Abstract
Common fragile sites are chromosomal loci prone to breakage and rearrangement, hypothesized to provide targets for foreign DNA integration. We cloned a simian virus 40 integration site and showed by fluorescent in situ hybridization analysis that the integration event had occurred within a common aphidicolin-induced fragile site on human chromosome 7, FRA7H. A region of 161 kb spanning FRA7H was defined and sequenced. Several regions with a potential unusual DNA structure, including high-flexibility, low-stability, and non-B-DNA-forming sequences were identified in this region. We performed a similar analysis on the published FRA3B sequence and the putative partial FRA7G, which also revealed an impressive cluster of regions with high flexibility and low stability. Thus, these unusual DNA characteristics are possibly intrinsic properties of common fragile sites that may affect their replication and condensation as well as organization, and may lead to fragility.
Fragile sites are specific chromosomal loci prone to breakage and characterized by constrictions, gaps, or breaks on chromosomes from cells exposed to specific tissue culture and chemical conditions (1). Fragile sites are classified as either rare or common, depending on their frequency within the population and their mode of induction. The instability of fragile sites at the molecular and cytogenetic levels can lead to disease manifestation by silencing adjacent gene(s) (2, 3) or by causing chromosomal rearrangements and disrupting gene expression (4). Several rare fragile sites have been characterized at the molecular level by positional cloning using families expressing these sites (4–9). The expression of these sites is associated with expanded CGG trinucleotide repeats or with an expanded 33-bp A+T-rich minisatellite repeats. The mechanism by which the repeat expansion is associated with the cytogenetic expression and chromosomal breakage of these sites is not well understood.
Common fragile sites (n < 100), on the other hand, are considered to be part of the normal chromosome structure. Most of these sites are induced by aphidicolin, an inhibitor of the elongation activity of DNA polymerases (10). Because common fragile sites are part of the normal chromosome, cloning of these sites could not use positional cloning approaches. Two strategies were described for the cloning of the fragile site at 3p14.2 (FRA3B). One was based on the observation that many chromosomal rearrangements and cancer breakpoints fall within chromosomal bands to which fragile sites have been mapped. The second strategy was based on the hypothesis that fragile sites are recombinogenic and that foreign DNA might preferentially integrate into these sites. These studies led to the identification of FRA3B, which spans a region greater than 250 kb (11–17). More recently, several yeast artificial chromosome (YAC) clones were found to span a common aphidicolin-induced fragile site on human chromosome 7, FRA7G (18). However, the precise region of FRA7G is yet to be defined.
Partial sequences of FRA3B (276 kb) revealed no expanded repeats or other features that could clearly point to the molecular basis of its fragility (12–14, 16). Thus, additional common fragile sites had to be cloned and sequenced to enable the identification of characteristics shared by common fragile sites that are implicated in the instability of these sites.
Here we report the identification and characterization of a common aphidicolin-induced fragile site, FRA7H, on human chromosome 7. Our approach was based on previous cytogenetic observations linking viral integration sites and fragile sites (19, 20).
MATERIALS AND METHODS
Cell Lines.
Two cell lines were used in this study: GM00847 (NIGMS, Camden, NJ), a simian virus 40 (SV40)-transformed human fibroblast cell line, and GM10791A (NIGMS), a chromosome 7 somatic cell hybrid.
Phage Genomic Library Construction.
Sau3A partially digested DNA (9–23 kb), extracted from the cell line GM00847, was used to contract a λ-Dash-II library (Stratagene).
Genomic Libraries Used for Contig Constructions.
A YAC library (21), a human P1-derived artificial chromosome (PAC) library (22), and a chromosome 7-specific cosmid library constructed from GM10791A were used.
Chromosome Preparation and Fragile Site Induction.
Cells were grown on coverslips and fragile sites were induced by growing the cells in M-199 medium in the presence of 0.4 μM aphidicolin and 0.5% ethanol for 24 h prior to chromosome fixation using standard procedures.
Fluorescent in Situ Hybridization (FISH).
Cosmid and YAC DNA were labeled with digoxigenin (DIG)-11-dUTP (Boehringer Mannheim) by nick-translation. FISH was performed as previously described (23).
Analysis of Hybridization Signals.
Green and red fluorescence signals were visualized simultaneously by using a Nikon B-2A filter cube. For weak signals a modified Chromatech HQ-FITC (Chroma Technology, Brattleboro, VT) filter set was used (excitation band, 460–500 nm; emission band, 520–600 nm). Images were captured with an intensified charge-coupled device imager (Paultek Imaging, Grass Valley, CA) and digitized with a frame grabber (Imascan/MONO-D, Imagraph, Chelmsford, MA).
