it cannot hydrogen bond and this destabilizes the helix N-terminal to the proline. In rhodopsin, the carbonyl of H211 hydrogen bonds to the carboxylate of E122 (which can only occur if H211 is not hydrogen bonded to P215, a residue highly conserved in GPCR [40]). The H211 carbonyl is important in rhodopsin activation, since its hydrogen bonding with E122 is disrupted upon rhodopsin activation [41]. We have previously shown that F110, which is analogous to E122 of rhodopsin, is important in FPR activation, since the F110A mutant exhibits very poor coupling to G protein, and very poor chemotaxis toward fMLF [42]. While FPR F110 cannot hydrogen bond to the carbonyl of FPR G209 (analogous to rhodopsin H211), aromatic rings do exhibit energetically favorably interactions with carbonyl oxygens [43]. F110 interaction with the G209 carbonyl and disruption of the F110–G209 interaction by ligand binding could be important in FPR activation. The fact that W190/N190 is predicted to exhibit lower helicity near G209 may alter its ability to be activated in response to ligand binding (Table 2).
Table 2.
Structure prediction by PSIPRED

Conf, confidence; Pred, predicted secondary structure; AA, amino acid; C, coil; H, helix; E, extended. Numbers in parentheses are the summation of the underlined confidence prediction. Underlined bold, identical between rhodopsin and FPR; underlined, similar between rhodopsin and FPR. The highly conserved proline is indicated in italics [40].
We had previously observed that the polymorphism W190/N192 exhibited slightly reduced (∼ 2 fold) affinity for formyl-Nle-Leu-Phe but a 4 fold higher affinity for several virally derived peptides when compared with either R190/K192 or R190/N192 [8]. Here we observed that the three W in the peptide WEDWVGWI are all essential for FPR binding, just as they are essential for antiviral activity [31]. This could be due to changes in structure of the peptide since WEDWVGAI was shown by NMR in DMSO–water solution to adopt a different structure than WEDWVGWI [31]. Nonetheless, the correlation between antiviral activity and ability to interact with FPR is an intriguing one, especially since FPR is able to interact with the proximal membrane region of many viruses [8].
We also evaluated several formyl-Nle derivatives of WEDWVGWI and formyl-MYVKWPWYVWL in order to identify possible formyl peptide ligands which exhibit markedly different activities toward the W190/N192 polymorphism. We also identified a similar peptide from blastp bacterial sequence data bases. Most of the peptides evaluated interacted similarly with R190/K192 and R190/N192 FPRs, but both formyl-MYVKWPWYVWL and formyl- MFTFEPFPTN activated R190/K192 ∼ 2fold more effectively than R190/N192. W190/N192 was activated much more poorly than either R190/K192 or R190/N192 by all the formyl peptides investigated here and exhibited much greater differences in EC50s than observed for formyl-Nle-Leu-Phe. This was true even though W190/N192 bound formyl-NleNleWEDWVGWI and formyl-MYVKWPWYVWL with similar affinity to that observed with R190/K192 or R190/N192. This implies that a W at position 190 interferes with the activation process but does not alter the binding constant appreciably with these peptides.
We also identified a formyl peptide derived from a protein in M. avium subsp. paratuberculosis, formyl-MFEDAVAWI, which activates R190/K192 and R190/N192 ∼ 10 fold better than W190/R190. This could be important since M. avium subsp. paratuberculosis is a facultative intracellular pathogen of macrophages which is associated with Johne's disease in cattle and may be associated with Crohn's disease in humans [44], [45].
Thus, the replacement of R190 by W190 produces profound effects on the activation of the receptor by six different formyl peptides composed of nine or more residues but a much lesser effect on the tri-peptide formyl-Nle-Leu-Phe. For formyl-MYVKWPWYVWL, formyl-NleNleWEDWVGWI, formyl-NleNleWEDAVGWI, and formyl-NleWEDWVGWI, the major difference between W190/N192 and R190/N192 or R190/K192 is the ability to activate the receptor and the difference in K i is small (~ 2 fold or less). For formyl-MFTFEPFPTN and formyl-MFEDAVAWF both K i and EC50 were markedly increased in W190/N192 compared to either R190/N192 or R190/K192. It should be remembered that K i does not reflect the binding energy used to promote conformational change in the receptor, since the latter is potential energy. The EC50 for downregulation, therefore, may be a better representation of the total binding energy.
