Abstract
Arsenic is effective in the treatment of acute promyelocytic leukemia. Paradoxically, it is also carcinogenic. In the process of elucidating a mechanism of arsenic resistance in a leukemia cell line, NB4, we discovered that arsenic exposure causes chromosomal abnormalities, with a preponderance of end-to-end fusions. These chromosomal end fusions suggested that telomerase activity may be inhibited by arsenic. We found that arsenic inhibits transcription of the hTERT gene, which encodes the reverse transcriptase subunit of human telomerase. This effect may in part be explained by decreased c-Myc and Sp1 transcription factor activities. Decreased telomerase activity leads to chromosomal end lesions, which promote either genomic instability and carcinogenesis or cancer cell death. These phenomena may explain the seemingly paradoxical carcinogenic and antitumor effects of arsenic.
Introduction
Hippocrates’ medicinal repertoire over 2000 years ago included arsenic, which was also recommended as an antileukemic agent in Sir William Osler’s 1892 first edition of The Principles and Practice of Medicine. Although arsenic has long been regarded as a carcinogen and toxin (1–4), clinical studies in China, subsequently confirmed elsewhere, demonstrate the dramatic effect of arsenic trioxide in the therapy of acute promyelocytic leukemia (APL) (5–8).
APL accounts for 10% of adult acute myeloid leukemia and presents with coagulopathy and a specific chromosomal translocation, t(15;17), that results in the fusion protein PML-RARα (9). The prognosis of APL dramatically improved after introduction of all-trans retinoic acid, which leads to as high as a 95% remission induction (9, 10). However, emergence of resistance to all-trans retinoic acid is rapid and salvage therapy is needed.
Arsenic trioxide has proven to be effective against APL that is refractory to all-trans retinoic acid and conventional chemotherapy (5–8). Although the exact mechanism of arsenic efficacy remains unknown, it appears to exert its antitumor effects by activating apoptosis (5, 7). Some reports attributed its effect to induction of reactive oxygen species (11–14). Arsenic also can induce degradation of the PML-RARα fusion protein, and this effect was thought to underlie arsenic anti-APL activity (15, 16). However, since the presence of PML-RARα fusion protein is neither necessary nor sufficient for the efficacy of arsenic (8, 17) and arsenic induces apoptosis in other cancer cell lines lacking PML-RARα (17), alternative mechanisms must be considered.
In this report, we find that arsenic potently inhibits the transcription of the reverse transcriptase subunit of the human telomerase gene (hTERT). Telomerase is an enzyme that maintains the length of chromosomal ends or telomeres, which otherwise would progressively shorten after each cell division (18). Although not universal, telomerase activity is frequently found in advanced cancer cells and is important for continuous cancer cell proliferation (19, 20). Since most cancer cells lacking telomerase showed sluggish growth and death, telomerase has become an attractive target for anticancer treatment (21, 22). There is, however, a dearth of small-molecule chemotherapeutics that specifically inhibit telomerase. Interestingly, cells lacking telomerase are paradoxically prone to genomic instability and carcinogenesis (23–26). In this report, we describe arsenic as a potent inhibitor of hTERT expression, and this effect appears at doses comparable to or lower than those clinically achievable. The effect may result from a diminished level or function of two transcription factors, c-Myc and Sp1, which are both important for hTERT expression (27, 28).
Methods
Cell culture and reagents.
All cell lines were cultured in 5% CO2 at 37°C in DMEM high-glucose medium except for NB4 (a gift of Robert Redner, University of Pittsburgh, Pittsburgh, Pennsylvania, USA), which was maintained in RPMI1640. The medium was supplemented with 10% FBS and 100 units/ml of penicillin plus 100 μg/ml streptomycin. Arsenic trioxide, 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT), and actinomycin D were purchased from Sigma Chemical Co. (St. Louis, Missouri, USA).
MTT cytotoxicity assay.
The viability of cells plated on culture dishes was measured by adding MTT reagent, which was dissolved in PBS, to a final concentration of 0.5 mg/ml. After 4 hours of incubation, cells were solubilized in 0.5 N HCl, 5% Triton X-100, and 45% 2-propanol (final concentration). The intensity of dissolved formazan crystal was measured at 590 nm.
Cytogenetic and telomere fluorescence in situ hybridization studies.