Because there are several aphidicolin-induced fragile sites on 7q their positions had to be carefully determined. The Image-Pro plus program (Media Cybernetics, Silver Spring, MD) was used to measure the fragile site-telomere distance relative to the short arm length. According to the Genome Database (GDB 6.0), this value should be ≈45% for FRA7H, ≈82% for FRA7G, and ≈11% for FRA7I. Analysis based on 100 random measurements indicated that the standard deviation (SD) was 5%. Thus, observed cytogenetic gaps at fragile sites of 35–55% (two SD) were considered to be FRA7H.
Sequencing.
Shotgun sequencing was performed as previously described (24).
Sequence Annotation.
Computer analysis was performed by using our newly developed automated tool, rummage (B.D., J. Weber, R. Schattevoy, and A.R., unpublished work), which combines 23 different programs, including censor, blast version 1.4, fasta version 2.0, grail, fexhb, and mzef. CpG islands were identified by using the following criteria: G+C > 50%, CpG ratio observed/expected >0.6, and length >200 bp. We also developed a tool, exonsampler, that filters and associates expressed sequence tag (EST) matches and verifies splice sites (J. Weber, B.D., and A.R., unpublished work). Repeat analysis was also performed, using RepEater (R. Gill-More and M. Amitai, personal communication). The following programs from the Genetics Computer Group (GCG) were used: findpatterns, map, bestfit, gap, composition, fetch, repeat, stemloop, and shuffle.
Computer Analysis of Helix Flexibility and Stability.
The flexibility parameter that we used measured potential local variations in the DNA structure, expressed as fluctuations in the twist angle (25). The analysis was performed in overlapping windows of 100 bp. Dinucleotide values were summed along the window and averaged by the window length. Windows with outstanding values, deviating significantly from the average, were considered as potential flexible regions. Helix stability was based on the sequence-dependent free energy values of the helix-to-coil transition, and it is expressed in kcal/mol (1 kcal = 4.18 kJ) (26). The analysis was performed as described for DNA flexibility.
Identification of Potential Non-B-DNA Sequences.
DNA sequences that potentially form a non-B-DNA structure under negative superhelical strain were manually scanned in the entire FRA7H region. We highlighted homopurine/homopyrimidine sequences of >30 nucleotides, other than poly(A) or poly(T), that readily form intramolecular triple-helix structures (27). We also searched for potential nuclear matrix attachment regions (MARs) by identifying relatively A+T-rich sequences (>70%) in which one strand consists exclusively of mixed As, Ts, and Cs, but not Gs (ATC sequences) (28). Only two stretches of ATC sequences, one of at least 18 nucleotides and the other of at least 16, separated by not more than 10 nucleotides were highlighted.
Non-B-DNA Structure Analysis.
Non-B-DNA structures were detected by chloroacetaldehyde (CAA) in vitro (29). In brief, supercoiled plasmid DNA was reacted with 2 μl of double-distilled CAA in a 100-μl reaction volume at 37°C with or without 2 mM MgCl2 at pH 7 (50 mM Tris⋅HCl). CAA-modified DNAs were end labeled with [32P]dATP at the SalI site (the 4.4-kb plasmid) or at the BamHI site (the 2.2-kb plasmid) and further cleaved at a distal KpnI site. DNA fragments were isolated and subjected to hydrazine and piperidine for the detection of modified adenines or formic acid and piperidine for the detection of modified cytosines. The cleavage products were resolved on a denaturing urea/polyacrylamide gel.
RESULTS AND DISCUSSION
Cloning of the SV40 Integration Site.
We have studied an SV40-transformed human fibroblast cell line, GM00847, in which several viral genomes have integrated in 7q31–35 in tandem (30, 31). Because there are two common fragile sites in this region, FRA7G in 7q31.2 and FRA7H in 7q32.3, we speculated that the viral genomes might have integrated into one of these fragile sites. Therefore, as the first step toward cloning a common fragile site, a phage genomic library was constructed from the GM00847 cell line and screened for the SV40 integration site by using SV40 DNA as a probe. Four clones were identified, all sharing the same human-SV40 junction fragment.
Identification of FRA7H and Association with the SV40 Integration Site.