The fact that the addition of formyl-Nle to WEDWVGWI had little effect (< 20%) on its affinity but the addition of formyl-Met to FTFEPFPTN enhanced the affinity ∼ 10,000 fold implies a marked difference in the mode of binding of the two peptides. One possibility is that WEDWVGWI occupies the formyl-Met (or formyl-Nle) binding pocket but FTFEPFPTN does not. The fact that formyl-MFTFEPFPTN binds with similar affinity to CHIPS may indicate that in the intact protein, some residue of the protein is able to interact with the formyl-Met binding site and substitute for it but not activate the receptor.
We had previously proposed that R205 in TMH V interacts with the C-terminus of fMLF [46] and is essential for binding this tri-peptide, but is much less essential in binding larger peptides like formyl-Nle-Leu-Phe-Nle-Tyr-Lys-FITC [39]. We have also shown that W190 quenches the fluorescence of formyl-Nle-Leu-Phe-Nle-Tyr-Lys-AlexaFluor, indicating W190 must reside close to the AlexaFluor moiety when this peptide is bound to FPR [8]. These findings are both consistent with the larger effects observed with W190/N192 with longer peptides since only peptides much longer than three residues are likely to interact near R or W190. In addition, provided that formyl-MFTFEPFPTN is a good model for CHIPS binding, this might indicate that the polymorphism W190/N192 would be more resistant to inhibition by CHIPS and thus those with this polymorphism might control infections with S. aureus better than those with the R190 polymorphism. Recent studies have indicated that a high percentage of normal individuals (24/24) express antibodies to CHIPS [47], so that a polymorphism that was more resistant to its effect might exhibit a selective advantage, since W190/N192 retains most of its ability to interact with formylated tri peptides.
Acknowledgments
I thank Heini Miettinen for supplying PGBSA FPR expressing plasmid. I thank Heini Miettinen, John Walters, and Jim Burrit for helpful discussions and critical reading of this manuscript.
These studies were supported by NIH grant 1R21DE016114-03.
References
- 1.Katanaev V.L. Signal transduction in neutrophil chemotaxis. Biochemistry (Mosc.) 2001;66:351. doi: 10.1023/a:1010293809553. [DOI] [PubMed] [Google Scholar]
- 2.Springer T.A. Traffic signals on endothelium for lymphocyte recirculation and leukocyte emigration. Annu. Rev. Physiol. 1995;57:827. doi: 10.1146/annurev.ph.57.030195.004143. [DOI] [PubMed] [Google Scholar]
- 3.Schiffman E., Corcorin B., Wahl B. N-formyl methionine peptides as chemoattractants for leukocytes. Proc. Natl. Acad. Sci. U. S. A. 1975;72:1059. doi: 10.1073/pnas.72.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaughan M.D., Sampson P.B., Honek J.F. Methionine in and out of proteins: targets for drug design. Curr. Med. Chem. 2002;9:385. doi: 10.2174/0929867023371102. [DOI] [PubMed] [Google Scholar]
- 5.Su S.B., Gong W.H., Gao J.L., Shen W.P., Grimm M.C., Deng X., Murphy P.M., Oppenheim J.J., Wang J.M. T20/DP178, an ectodomain peptide of human immunodeficiency virus type 1 gp41, is an activator of human phagocyte N-formyl peptide receptor. Blood. 1999;93:3885. [PubMed] [Google Scholar]
- 6.Su S.B., Gao J., Gong W., Dunlop N.M., Murphy P.M., Oppenheim J.J., Wang J.M. T21/DP107, A synthetic leucine zipper-like domain of the HIV-1 envelope gp41, attracts and activates human phagocytes by using G-protein-coupled formyl peptide receptors. J. Immunol. 1999;162:5924. [PubMed] [Google Scholar]