Preparations for cytogenetic analysis were made by exposing the cell lines to colcemid at 5 ng/ml for 16−18 hours to accumulate metaphases, followed by hypotonic treatment at 37°C for 30 minutes, and fixation in 3:1 methanol/glacial acetic acid. Air-dried slides were made and solid-stained for analysis of breakage and fusion rearrangements. Where sufficient material was available, a slide was also G-banded by standard techniques to examine the karyotype of the cell lines, and to identify chromosomes involved in fusion events. Ten to twenty well-spread metaphases were counted and the number of dicentrics, tricentrics, rings, and acentric fragments was recorded. Less commonly observed events, such as chromosomal breakage, multicentric chromosomes, or chromosome end-clusters, were also recorded. Fusion events were calculated as sum of numbers of dicentrics, 2 × tricentrics, and 3 × quadricentrics. Fluorescence in situ hybridization (FISH) was performed on metaphase spread using the peptide nucleic acid–telomere probe (Applied Biosystems, Foster City, California, USA; kindly provided by Carol Greider, the Johns Hopkins University, Baltimore, Maryland, USA) according to the manufacturer’s instructions. A normal lymphocyte metaphase control was included on the same slide with the arsenic-treated cells.
Telomerase activity assay.
Telomere repeat amplification protocol (TRAP) was implemented with a TRAPeze kit according to the manufacturer’s instructions (Intergen Co., Purchase, New York, USA). Five hundred cells were assayed in each sample.
Telomere length measurement.
Genomic DNA was digested with RsaI and HinfI, followed by separation on 0.6% agarose gel and hybridization with (TTAGGG)6 probes.
Real-time PCR.
Real-time PCR was performed using 7700 model ABI PRISM sequence detector (Applied Biosystems). The sequences of the forward primer, reverse primer, and TaqMan probe for hTERT were 5′-TACGTCGTGGGAGCCAGAAC-3′, 5′-CCTTCACCCTCGAGGTGAGA-3′, and 5′-TTCCGCAGAGAAAAGAGGGCCGA-3′, respectively. The sequences of the forward primer, reverse primer, and TaqMan probe for human c-Myc were 5′-TCAAGAGGTGCCACGTCTCC-3′, 5′-TCTTGGCAGCAGGATAGTCCTT-3′, and 5′-CAGCACAACTACGCAGCGCCTCC-3′, respectively. One hundred nanograms total RNA from NB4 cells and 500 ng total RNA from other cell lines were used in 25 μl reaction mixture per well using TaqMan one-step RT-PCR kit (Applied Biosystems). Internal control of human phosphoprotein (huPo) or β-actin tagged with VIC dye (Applied Biosystems) was included in every well as control. The reaction conditions were 55°C for 30 minutes, followed by 95°C for 10 minutes, and 40 cycles of 95°C for 15 seconds and 60°C for 1 minute.
Nuclear run-on reaction.
The experiment was performed as described earlier (29). Twenty-five million freshly isolated nuclei from NB4 cells either treated or untreated with 0.75 μM arsenic for 8 days were used in the reaction. After 20 minutes of in vitro transcription reaction at 27°C, [32P]UTP-labeled RNA from these two populations was treated with DNase I and proteinase K, followed by phenol-chloroform extraction. RNA with an equal amount of radioactivity was added onto nylon membranes slot-blotted with an excess amount of cDNA fragments of hTERT, c-Myc, and huPo. After 40 hours of hybridization at 42°C, the blots were washed and developed.
Plasmid construction and transfection.
The insert of pSG5-Sp1 is a 3.1-kb MfeI-SalI fragment of pPac-Sp1 (30). pSG5-Luc was created by subcloning a 1.7-kb HindIII-XbaI fragment of Luc+ from pGL3-Basic vector (Promega Corp., Madison, Wisconsin, USA). pMT-Luc and pMT-Sp1 were created by subcloning a 1.7-kb KpnI-XbaI Luc+ fragment from pGL3-Basic and a 3.4-kb MfeI-HindIII Sp1 fragment from pPac-Sp1 into pMT-CB6+ vector, respectively. pHTR-Luc, which contained an 800-bp promoter region of hTERT fused with luciferase gene, is a gift from Riccardo Dalla-Favera (Columbia University, New York, New York, USA) (28). One hundred thousand NIH-3T3 cells per well were plated into six-well plates 16 hours before transfection. Two micrograms total plasmid was transfected per well using SuperFect reagent (QIAGEN Inc., Valencia, California, USA). Fresh medium with or without 5 μM arsenic was added 3 hours later. Luciferase activity was determined 44 hours after transfection.
Gel shift assay.