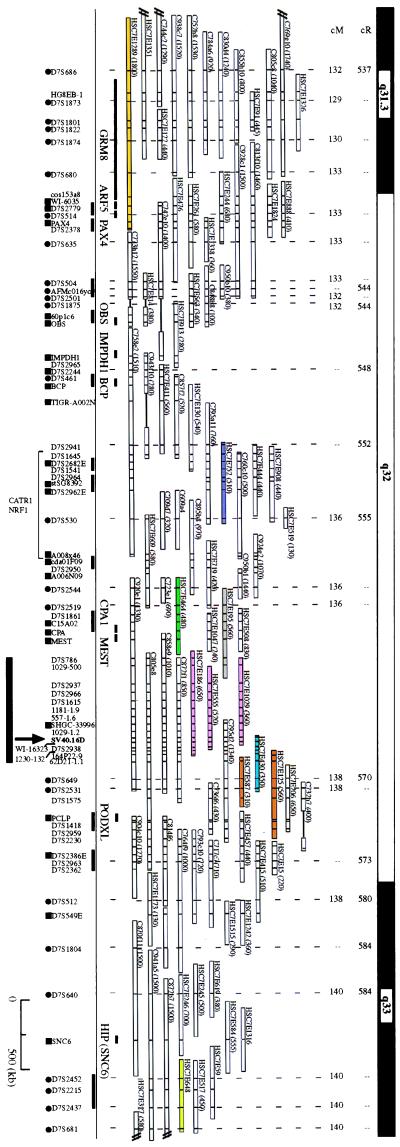
A repeat-free 2.2-kb EcoRI fragment adjacent to the human-SV40 junction fragment was used to screen a YAC library. Nine unique clones were identified and used to construct a YAC contig of ≈8 Mb covering the entire 7q32 region (Fig. 1). These YAC clones were mapped to distinct cytogenetic bands in chromosome 7 by using FISH (32). The order of the clones and DNA markers was consistent with the radiation hybrid mapping from the Massachusetts Institute of Technology/Whitehead Genome Center and the genetic data from the Genethon linkage map.
Figure 1.
A physical map across the 7q32 and FRA7H region. The order and extent of overlap of YAC clones was based on their DNA marker content. Distances from 7pter are shown in centirads (cR) and centimorgans (cM). Solid bar links DNA markers for which the local order could not be determined. •, genetic markers; ▪, ESTs/genes. Markers with no denotation are sequence-tagged sites (STSs) or unique DNA probes. Vertical bars within each YAC clone indicate the presence of a marker. YAC clones used for FISH analysis are colored. YAC clones that cover the same genomic region are marked by the same color. Solid box indicates the region shown in greater detail in Fig. 3. Information on this contig can be found in the Genome Database and at http://www.genet.sickkids.on.ca/chromosome7/.
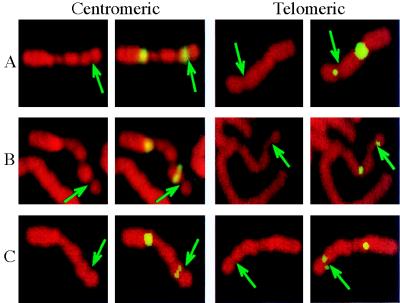
To test if the SV40 integration site was at the same region as FRA7H and to clone the FRA7H genomic region, we performed FISH analysis using YAC clones containing the integration region as probes, on metaphase chromosomes induced to exhibit fragile sites (Fig. 2). The expression of fragile sites in the studied cell line (GM00847) is due to aphidicolin induction and is not observed in untreated cells. Four YAC clones harboring the SV40 integration site—HSC7E186, HSC7E555, HSC7E1029 (all three pink), and HSC7E430 (light blue) (Fig. 1 and Table 1)—appeared to “span” the FRA7H gap. Hence, on different chromosomes from the same preparation their hybridization signals appeared centromeric or telomeric to the FRA7H gap, or crossed the FRA7H gap (“on”). The distant centromeric YAC clones HSC7E1289 (orange) and HSC7E752 (purple) showed hybridization signals only centromeric to FRA7H, and the distant telomeric YAC HSC7E648 (light green) showed signals only telomeric to FRA7H (Fig. 1 and Table 1). Thus, these clones are located outside the fragile region. To exclude the possibility that the analyzed fragile site in the GM00847 cell line is caused by the integrated SV40 genomes, we performed FISH analysis on the somatic cell hybrid GM10791A, which contains one copy of normal chromosome 7, with no SV40 sequences. FISH analysis using HSC7E186 (pink) showed hybridization signals spanning the FRA7H site (Fig. 2B): 2 signals were on the site and 6 were centromeric and 10 telomeric to the site, providing evidence that the analyzed FRA7H site spans the same genomic region in the two cell lines. Because the YAC clones spanning FRA7H contain the SV40 integration site, our observations suggest that the SV40 integration event occurred within the FRA7H site, supporting the hypothesis that fragile sites in the human genome might represent target sites for viral integration. FISH results with additional YAC clones (HSC7E125/HSC7E587, HSC7E464, and HSC7E195) flanking the FRA7H region showed inconsistency with the physical map over a region of about 1.5 Mb (Table 1, discussed below).