- 7.Bellner L., Thoren F., Nygren E., Liljeqvist J.A., Karlsson A., Eriksson K. A proinflammatory peptide from herpes simplex virus type 2 glycoprotein G affects neutrophil, monocyte, and NK cell functions. J. Immunol. 2005;174:2235. doi: 10.4049/jimmunol.174.4.2235. [DOI] [PubMed] [Google Scholar]
- 8.Mills J.S. Peptides derived from HIV-1, HIV-2, Ebola virus, SARS coronavirus and coronavirus 229E exhibit high affinity binding to the formyl peptide receptor. Biochim. Biophys. Acta. 2006;1762:693. doi: 10.1016/j.bbadis.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabiet M.J., Huet E., Boulay F. The N-formyl peptide receptors and the anaphylatoxin C5a receptors: an overview. Biochimie. 2007 doi: 10.1016/j.biochi.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haas P.J., de Haas C.J., Kleibeuker W., Poppelier M.J., van Kessel K.P., Kruijtzer J.A., Liskamp R.M., van Strijp J.A. N-terminal residues of the chemotaxis inhibitory protein of Staphylococcus aureus are essential for blocking formylated peptide receptor but not C5a receptor. J. Immunol. 2004;173:5704. doi: 10.4049/jimmunol.173.9.5704. [DOI] [PubMed] [Google Scholar]
- 11.Postma B., Poppelier M.J., van Galen J.C., Prossnitz E.R., van Strijp J.A., de Haas C.J., van Kessel K.P. Chemotaxis inhibitory protein of Staphylococcus aureus binds specifically to the C5a and formylated peptide receptor. J. Immunol. 2004;172:6994. doi: 10.4049/jimmunol.172.11.6994. [DOI] [PubMed] [Google Scholar]
- 12.Prat C., Bestebroer J., de Haas C.J., van Strijp J.A., van Kessel K.P. A new staphylococcal anti-inflammatory protein that antagonizes the formyl peptide receptor-like 1. J. Immunol. 2006;177:8017. doi: 10.4049/jimmunol.177.11.8017. [DOI] [PubMed] [Google Scholar]
- 13.Sahagun-Ruiz A., Colla J.S., Juhn J., Gao J.L., Murphy P.M., McDermott D.H. Contrasting evolution of the human leukocyte N-formylpeptide receptor subtypes FPR and FPRL1R. Genes Immun. 2001;2:335. doi: 10.1038/sj.gene.6363787. [DOI] [PubMed] [Google Scholar]
- 14.Daniel M.A., McDonald G., Offenbacher S., Van Dyke T.E. Defective chemotaxis and calcium response in localized juvenile periodontitis neutrophils. J. Periodontol. 1993;64:617. doi: 10.1902/jop.1993.64.7.617. [DOI] [PubMed] [Google Scholar]
- 15.Lavine W.S., Maderazo E.G., Stolman J., Ward P.A., Cogen R.B., Greenblatt I., Robertson P.B. Impaired neutrophil chemotaxis in patients with juvenile and rapidly progressing periodontitis. J. Periodontal Res. 1979;14:10. doi: 10.1111/j.1600-0765.1979.tb00213.x. [DOI] [PubMed] [Google Scholar]
- 16.Perez H.D., Kelly E., Elfman F., Armitage G., Winkler J. Defective polymorphonuclear leukocyte formyl peptide receptor(s) in juvenile periodontitis. J. Clin. Invest. 1991;87:971. doi: 10.1172/JCI115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibata K., Warbington M.L., Gordon B.J., Kurihara H., Van Dyke T.E. Nitric oxide synthase activity in neutrophils from patients with localized aggressive periodontitis. J. Periodontol. 2001;72:1052. doi: 10.1902/jop.2001.72.8.1052. [DOI] [PubMed] [Google Scholar]
- 18.Sigusch B., Eick S., Pfister W., Klinger G., Glockmann E. Altered chemotactic behavior of crevicular PMNs in different forms of periodontitis. J. Clin. Periodontol. 2001;28:162. doi: 10.1034/j.1600-051x.2001.028002162.x. [DOI] [PubMed] [Google Scholar]
- 19.Van Dyke T.E., Levine M.J., Tabak L.A., Genco R.J. Reduced chemotactic peptide binding in juvenile periodontitis: a model for neutrophil function. Biochem. Biophys. Res. Commun. 1981;100:1278. doi: 10.1016/0006-291x(81)91962-8. [DOI] [PubMed] [Google Scholar]
- 20.P. Maney, J.S. Mills, J.D. Walters, Formylpeptide Receptor Polymorphisms in African-Americans With Aggressive Periodontitis., 2006, p. 2335.