Nuclear lysate of NB4 cells either treated or untreated with 0.75 μM arsenic for 8 days was extracted and dissolved in 50 mM Hepes (pH 7.8), 50 mM KCl, 400 mM NaCl, 0.1 mM EDTA, 10% glycerol, 3 mM DTT, and protease inhibitor made from 1 complete protease inhibitor cocktail tablet (Roche Molecular Biochemicals, Indianapolis, Indiana, USA) in 50 ml reaction. In 10 μl of binding reaction, 2.5 μl of nuclear extract (0.625 μg for SP1 and 2.5 μg for Oct-1) was incubated with binding buffer containing 4% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 5 mM NaCl, 10 mM Tris-Cl (pH 7.5), 0.02 N NaOH, and 0.5 μg poly(dI-dC) × poly(dI-dC) with or without corresponding antibody (Santa Cruz Biotechnology Inc., Santa Cruz, California, USA) for 30 minutes on ice. End-labeled oligonucleotide was then added and incubated on ice for another 30 minutes before loading on 5% nondenaturing polyacrylamide gel. The sequences of oligonucleotide for Sp1 and Oct-1 binding sites were 5′-CTGCGGGGCGGGGCAGACCCCGCCCGTCTGACG-3′ (31) and 5′-TGTCGAATGCAAATCACTAGAA-3′ (Santa Cruz Biotechnology Inc.), respectively.
Results
Arsenic causes chromosomal end-to-end fusion and diminishes telomerase activity.
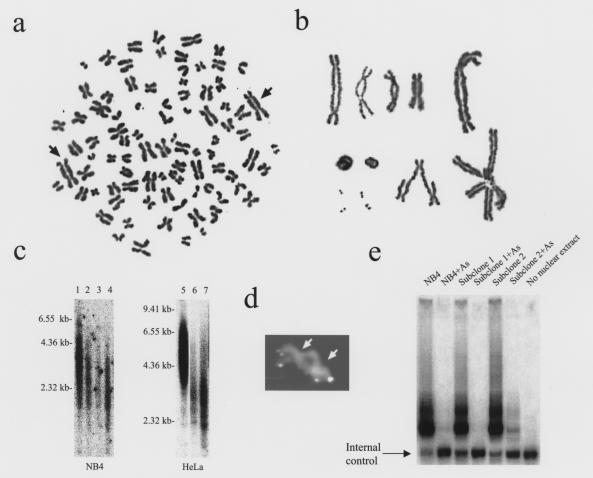
During our screening for arsenic-resistant NB4 clones, cells that remained after a 3-week exposure to 0.75 μM arsenic dramatically enlarged. No clones out of 2 × 108 cells survived 1 month of continuous exposure to arsenic at this dose. Karyotyping of these arsenic-treated, morphologically abnormal, enlarged cells showed striking chromosomal end-to-end fusion (Figure 1, a and b). From 80 karyotypes, we observed more prominent polyploidy in 32 cells and an average of 2.4 fusion events per cell. The chromosomal fusions are associated with attrition of telomeres, since no signal was seen at the fusion junction in dicentric chromosomes by FISH using a telomere-specific probe (Figure 1d). In addition, Southern blot analysis of NB4 cells reveals a shortening of telomeres with exposure to arsenic (Figure 1c). By contrast, untreated NB4 cells showed the characteristic t(15;17) chromosomal translocation without end-to-end fusions or breakage and had a consistent near-triploid range of N = 75−80. Further cytogenetic analysis of NB4 cells and two of its subclones exposed to various doses of arsenic (0.75 μM to 1.0 μM) for 2 weeks showed a dose-related increase of chromosomal end-to-end fusions (data not shown).
Figure 1.
Chromosomal abnormalities and telomerase activity of NB4 cells treated with arsenic. (a) A representative entire metaphase from NB4 cells treated with arsenic at 0.75 μM for 3 weeks. The arrows indicate dicentric fusion chromosomes. (b) Representative dicentric chromosomal fusions as well as other abnormal fused chromosomes and fragments from cells treated with arsenic at 0.75 μM for 3 weeks. (c) Southern blot of digested genomic DNA from NB4 (left) and HeLa cells (right) showed decreased telomere length after arsenic exposure. Lane 1: untreated NB4 cells; lanes 2−4: 0.25 μM arsenic for 4, 5, and 6 weeks, respectively. Lane 5: untreated HeLa cells; lanes 6 and 7: 1 μM arsenic for 3 and 4 weeks, respectively. (d) Dicentric chromosome after FISH with a telomere-specific probe. Arrows indicate centromeres. FISH signals are clearly visible at both ends of the fusion chromosome. Among 16 fusions studied, no hybridization is seen between the centromeres where fusion occurred. In contrast, 95−97% of chromosome ends of a normal lymphocyte control display intense signal (data not shown). (e) Telomerase activity of NB4 cells treated with arsenic. Pooled NB4 cells and independently isolated subclones showed dramatically decreased telomerase activity after 8 days of 0.75 μM arsenic. The internal control represents a band that should appear in every TRAP assay to ensure a reliable PCR reaction. Because of competition of components, higher telomerase activities result in fainter internal control bands.
