Abstract
As part of our studies of lethal viral mutagens, a series of 5-substituted cytidine analogues were synthesized and evaluated for antiviral activity. Among the compounds examined, 5-nitrocytidine was effective against poliovirus (PV) and coxsackievirus B3 (CVB3) and exhibited greater activity than the clinically employed drug ribavirin. Instead of promoting viral mutagenesis, 5-nitrocytidine triphosphate inhibited PV RNA-dependent RNA polymerase (Kd = 1.1 ± 0.1 μM), and this inhibition is sufficient to explain the observed antiviral activity.
Ribonucleoside analogues that enhance the basal mutation frequency of RNA viruses constitute a promising new class of antiviral therapeutics. Such compounds, termed lethal mutagens, accelerate viral mutagenesis to intolerable levels, resulting in “error catastrophe” and loss of viral viability.1–9 The mechanism of antiviral activity for mutagenic ribonucleoside analogues typically involves (i) in vivo conversion to ribonucleotides facilitated by host cell enzymes, (ii) misincorporation into the viral genome by error-prone viral RNA-dependent RNA polymerases (RdRPa), and (iii) indiscriminate nucleotide templating during genomic replication. Over successive rounds of replication, the accrual of excessive mutations forces the virus into “error catastrophe”, and viral viability is lost. Previously, we demonstrated that ribavirin (1), a clinically employed antiviral drug, functions as a lethal mutagen against poliovirus (PV)8 and hepatitis C virus.10 Inspired by the known lethal mutagen for HIV, 5-hydroxy-2′-deoxycytidine (2),6,9 we report here the antiviral evaluation activity of a suite of 5-substituted cytidine analogues (4–7).

Hydroxylated cytosines, such as 5-hydroxycytosine, are hallmarks of oxygen radical induced DNA damage and early causative factors in genomic mutagenesis.11 The addition of a hydroxyl moiety to the 5-position of cytosine alters the relative distribution of amino to imino nucleobase tautomers, thereby increasing the abundance of imino 5-hydroxycytosine, which can base-pair with adenine.12–14 Mispairing of this oxidative lesion with A during DNA replication promotes transition mutations and consequential scrambling of the encoded genetic message. Oxidative DNA damage can also occur by the introduction of oxidized deoxycytidine triphosphates (dCTPs) into the genome during replication. The 5′-triphosphate of 2, a product of dCTP oxidation, is incorporated into DNA by Klenow DNA polymerase I.15,16 Fortunately, the integrity of DNA is maintained by complex networks of repair machinery that target such forms of DNA damage.17,18
On the basis of its mutagenic capacity toward genomic DNA, 5-hydroxy-2′-deoxycytidine (2) has been evaluated as an antiviral lethal mutagen against the HIV retrovirus.9 Treatment with 2 confers significant reductions in viral titer, including an increase in G to A substitutions in the gene-encoding reverse transcriptase (RT).9 In addition, the 5′-triphosphate of 2 functions as a substrate for HIV RT and is incorporated opposite G and A in the DNA template.9,19 To build upon these results, we hypothesized that ribonucleoside 4 might function as an analogous antiviral lethal mutagen against RNA viruses. To test this hypothesis, we synthesized 5-hydroxycytidine (4) and related analogues 5–7 and evaluated the antiviral activity of these compounds against the RNA viruses poliovirus and coxsackievirus B3 (CVB3).
An improved synthesis of 5-nitrocytidine (6) and 5-aminocytidine (7) is shown in Scheme 1. Readily prepared 5-nitrocytosine (10)20 was persilylated by reaction with HMDS and catalytic TMSCl to provide 11. Vorbrüggen coupling conditions21,22 afforded benzoyl-protected 5-nitrocytidine 13.23 Hydrogenation of 1323 provided the protected 5-aminocytidine 14. Saponification of esters 13 and 14 as previously described23 delivered 6 and 7 in 40% and 26% overall yields. This approach is more rapid than an earlier reported syntheses of 6 and 7 via the common intermediate 13.23 Compound 7 has also been synthesized by amination of 5-bromocytidine (5) with ammonia. However, these approaches suffer from low yields or require separation of the 5- and 6-amino regioisomers.24,25
Scheme 1a.