- 21.Nibali L., Parkar M., Brett P., Knight J., Tonetti M.S., Griffiths G.S. NADPH oxidase (CYBA) and FcgammaR polymorphisms as risk factors for aggressive periodontitis: a case-control association study. J. Clin. Periodontol. 2006;33:529. doi: 10.1111/j.1600-051X.2006.00952.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Syed R., Uygar C., Pallos D., Gorry M.C., Firatli E., Cortelli J.R., VanDyke T.E., Hart P.S., Feingold E., Hart T.C. Evaluation of human leukocyte N-formylpeptide receptor (FPR1) SNPs in aggressive periodontitis patients. Genes Immun. 2003;4:22. doi: 10.1038/sj.gene.6363900. [DOI] [PubMed] [Google Scholar]
- 23.Heit B., Jones G., Knight D., Antony J.M., Gill M.J., Brown C., Power C., Kubes P. HIV and other lentiviral infections cause defects in neutrophil chemotaxis, recruitment, and cell structure: immunorestorative effects of granulocyte-macrophage colony-stimulating factor. J. Immunol. 2006;177:6405. doi: 10.4049/jimmunol.177.9.6405. [DOI] [PubMed] [Google Scholar]
- 24.Overbaugh J., Luciw P.A., Hoover E.A. Models for AIDS pathogenesis: simian immunodeficiency virus, simian-human immunodeficiency virus and feline immunodeficiency virus infections. AIDS. 1997;11:S47–S54. Suppl A. [PubMed] [Google Scholar]
- 25.Kubes P., Heit B., G.van M., Johnston J.B., Knight D., Khan A., Power C. In vivo impairment of neutrophil recruitment during lentivirus infection. J. Immunol. 2003;171:4801. doi: 10.4049/jimmunol.171.9.4801. [DOI] [PubMed] [Google Scholar]
- 26.Ueda H., Howard O.M., Grimm M.C., Su S.B., Gong W., Evans G., Ruscetti F.W., Oppenheim J.J., Wang J.M. HIV-1 envelope gp41 is a potent inhibitor of chemoattractant receptor expression and function in monocytes. J. Clin. Invest. 1998;102:804. doi: 10.1172/JCI3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosch B.J., van der Z.R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambert D.M., Barney S., Lambert A.L., Guthrie K., Medinas R., Davis D.E., Bucy T., Erickson J., Merutka G., Petteway S.R., Jr. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2186. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wild C.T., Shugars D.C., Greenwell T.K., McDanal C.B., Matthews T.J. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9770. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malashkevich V.N., Chan D.C., Chutkowski C.T., Kim P.S. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc. Natl. Acad. Sci. U. S. A. 1998;95:9134. doi: 10.1073/pnas.95.16.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giannecchini S., Di Fenza A., D'Ursi A.M., Matteucci D., Rovero P., Bendinelli M. Antiviral activity and conformational features of an octapeptide derived from the membrane-proximal ectodomain of the feline immunodeficiency virus transmembrane glycoprotein. J. Virol. 2003;77:3724. doi: 10.1128/JVI.77.6.3724-3733.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 33.Clark R.B., Knoll B.J., Barber R. Partial agonists and G protein-coupled receptor desensitization. Trends Pharmacol. Sci. 1999;20:279. doi: 10.1016/s0165-6147(99)01351-6. [DOI] [PubMed] [Google Scholar]
- 34.Suvorova E.S., Gripentrog J.M., Miettinen H.M. Different endocytosis pathways of the C5a receptor and the N-formyl peptide receptor. Traffic. 2005;6:100. doi: 10.1111/j.1600-0854.2004.00256.x. [DOI] [PubMed] [Google Scholar]