Chromosomal end-to-end fusions have been observed in telomerase-deficient murine cells (23). To determine the basis for the generation of end-to-end chromosomal fusions in our experiment, we measured telomerase activity using the TRAP assay in cells exposed to arsenic (19). Telomerase activity was severely diminished in NB4 cells after 8 days of 0.75 μM arsenic treatment (Figure 1e), although significant numbers of fusion chromosomes were not observed until 2−3 weeks (data not shown). To determine whether arsenic could inhibit telomerase activity directly in vitro, 10 μM arsenic was added to the TRAP reaction mixture; no effect on telomerase activity was observed (data not shown).
Arsenic decreases hTERT mRNA levels in different cell types.
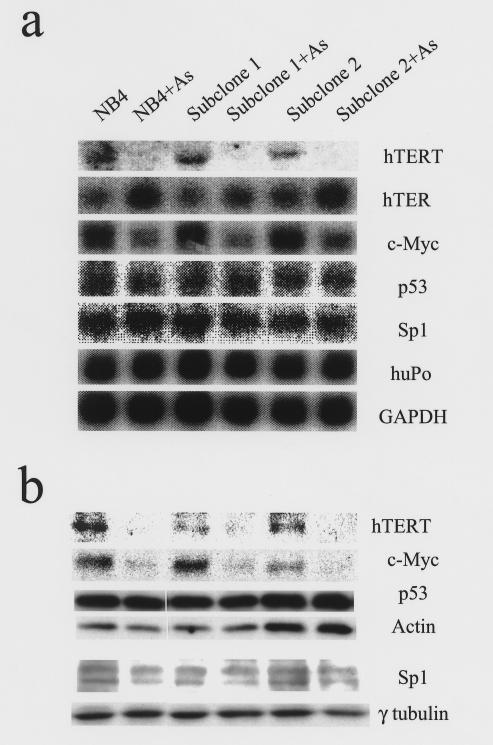
The suppression of telomerase activity by arsenic correlated with a dramatic decrease in hTERT mRNA and protein, indicating that arsenic inhibits the expression of hTERT (Figure 2, a and b). Moreover, there is a dose-related suppression of hTERT mRNA expression as measured by real-time PCR (Figure 3a). In contrast to hTERT mRNA levels, the RNA component of telomerase hTER was elevated in the presence of arsenic (Fig. 2a).
Figure 2.
mRNA and protein levels of hTERT, hTER, c-myc, p53, and Sp1 in NB4 cells after arsenic treatment. (a) Northern blot analysis shows diminished hTERT and c-myc mRNA after 8 days of 0.75 μM arsenic treatment. By contrast, the RNA component of telomerase, hTER, is elevated in arsenic-treated samples. Levels of p53 and Sp1 mRNA were unchanged by arsenic. Human phosphoprotein (huPo) and GAPDH served as loading controls. (b) Immunoblotting shows a corresponding decrease in protein levels of hTERT and c-Myc after arsenic treatment, but p53 and Sp1 protein levels remained unaltered. Actin and γ-tubulin served as loading controls.
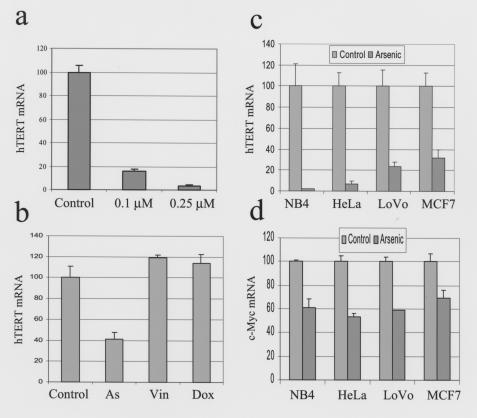
Figure 3.
Arsenic specifically suppresses hTERT mRNA levels independent of cell type. The expression of hTERT and c-myc was measured by real-time PCR and normalized to actin in b and human phosphoprotein in a, c, and d. hTERT mRNA levels are expressed as percent of untreated control. (a) Arsenic diminished hTERT mRNA expression in a dose-dependent fashion. Experiments were performed after 12 days of arsenic exposure. (b) Two days of arsenic (0.75 μM) but not vincristine (Vin; 0.075 nM) or doxorubicin (Dox; 50 nM) diminishes hTERT mRNA levels. (c) Cell lines other than NB4 also had decreased hTERT mRNA levels after arsenic treatment. NB4 cells were especially sensitive as 0.75 μM for 8 days had resulted in more than 98% inhibition of hTERT expression. The other cell lines were treated with 2 μM arsenic for 14 days. (d) c-myc expression was also decreased in all four cell lines with arsenic exposure.