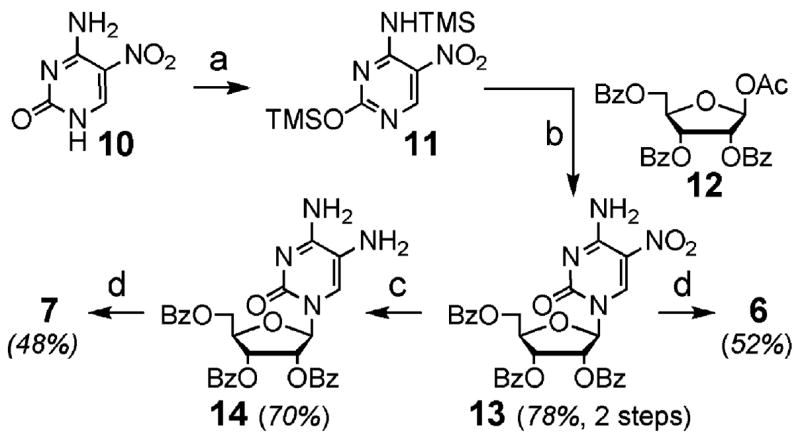
a (a) TMSCl, HMDS; (b) SnCl4, MeCN; (c) 10% Pd/C, AcOH, THF; (d) NaOH(aq), EtOH.
The cytotoxicity of ribavirin (1), 5-hydroxy-2′-deoxycytidine (2), 5-bromo-2′-deoxycytidine (3), and ribonucleoside analogues 4–7 was evaluated in HeLa S3 cells (Figure 1). 5-Hydroxycytidine (4) was the most toxic ribonucleoside, with associated host cell viability ranging from 31% to 40% across the four concentrations tested. Interestingly, 2 was significantly less cytotoxic, with >73% cell viability observed at all of the concentrations examined. The least cytotoxic ribonucleoside proved to be 5-nitrocytidine (6), a compound that yielded cell viabilities greater than 76% for all concentrations tested.
Figure 1.

(A) Cytotoxicity to HeLa S3 cells after treatment with 1–7 for 7 h, followed by recovery without compounds for 24 h. (B, C) Antiviral effects of compounds against poliovirus (B) and coxsackievirus B3 (C). HeLa S3 cells were incubated with 1–7 for 1 h at the concentrations shown and subsequently infected with 106 PFU of PV or CVB3. Fifteen minutes after the infection, fresh media containing 1–7 was added, and the infection progressed for 6 h. Cell-associated virus was titered with plaque assays.
The antiviral activity of 1–7 was evaluated against PV and CVB3 in cell culture (Figure 1). In these experiments, HeLa S3 cells were pretreated with 1–7 for 1 h, followed by administration of a high multiplicity of infection (MOI) dose of either virus. After rapid association of virus with the host cells (15 min), fresh media containing 1–7 was added at the concentrations shown. The infection was allowed to progress for an additional 6 h, and cell-associated virus was subsequently titered by plaque assay as previously described.8,26 As expected, the antiviral drug ribavirin (1) elicited a dose-dependent reduction in viral titer in both PV and CVB3 infected cells (Figure 1). Both 2′-deoxycytidines (2 and 3) failed to reduce the titer of either virus at all concentrations tested. Surprisingly, 5-hydroxycytidine (4) also failed to significantly affect PV or CVB3 titer at any concentration. Interestingly, 5-nitrocytidine (6) and 5-aminocytidine (7) substantially decreased viral titer in PV and CVB3-infected cells, with 6 surpassing the antiviral activity of 1. Compared to treatment with ribavirin, virally infected HeLa S3 cells treated with 6 produced 33-fold and 12-fold less viable PV and CVB3, respectively, at the highest concentration tested.