- 35.de Haas C.J., Veldkamp K.E., Peschel A., Weerkamp F., van Wamel W.J., Heezius E.C., Poppelier M.J., van Kessel K.P., van Strijp J.A. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J. Exp. Med. 2004;199:687. doi: 10.1084/jem.20031636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Postma B., Kleibeuker W., Poppelier M.J., Boonstra M., van Kessel K.P., van Strijp J.A., de Haas C.J. Residues 10–18 within the C5a receptor N terminus compose a binding domain for chemotaxis inhibitory protein of Staphylococcus aureus. J. Biol. Chem. 2005;280:2020. doi: 10.1074/jbc.M412230200. [DOI] [PubMed] [Google Scholar]
- 37.Palczewski K., Kumasaka T., Hori T., Behnke C.A., Motoshima H., Fox B.A., Le T., Teller D.C., Okada T., Stenkamp R.E., Yamamoto M., Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 38.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 39.Miettinen H.M., Mills J.S., Gripentrog J.M., Dratz E.A., Granger B.L., Jesaitis A.J. The ligand binding site of the formyl peptide receptor maps in the transmembrane region. J. Immunol. 1997;159:4045. [PubMed] [Google Scholar]
- 40.Eilers M., Hornak V., Smith S.O., Konopka J.B. Comparison of class A and D G protein-coupled receptors: common features in structure and activation. Biochemistry. 2005;44:8959. doi: 10.1021/bi047316u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patel A.B., Crocker E., Reeves P.J., Getmanova E.V., Eilers M., Khorana H.G., Smith S.O. Changes in interhelical hydrogen bonding upon rhodopsin activation. J. Mol. Biol. 2005;347:803. doi: 10.1016/j.jmb.2005.01.069. [DOI] [PubMed] [Google Scholar]
- 42.Jones B.E., Miettinen H.M., Jesaitis A.J., Mills J.S. Mutations of F110 and C126 of the formyl peptide receptor interfere with G-protein coupling and chemotaxis. J. Periodontol. 2003;74:475. doi: 10.1902/jop.2003.74.4.475. [DOI] [PubMed] [Google Scholar]
- 43.Thomas K.A., Smith G.M., Thomas T.B., Feldmann R.J. Electronic distributions within protein phenylalanine aromatic rings are reflected by the three-dimensional oxygen atom environments. Proc. Natl. Acad. Sci. U. S. A. 1982;79:4843. doi: 10.1073/pnas.79.16.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Autschbach F., Eisold S., Hinz U., Zinser S., Linnebacher M., Giese T., Loffler T., Buchler M.W., Schmidt J. High prevalence of Mycobacterium avium subspecies paratuberculosis IS900 DNA in gut tissues from individuals with Crohn's disease. Gut. 2005;54:944. doi: 10.1136/gut.2004.045526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greenstein R.J. Is Crohn's disease caused by a mycobacterium? Comparisons with leprosy, tuberculosis, and Johne's disease. Lancet Infect. Dis. 2003;3:507. doi: 10.1016/s1473-3099(03)00724-2. [DOI] [PubMed] [Google Scholar]
- 46.Mills J.S., Miettinen H.M., Cummings D., Jesaitis A.J. Characterization of the binding site on the formyl peptide receptor using three receptor mutants and analogs of Met-Leu-Phe and Met-Met-Trp-Leu-Leu. J. Biol. Chem. 2000;275:39012. doi: 10.1074/jbc.M003081200. [DOI] [PubMed] [Google Scholar]
- 47.Wright A.J., Higginbottom A., Philippe D., Upadhyay A., Bagby S., Read R.C., Monk P.N., Partridge L.J. Characterisation of receptor binding by the chemotaxis inhibitory protein of Staphylococcus aureus and the effects of the host immune response. Mol. Immunol. 2007;44:2507. doi: 10.1016/j.molimm.2006.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]