To determine whether the inhibition of hTERT expression is specific to arsenic, two chemotherapeutic agents, vincristine and doxorubicin, with different well-documented mechanisms of cytotoxicity were tested at concentrations that maintain sufficient cell viability so that cell death is not a confounding factor. Treatment of NB4 cells for 48 hours with 0.075 nM vincristine, 25 nM doxorubicin, or 0.75 μM arsenic resulted in about 80% viability (determined by MTT assay), as compared with untreated control (data not shown). While the cells treated with vincristine or doxorubicin did not have significant changes in hTERT mRNA level as measured by real-time PCR, arsenic treatment resulted in a marked decrease in hTERT expression (Figure 3b). Although most cells died at a higher concentration of vincristine (0.25 nM), hTERT mRNA levels remained unsuppressed (data not shown). Furthermore, treatment of NB4 cells at concentrations of arsenic as low as 0.1 μM for 12 days also significantly suppressed hTERT expression (Figure 3a), demonstrating the potency of arsenic in inhibiting telomerase. To determine whether arsenic could inhibit telomerase expression in other cell lines, HeLa (cervical cancer), HepG2 (hepatoma), LoVo (colon cancer), and MCF7 (breast cancer) cells were tested. After 14 days of exposure to 2 μM arsenic, all cell lines, except for HepG2 (data not shown), had decreased hTERT expression (Figure 3c). In contrast to HepG2 cells that were inhibited by 30% after 1 week of 2 μM arsenic exposure, there was about 80% growth inhibition in HeLa, LoVo, and MCF7 (determined by MTT assay; data not shown). It is notable that in addition to NB4 cells, arsenic also shortens telomeres in HeLa cells treated with arsenic (Figure 1c). These observations suggest a relationship between telomerase inhibition and viability of these cell lines. Hence, inhibition of hTERT expression is relatively specific to arsenic, can be seen in cancer cell lines other than APL, and is not simply due to cell death.
Arsenic inhibits hTERT transcription.
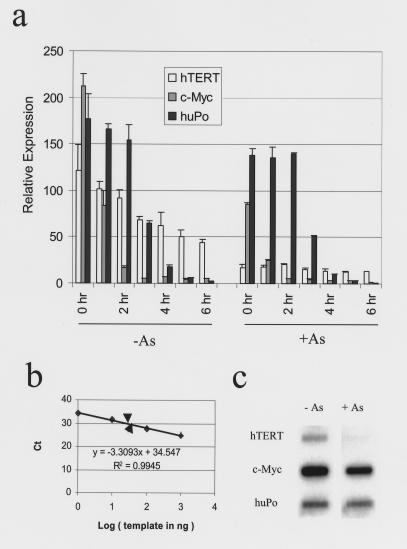
Because arsenic inhibits hTERT expression, we sought to determine whether arsenic inhibits transcription of hTERT or shortens the half-life of the hTERT mRNA. The half-life of hTERT mRNA is relatively long (4 hours) and is not shortened by exposure of cells to arsenic (Figure 4a). Though hTERT expression is low, especially after arsenic treatment, the cycles of threshold (Ct) are still within the linear range of the standard curve of real-time PCR, assuring the validity of the measurement (Figure 4b). Furthermore, nuclear run-on studies demonstrated severely diminished transcription of hTERT in arsenic-treated NB4 cells as compared with control cells (Figure 4c). These studies indicate that arsenic inhibits hTERT at the transcription level.
Figure 4.
Arsenic inhibits the transcription of hTERT. (a) Half-life of hTERT mRNA is unaltered by arsenic. Control and arsenic-exposed (0.75 μM arsenic for 2 days) NB4 cells were treated with actinomycin D (5 μg/ml). RNA was collected at the time points indicated and real-time PCR was performed to detect the expression of hTERT, c-myc, and huPo. Both c-myc and huPo mRNA decayed in control and arsenic-treated samples and thus served as positive controls for this assay. hTERT mRNA decay was not increased by arsenic. (b) hTERT expression in control (lower arrowhead) and arsenic-treated (upper arrowhead) samples was well within the linear range on the standard curve of real-time PCR. (c) Nuclear run-on reaction showed diminished transcription of hTERT in NB4 cells treated with 0.75 μM for 8 days. c-myc transcription rate also decreased to 40%. huPo served as control.
Arsenic inhibits c-Myc transcription and Sp1 DNA binding activity.