To probe the antiviral mechanism of action of 6, we synthesized its 5′-triphosphate 9 and evaluated the ability of 9 to function as a substrate for PV RdRP in a primer-extension assay.27 Four distinct primer templates were utilized to probe the in vitro incorporation of 9 opposite each RNA nucleobase mediated by purified PV RdRP.28 For comparison, the incorporation of the structurally related 5-bromocytidine triphosphate 8 and natural nucleotides opposite each templating base was examined. Both 8 and 9 were incorporated opposite guanine and adenine (Figure 2). However, an unusually long, biologically irrelevant time scale (15 min) was employed so that inefficient incorporation by PV RdRP could be detected. Neither nucleotide was incorporated opposite cytosine or uracil. Compared with incorporation of CTP into S/S–G (kpol = 157 ± 8 s−1, Kd = 19.2 ± 3.2 μM),8 5-nitrocytidine triphosphate 9 was added approximately 1900-fold more slowly (kpol = 0.082 ± 0.005 s−1) and bound the polymerase with an 18-fold higher affinity (Kd = 1.08 ± 0.08 μM). The slow rate of incorporation of 9 into viral RNA by PV RdRP, coupled with a negative result in a previously described guanidine-resistance assay for poliovirus mutagenesis (data not shown),8,29 suggests that the antiviral activity observed for 5-nitrocytidine (6) does not result from lethal mutagenesis. Alternatively, triphosphate 9, owing to its 18-fold higher affinity for the RdRP (compared with CTP), more likely functions as an inhibitor of this enzyme.
Figure 2.
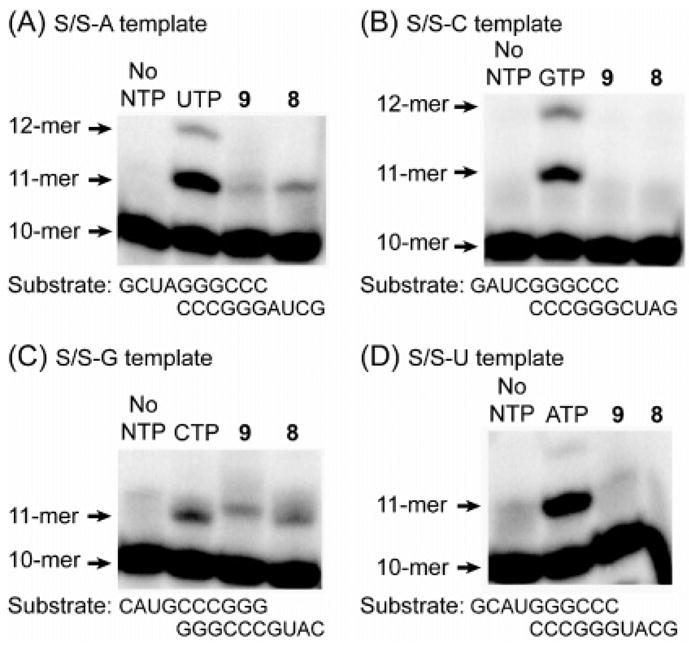
Incorporation of nucleotides derived from triphosphates 8 and 9 into symmetrical RNA substrates (S/S–N) in vitro. The 10-mer substrates were end-labeled with 32P, complexed with PV RdRP, and treated with the correct NTP, 8, or 9. An extended time period (15 min) was used to detect incorporation of 8 and 9. Products were separated by denaturing PAGE.
For triphosphate 9 to confer an antiviral effect by inhibiting PV RdRP, 6 must be metabolized to 9 in living HeLa cells. To examine the presence of triphosphate 9 in cell extracts, HeLa S3 cells were incubated with nucleoside 6, the intracellular nucleotides were extracted, and phosphorylated metabolites were analyzed by reverse-phase (RP) HPLC.30 As shown in Figure 3, analysis of the intracellular nucleotide pool revealed the presence of 9 from the similarity in retention time and UV absorbance compared with an authentic standard (details in the Supporting Information). Co-injection of crude intracellular material spiked with a known amount of 9 as a standard revealed an enhancement in signal intensity, confirming that 9 is formed from 6 in HeLa S3 cells.
Figure 3.