Because the hTERT promoter contains binding sites for c-Myc and Sp1 (32), and these transcription factors can cooperatively activate hTERT expression (27), we tested whether these transcription factors were affected in arsenic-treated cells. c-myc mRNA and protein levels decreased in arsenic-treated NB4 cells (Figure 2, a and b). We also observed a decrease in c-myc RNA levels with arsenic exposure in the various cell lines tested, except for HepG2 (Figure 3d and data not shown). Nuclear run-on and RNA stability studies indicate that c-myc transcription is also inhibited by arsenic (Figure 4, a and c). By contrast, Sp1 mRNA and protein were not changed by arsenic exposure (Figure 2, a and b). Another candidate for investigation is p53, which sequesters Sp1 and in turn inhibits hTERT expression (33). However, in our study, arsenic did not affect p53 mRNA or protein levels (Figure 2, a and b).
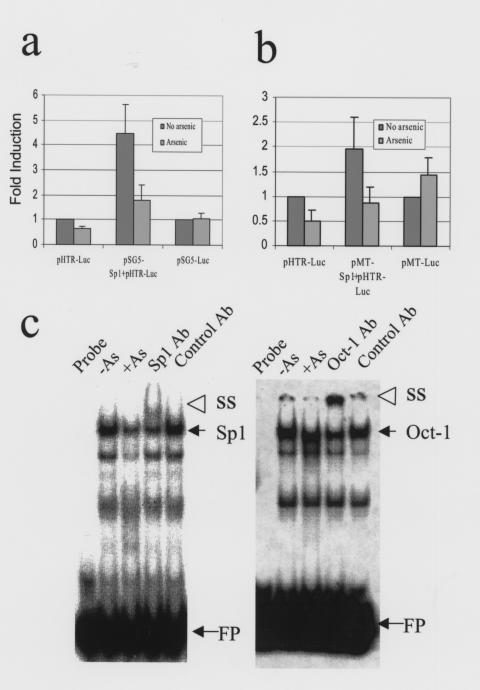
Because Sp1 activates the transcription of both c-myc and hTERT, we sought to determine whether Sp1 activity could be inhibited by arsenic. We performed luciferase assays using NIH-3T3 cells cotransfected with pSG5-Sp1 and a reporter plasmid, pHTR-Luc, which contains an 800-bp hTERT promoter with c-Myc and Sp1 binding sites in it. Overexpression of Sp1 increased hTERT promoter activity fourfold (Figure 5a). Arsenic significantly inhibited the induction of hTERT promoter by Sp1 (Figure 5a). This inhibition was not due to an effect of arsenic on the SV40 early promoter in pSG5-Sp1, since arsenic did not inhibit the activity of pSG5-Luc (Figure 5a). To further substantiate these observations, we have also used a plasmid pMT-Sp1 with the arsenic-inducible sheep metallothionein promoter driving Sp1 expression. In this case, we circumvented any potential inhibition of the promoter by arsenic, and we again observed an arsenic-dependent inhibition of Sp1 induction of the hTERT promoter (Figure 5b). Hence, arsenic could inhibit Sp1-mediated hTERT transcription at a functional level.
Figure 5.
Arsenic inhibits Sp1 function in hTERT transcription. (a) Cotransfection with pSG5-Sp1 and the reporter, pHTR-Luc, activates hTERT transcription fourfold compared with pHTR-Luc only. A 44-hour arsenic (5 μM) exposure significantly inhibits pHTR-Luc alone and the combination of pSG5-Sp1 and pHTR-Luc. The inherent activities of both the hTERT promoter (reporter plasmid only) and pSG5-Luc without arsenic were set as 1.0. While arsenic inhibited pSG5-Sp1 activation of the hTERT promoter, it could not inhibit pSG5-Luc. (b) Cotransfection with pMT-Sp1 and pHTR-Luc recapitulates the result in a. The inherent activities of both the hTERT promoter and pSG5-Luc without arsenic were set as 1.0. (c) Sp1 (left panel) but not Oct-1 (right panel) DNA binding activity is diminished by 0.75 μM arsenic exposure for 8 days. SS, supershift band; FP, free probe.
We further observed that Sp1 DNA binding activity was decreased in arsenic-treated cells, as determined by gel shift assays (Figure 5c, left panel). The transcription factor Oct-1, on the other hand, was minimally affected by arsenic (Figure 5c, right panel), indicating that arsenic exposure does not cause a general decline in transcription factor function. Sp1 DNA binding activity, however, was not inhibited in vitro through the direct addition of arsenic to gel shift assays (data not shown). These observations suggest that arsenic inhibits Sp1 indirectly, perhaps through the generation of reactive oxygen species, which in turn oxidizes and inactivates Sp1 (34). We surmise that together with the reduction in c-myc gene expression, which also depends on Sp1 activity (35), decreased Sp1 activity contributes to the marked decrease in hTERT transcription.