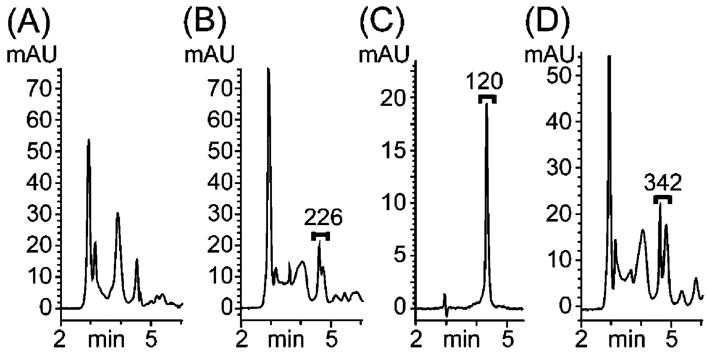
Analysis of HeLa cell extracts by RP HPLC. (A) Untreated cells were lysed, nucleotides were extracted, and crude cellular material was injected. (B) Cell extract after treatment with 6 (2 mM) for 3 h. The bracketed region integrating for 226 mAU s−1 includes 9 (see Supporting Information). (C) Analysis of 9 (0.56 nmol) as a standard (120 mAU s−1). (D) Co-injection of the material shown in panel B spiked with 9 (0.56 nmol). The area under the bracket (342 mAU s−1) is the sum of materials from panels B and C analyzed separately.
To determine if 5-nitrocytidine (6) affects the kinetics of viral replication, a luciferase-based reporter assay for PV replication was performed. HeLa S3 cells were transfected with the subgenomic replicon pRLucRA, which contains the wild-type PV sequence with the capsid-coding region replaced by the luciferase reporter gene.31,32 Cells were treated with 6, 1, or a vehicle control (DMSO, 1%). Additionally, guanidine hydrochloride, a reversible inhibitor of PV replication,29 provided another control. As shown in Figure 4, cells treated with 6 showed a lag in replication kinetics compared to the vehicle and ribavirin.
Figure 4.
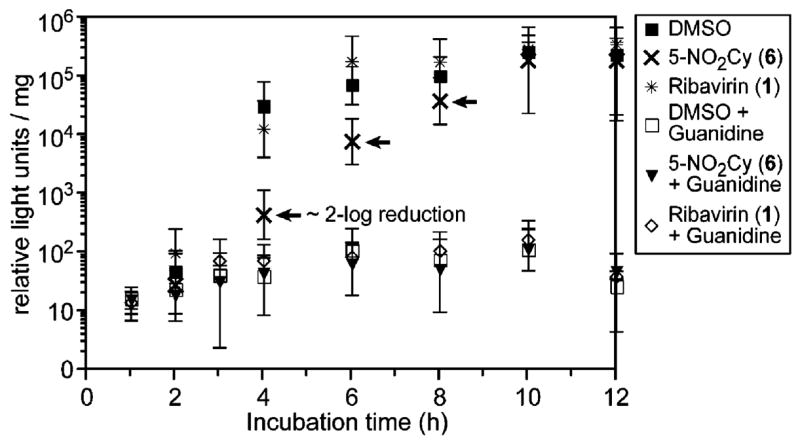
Replication of PV quantified by a luciferase reporter assay. HeLa S3 cells were transfected with pRLucRA. Cells were treated at 37 °C with DMSO (1%) vehicle control, 6 (1 mM), or 1 (2 mM) in the presence or absence of 3 mM guanidine hydrochloride. Arrows illustrate the lag in replication induced by 6.
Given the high affinity of the 5-nitrocytidine triphosphate (9) for PV RdRP, inhibition of this enzyme by metabolite 9 is a potential mechanism of antiviral activity. To further measure inhibition of PV RdRP in the presence of 9, we performed an additional primer-extension assay to detect “stalling” of this RdRP (Figure 5).27 The primer template utilized in this experiment places G (a templating nucleotide for 9) in the second position. As expected, extension of the nucleotide to the +3 product (13-mer) is rapidly achieved by addition of three nucleotides (GTP, CTP, and ATP) to corresponding templating bases (C, G, and U). Conversely, template extension is not observed in the presence of 9 alone. However, replacing CTP with 9 in the presence of GTP and ATP results in significant “stalling” at the +1 product, signifying rapid GTP incorporation opposite C and slow incorporation of 9 opposite templating G. In the context of the ~1800 possible CTP incorporations that occur during PV replication, this observed RdRP “stalling” would greatly diminish the efficiency of viral replication, and this inhibition is sufficient to explain the observed antiviral activity of 6.
Figure 5.
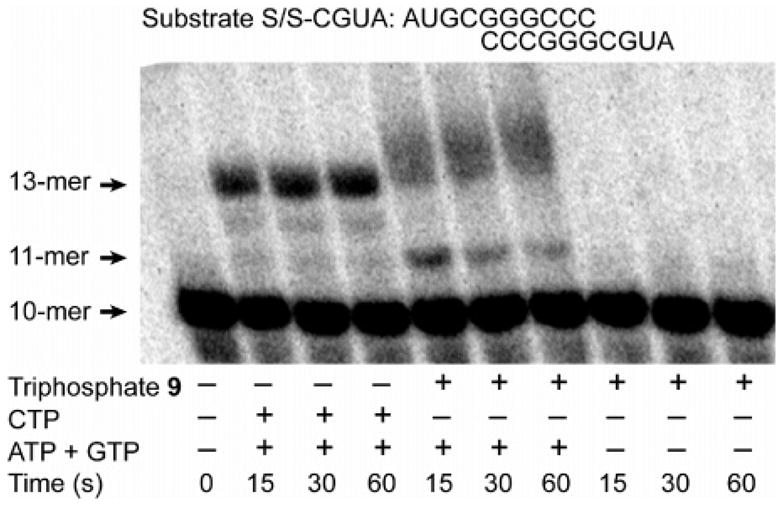
Incorporation of nucleotides derived from CTP, ATP, GTP, and 9 opposite complementary bases of the symmetrical substrate S/S–CGUA in vitro. The 10-mer substrates were end-labeled with 32P and treated with PV RdRP and nucleotides. Products were separated by denaturing PAGE.
In conclusion, we synthesized a 5-nitro derivative of the ribonucleoside cytidine (6) and demonstrated that this compound is a metabolic precursor to a potent inhibitor of PV RdRP. Compound 6 is phosphorylated intracellularly to the 5′-triphosphate (9), and this metabolite decreases the kinetics of nucleotide incorporation during replication of PV in vitro. This diminished viral replication yields a potent antiviral response in human cells infected with polio- and coxsackieviruses. Interestingly, a deoxy analogue of 6, 5-nitro-2′-deoxycytidine, exhibits antiviral activity against herpes simplex viruses 1 and 233 and HIV-1,34 although its mechanism of antiviral activity has not been fully elucidated. 5-Nitrocytidine (6) represents a promising lead for the development of novel antiviral therapeutics.
Supplementary Material
Supporting Information Available: Experimental procedures, supporting spectra, and characterization data for synthetic compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We thank Dr. N. Chapman and Dr. S. Tracy (University of Nebraska Medical Center) for CVB3 gene constructs and Dr. Zhi Hong (Valeant Pharmaceuticals) for the gift of ribavirin. We gratefully acknowledge the NIH (Grant AI054776 to B.R.P. and C.E.C.) and the American Heart Association (Grant 0340028N to C.E.C. and a predoctoral fellowship to D.A.H.) for financial support.
Footnotes
Abbreviations: RdRP, RNA-dependent RNA polymerase; PV, poliovirus; CVB3, coxsackievirus B3; HIV, human immunodeficiency virus; HCV, hepatitis C virus; RT, reverse transcriptase; S/S, symmetrical RNA substrate; AcOH, acetic acid; dCTPs, deoxycytidine triphosphates; MeCN, acetonitrile; MOI, multiplicity of infection; PFU, plaque-forming unit; PAGE, polyacrylamide gel electrophoresis; TMSCl, trimethylsilyl chloride.
Note Added after ASAP Publication. This manuscript was released ASAP on September 15, 2006, with an error in the caption (part D) of Figure 3. The correct version was posted on September 19, 2006.