Discussion
Pharmacokinetic studies showed that plasma arsenic levels peaked around 7 μM and then dropped to less than 1 μM after daily intravenous infusion into human subjects (6). These levels are comparable to those used in our in vitro studies, which demonstrate a dramatic inhibition of hTERT expression by arsenic. High levels of arsenic (10 μM for 2 days), however, are thought to induce reactive oxygen species that in turn induce multilocus chromosomal deletion rather than end-to-end fusion (13, 14). Since chronic arsenic exposure and numerous cell doublings are required for telomere attrition, the cytotoxicity and multilocus deletion occurring after short-term exposure to high levels of arsenic are likely due to alternative mechanisms. Nevertheless, reactive oxygen species might play a role in inhibition of hTERT transcription because of its potential oxidative inactivation of Sp1 (34). While our findings suggest a role of Sp1 in the inhibition of hTERT expression, the fact that arsenic did not inhibit the SV40 minimal promoter, which contains several Sp1 sites, is seemingly contradictory. Because the SV40 minimal promoter is complex, we do not understand why diminished Sp1 activity would not affect the SV40 promoter in this context. Since we have not exhaustively examined the global effect of arsenic on gene expression, the inhibitory effect of arsenic on other genes may also contribute to the decrease in hTERT expression and telomere maintenance.
An important implication of our finding is that the inhibition of hTERT by arsenic may explain the seemingly paradoxical role of arsenic in tumorigenesis and antitumor therapy. Likewise, telomerase plays a paradoxical role in tumorigenesis. For most advanced tumors, telomere maintenance is essential for continued proliferation and avoidance of senescence (21, 22). Similarly, the importance of telomerase activity in tumorigenesis is also recapitulated in vivo in telomerase-deficient mice, which have decreased propensity for the tumor development in the p16/p19ARF-null background (36). For noncancerous cells, however, loss of telomeres could lead to genomic instability and cancer formation (23–26). Consistent with this observation is the increased tumorigenesis in mice deficient for both p53 and telomerase (37). In this case, loss of p53 suppresses apoptosis that is triggered by telomerase deficiency. While our observations suggest that inhibition of telomerase expression by arsenic contributes to chromosomal end-to-end fusions, there may be other mechanisms that contribute to these chromosomal abnormalities. It is unlikely, however, that loss of TRF2, which binds to and protects telomeres, could contribute to arsenic-induced end-to-end chromosomal fusions. Loss of TRF2 would lead to chromosomal end fusions, but in this case telomere length would not be affected. Both our FISH and Southern blot analyses indicate that arsenic shortens telomere length. Thus, inhibition of telomerase expression by arsenic may account for both its tumorigenic potential and its effectiveness as an antineoplastic agent.
Acknowledgments
This work was supported by NIH grant CA-51497. We thank R. Dalla-Favera, L. Gardner, C. Greider, A. Lee, Q. Li, and R. Redner for comments and reagents.
References
- 1.Leonard A, Lauwerys RR. Carcinogenicity, teratogenicity and mutagenicity of arsenic. Mutat Res. 1980;75:49–62. doi: 10.1016/0165-1110(80)90027-5. [DOI] [PubMed] [Google Scholar]
- 2.Hertz-Picciotto I, Smith AH, Holtzman D, Lipsett M, Alexeeff G. Synergism between occupational arsenic exposure and smoking in the induction of lung cancer. Epidemiology. 1992;3:23–31. doi: 10.1097/00001648-199201000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Chappell WR, et al. Inorganic arsenic: a need and an opportunity to improve risk assessment. Environ Health Perspect. 1997;105:1060–1067. doi: 10.1289/ehp.971051060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitchin KT. Recent advances in arsenic carcinogenesis: modes of action, animal model systems, and methylated arsenic metabolites. Toxicol Appl Pharmacol. 2001;172:249–261. doi: 10.1006/taap.2001.9157. [DOI] [PubMed] [Google Scholar]
- 5.Chen GQ, et al. Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (APL). I. As2O3 exerts dose-dependent dual effects on APL cells. Blood. 1997;89:3345–3353. [PubMed] [Google Scholar]
- 6.Shen ZX, et al. Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (APL). II. Clinical efficacy and pharmacokinetics in relapsed patients. Blood. 1997;89:3354–3360. [PubMed] [Google Scholar]
- 7.Shao W, et al. Arsenic trioxide as an inducer of apoptosis and loss of PML/RARα protein in acute promyelocytic leukemia cells. J Natl Cancer Inst. 1998;90:124–133. doi: 10.1093/jnci/90.2.124. [DOI] [PubMed] [Google Scholar]