References
- 1.Vignuzzi M, Stone JK, Andino R. Ribavirin and lethal mutagenesis of poliovirus: molecular mechanisms, resistance and biological implications. Virus Res. 2005;107:173–181. doi: 10.1016/j.virusres.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Anderson JP, Daifuku R, Loeb LA. Viral error catastrophe by mutagenic nucleosides. Annu Rev Microbiol. 2004;58:183–205. doi: 10.1146/annurev.micro.58.030603.123649. [DOI] [PubMed] [Google Scholar]
- 3.Graci JD, Cameron CE. Challenges for the development of ribonucleoside analogues as inducers of error catastrophe. Antiviral Chem Chemother. 2004;15:1–13. doi: 10.1177/095632020401500101. [DOI] [PubMed] [Google Scholar]
- 4.Graci JD, Cameron CE. Quasispecies, error catastrophe, and the antiviral activity of ribavirin. Virology. 2002;298:175–180. doi: 10.1006/viro.2002.1487. [DOI] [PubMed] [Google Scholar]
- 5.Crotty S, Cameron C, Andino R. Ribavirin’s antiviral mechanism of action: lethal mutagenesis? J Mol Med. 2002;80:86–95. doi: 10.1007/s00109-001-0308-0. [DOI] [PubMed] [Google Scholar]
- 6.Loeb LA, Mullins JI. Lethal mutagenesis of HIV by mutagenic ribonucleoside analogs. AIDS Res Hum Retroviruses. 2000;16:1–3. doi: 10.1089/088922200309539. [DOI] [PubMed] [Google Scholar]
- 7.Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci USA. 2001;98:6895–6900. doi: 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crotty S, Maag D, Arnold JJ, Zhong W, Lau JYN, Hong Z, Andino R, Cameron CE. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 9.Loeb LA, Essigmann JM, Kazazi F, Zhang J, Rose KD, Mullins JI. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc Natl Acad Sci USA. 1999;96:1492–1497. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maag D, Castro C, Hong Z, Cameron CE. Hepatitis C virus RNA-dependent RNA polymerase (NS5B) as a mediator of the antiviral activity of ribavirin. J Biol Chem. 2001;276:46094–46098. doi: 10.1074/jbc.C100349200. [DOI] [PubMed] [Google Scholar]
- 11.Valko M, Izakovic M, Mazur M, Rhodes CJ, Telser J. Role of oxygen radicals in DNA damage and cancer incidence. Mol Cell Biochem. 2004;266:37–56. doi: 10.1023/b:mcbi.0000049134.69131.89. [DOI] [PubMed] [Google Scholar]
- 12.Feig DI, Sowers LC, Loeb LA. Reverse chemical mutagenesis. Identification of the mutagenic lesions resulting from reactive oxygen species-mediated damage to DNA. Proc Natl Acad Sci USA. 1994;91:6609–6613. doi: 10.1073/pnas.91.14.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suen W, Spiro TG, Sowers LC, Fresco JR. Identification by UV resonance Raman spectroscopy of an imino tautomer of 5-hydroxy-2′-deoxycytidine, a powerful base analog transition mutagen with a much higher unfavored tautomer frequency than that of the natural residue 2′-deoxycytidine. Proc Natl Acad Sci USA. 1999;96:4500–4505. doi: 10.1073/pnas.96.8.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.La Francois CJ, Jang YH, Cagin T, Goddard WA, Sowers LC. Conformation and proton configuration of pyrimidine deoxynucleoside oxidation damage products in water. Chem Res Toxicol. 2000;13:462–470. doi: 10.1021/tx990209u. [DOI] [PubMed] [Google Scholar]
- 15.Purmal AA, Kow YW, Wallace SS. 5-Hydroxypyrimidine deoxynucleoside triphosphates are more efficiently incorporated into DNA by exonuclease-free Klenow fragment than 8-oxopurine deoxynucleoside triphosphates. Nucleic Acids Res. 1994;22:3930–3935. doi: 10.1093/nar/22.19.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purmal AA, Kow YW, Wallace SS. Major oxidative products of cytosine, 5-hydroxycytosine and 5- hydroxyuracil, exhibit sequence context-dependent mispairing in-vitro. Nucleic Acids Res. 1994;22:72–78. doi: 10.1093/nar/22.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharova NP. How does a cell repair damaged DNA? Biochemistry (Moscow) 2005;70:275–291. doi: 10.1007/s10541-005-0113-4. [DOI] [PubMed] [Google Scholar]
- 18.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radical Biol Med. 2002;33:1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 19.Wuenschell GE, Valentine MR, Termini J. Incorporation of oxidatively modified 2′-deoxynucleotide triphosphates by HIV-1 RT on RNA and DNA templates. Chem Res Toxicol. 2002;15:654–661. doi: 10.1021/tx010167l. [DOI] [PubMed] [Google Scholar]
- 20.Andresen G, Gundersen LL, Lundmark M, Rise F, Sundell S. Regioselective addition of Grignard reagents to a 2-oxopurinium salt. Tetrahedron. 1995;51:3655–3664. [Google Scholar]
- 21.Niedballa U, Vorbruggen H. A general synthesis of pyrimidine nucleosides. Angew Chem, Int Ed Engl. 1970;9:461–462. doi: 10.1002/anie.197004612. [DOI] [PubMed] [Google Scholar]
- 22.Rajeev KG, Broom AD. 5,6-Diaminocytidine, a versatile synthon for pyrimidine-based bicyclic nucleosides. Org Lett. 2000;2:3595–3598. doi: 10.1021/ol0064765. [DOI] [PubMed] [Google Scholar]
- 23.Fox JJ, Van Praag D. Pyrimidine nucleosides. VIII. Synthesis of 5-nitrocytidine and related nucleosides. J Org Chem. 1961;26:526–532. [Google Scholar]
- 24.Fukuhara TK, Visser DW. Cytidine derivatives. J Am Chem Soc. 1955;77:2393–2395. [Google Scholar]
- 25.Goldman D, Kalman TI. Formation of 5- and 6-aminocytosine nucleosides and nucleotides from the corresponding 5-bromocytosine derivatives: synthesis and reaction mechanism. Nucleosides Nucleotides. 1983;2:175–187. [Google Scholar]
- 26.Harki DA, Graci JD, Korneeva VS, Ghosh SKB, Hong Z, Cameron CE, Peterson BR. Synthesis and antiviral evaluation of a mutagenic and non-hydrogen bonding ribonucleoside analogue: 1-beta-D-ribofuranosyl-3-nitropyrrole. Biochemistry. 2002;41:9026–9033. doi: 10.1021/bi026120w. [DOI] [PubMed] [Google Scholar]
- 27.Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub) J Biol Chem. 2000;275:5329–5336. doi: 10.1074/jbc.275.8.5329. [DOI] [PubMed] [Google Scholar]
- 28.Gohara DW, Ha CS, Ghosh SKB, Arnold JJ, Wisniewski TJ, Cameron CE. Production of “authentic” poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expression Purif. 1999;17:128–138. doi: 10.1006/prep.1999.1100. [DOI] [PubMed] [Google Scholar]
- 29.Pfister T, Wimmer E. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J Biol Chem. 1999;274:6992–7001. doi: 10.1074/jbc.274.11.6992. [DOI] [PubMed] [Google Scholar]
- 30.Pogolotti AL, Jr, Santi DV. High-pressure liquid chromatography–ultraviolet analysis of intracellular nucleotides. Anal Biochem. 1982;126:335–345. doi: 10.1016/0003-2697(82)90524-3. [DOI] [PubMed] [Google Scholar]
- 31.Herold J, Andino R. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J Virol. 2000;74:6394–6400. doi: 10.1128/jvi.74.14.6394-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pathak HB, Ghosh SKB, Roberts AW, Sharma SD, Yoder JD, Arnold JJ, Gohara DW, Barton DJ, Paul AV, Cameron CE. Structure–function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol). A surface of the primary oligomerization domain functions in capsid precursor processing and VPg uridylylation. J Biol Chem. 2002;277:31551–31562. doi: 10.1074/jbc.M204408200. [DOI] [PubMed] [Google Scholar]
- 33.De Clercq E, Balzarini J, Descamps J, Huang GF, Torrence PF, Bergstrom DE, Jones AS, Serafinowski P, Verhelst G, Walker RT. Antiviral, antimetabolic, and cytotoxic activities of 5-substituted 2′-deoxycytidines. Mol Pharmacol. 1982;21:217–223. [PubMed] [Google Scholar]
- 34.Colacino E, Sindona G, Gosselin G, Mathe C. Synthesis and biological evaluation of some 5-nitro- and 5-amino derivatives of 2′-deoxycytidine, 2′,3′-dideoxyuridine, and 2′,3′-dideoxycytidine. Nucleosides Nucleotides Nucleic Acids. 2003;22:2013–2026. doi: 10.1081/NCN-120026403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Available: Experimental procedures, supporting spectra, and characterization data for synthetic compounds. This material is available free of charge via the Internet at http://pubs.acs.org.