- 8.Soignet SL, et al. Complete remission after treatment of acute promyelocytic leukemia with arsenic trioxide. N Engl J Med. 1998;339:1341–1348. doi: 10.1056/NEJM199811053391901. [DOI] [PubMed] [Google Scholar]
- 9.Warrell RP, De The H, Wang ZY, Degos L. Acute promyelocytic leukemia. N Engl J Med. 1993;329:177–189. doi: 10.1056/NEJM199307153290307. [DOI] [PubMed] [Google Scholar]
- 10.Fenaux P, Chastang C, Chomienne C, Degos L. Tretinoin with chemotherapy in newly diagnosed acute promyelocytic leukaemia. European APL group. Lancet. 1994;343:1033. doi: 10.1016/s0140-6736(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 11.Chen YC, Lin-Shiau SY, Lin JK. Involvement of reactive oxygen species and caspase 3 activation in arsenite-induced apoptosis. J Cell Physiol. 1998;177:324–333. doi: 10.1002/(SICI)1097-4652(199811)177:2<324::AID-JCP14>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 12.Ozaki M, Deshpande SS, Angkeow P, Suzuki S, Irani K. Rac1 regulates stress-induced, redox-dependent heat shock factor activation. J Biol Chem. 2000;275:35377–35383. doi: 10.1074/jbc.M005287200. [DOI] [PubMed] [Google Scholar]
- 13.Hei TK, Liu SX, Waldren C. Mutagenicity of arsenic in mammalian cells: roles of reactive oxygen species. Proc Natl Acad Sci USA. 1998;95:8103–8107. doi: 10.1073/pnas.95.14.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu SX, Athar M, Lippai I, Waldren C, Hei TK. Induction of oxyradicals by arsenic: implication for mechanism of genotoxicity. Proc Natl Acad Sci USA. 2000;98:1643–1648. doi: 10.1073/pnas.031482998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller S, Matunis MJ, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sternsdorf T, et al. PIC-1/SUMO-1-modified PML-retinoic acid receptor α mediates arsenic trioxide-induced apoptosis in acute promyelocytic leukemia. Mol Cell Biol. 1999;19:5170–5178. doi: 10.1128/mcb.19.7.5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu XH, et al. Apoptosis and growth inhibition in malignant lymphocytes after treatment with arsenic trioxide at clinically achievable concentrations. J Natl Cancer Inst. 1999;91:772–778. doi: 10.1093/jnci/91.9.772. [DOI] [PubMed] [Google Scholar]
- 18.Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 19.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 20.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Mar V, Zhou W, Harrington L, Robinson MO. Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 1999;13:2388–2399. doi: 10.1101/gad.13.18.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn WC, et al. Inhibition of telomerase limits the growth of human cancer cells. Nat Med. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 23.Rudolph KL, et al. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- 24.Hackett JA, Feldser DM, Greider CW. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001;106:275–286. doi: 10.1016/s0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- 25.Romanov SR, et al. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic change. Nature. 2001;409:633–637. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- 26.Blasco MA, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 27.Kyo S, et al. Sp1 cooperate with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT) Nucleic Acids Res. 2000;28:669–677. doi: 10.1093/nar/28.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu KJ, et al. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 29.Osthus RC, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 30.Courey AJ, Tijan R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 31.Leggett RW, Armstron SA, Barry D, Mueller CR. Sp1 is phosphrylated and its DNA binding activity down-regulated upon terminal differentiation of the liver. J Biol Chem. 1995;270:25879–25884. doi: 10.1074/jbc.270.43.25879. [DOI] [PubMed] [Google Scholar]
- 32.Wick M, Zubov D, Hagen G. Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT) Gene. 1999;232:97–106. doi: 10.1016/s0378-1119(99)00108-0. [DOI] [PubMed] [Google Scholar]
- 33.Xu D, et al. Downregulation of telomerase reverse transcriptase mRNA expression by wild type p53 in human tumor cells. Oncogene. 2000;19:5123–5133. doi: 10.1038/sj.onc.1203890. [DOI] [PubMed] [Google Scholar]
- 34.Ammendola R, Mesuraca M, Russo T, Cimino F. The DNA-binding efficiency of Sp1 is affected by redox changes. Eur J Biochem. 1994;225:483–489. doi: 10.1111/j.1432-1033.1994.t01-1-00483.x. [DOI] [PubMed] [Google Scholar]
- 35.Majello B, De Luca P, Suske G, Lania L. Differential transcriptional regulation of c-myc promoter through the same DNA binding sites targeted by Sp1-like proteins. Oncogene. 1995;10:1841–1848. [PubMed] [Google Scholar]
- 36.Greenberg RA, et al. Short dysfunctional telomeres impair tumorigenesis in the INK4aΔ2/3 cancer-prone mouse. Cell. 1999;97:515–525. doi: 10.1016/s0092-8674(00)80761-8. [DOI] [PubMed] [Google Scholar]
- 37.Chin L, et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]