Figure 2.
Examples of hybridization signals centromeric and telomeric to FRA7H. FISH analysis was performed on metaphase chromosomes expressing FRA7H from two cell lines: GM00847 (A and C) and GM10791A (B). FISH experiments were performed with fluorescein isothiocyanate (FITC)-labeled YAC HSC7E186 (A and B) or cosmid 141e11 (C) cohybridized with a probe for chromosome 7 centromere. Propidium staining (left panel in each pair) and FISH with FITC-labeled probes (right panel in each pair) are shown. Arrows point to FRA7H.
Table 1.
FISH analysis of YAC hybridization signals on chromosomes expressing FRA7H
| YAC clone HSC7E- | Color | No. of signals
|
||
|---|---|---|---|---|
| Centromeric | On | Telomeric | ||
| 1289 | Orange | 15 | — | — |
| 752 | Purple | 9 | — | — |
| 464 | Dark green | 15 | 7 | — |
| 195 | Gray | 7 | 6 | 6 |
| 1029/555/186 | Pink | 18 | 6 | 17 |
| 430 | Light blue | 11 | — | 2 |
| 125/587 | Brown | 20 | — | — |
| 648 | Light green | — | 7 | 9 |
Analysis of hybridization signals on metaphase chromosomes expressing FRA7H from cell line GM00847. The clones are ordered in accordance with the physical map, centromeric (top of the table) to telomeric (bottom). The results of clones covering the same genomic region were combined. The colors mark YAC clones in Fig. 1.
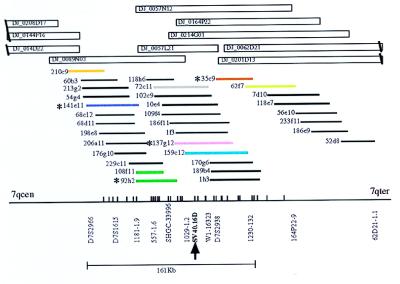
To define the FRA7H region more precisely, FISH analysis on the GM00847 cell line was performed, using cosmid clones mapped to the YACs spanning FRA7H (Fig. 3). Six of these clones—141e11 (purple), 108f11/92h2 (dark green), 72c11 (gray), 137 g12 (pink), 159c12, (light blue), and 35c9 (brown)—showed hybridization signals centromeric, telomeric, and crossing FRA7H (Fig. 2C and Table 2). FISH analysis with one of these cosmids, 137g12 (pink), was performed also on the somatic cell hybrid GM10791A, which is the cell line used for construction of the cosmid library. Again, the hybridization signals spanned FRA7H: one signal crossed the site, five were centromeric, and five were telomeric. This result further indicates that in both cell lines the analyzed fragile site is in the same genomic region, estimated to span ≈160 kb. However, clones 62f7 and 210c9 showed FISH results inconsistent with the physical map (Fig. 3 and Table 2; discussed below).
Figure 3.
A cosmid and PAC map covering the FRA7H region. P1-derived artificial chromosome (PAC) clones (name DJ_plate_row_column) and cosmids clones covering the region between markers D7S786 and 62D21–1.1 (see Fig. 1 for position within 7q32) are shown. Vertical bars along the horizontal baseline represent EcoRI sites. The region representing the 161-kb sequence contig is shown below. Asterisks indicate cosmids that were sequenced. Cosmids that were used for the FISH analysis are marked by colors. Clones covering the same region are marked by the same color.
Table 2.
FISH analysis of cosmids hybridization signals on chromosomes expressing FRA7H
| Cosmid clone | Color | No. of signals
|
||
|---|---|---|---|---|
| Centromeric | On | Telomeric | ||
| 210c9 | Orange | — | — | 11 |
| 141e11 | Purple | 5 | 3 | 4 |
| 108f11/92h2 | Dark green | 23 | 1 | 2 |
| 72c11 | Gray | 6 | 3 | 2 |
| 137g12 | Pink | 9 | 4 | 7 |
| 159c12 | Blue | 4 | — | 5 |
| 35c9 | Brown | 3 | 2 | 3 |
| 62f7 | Light green | 10 | — | — |
Analysis of hybridization signals on metaphase chromosomes expressing FRA7H from cell line GM00847. The clones are ordered in accordance with the physical map, centromeric (top of the table) to telomeric (bottom). The results of clones covering the same genomic region were combined. The colors mark cosmid clones in Fig. 3.
Sequence Analysis of the FRA7H Region.
To identify features at the nucleotide level that might provide insight into the molecular basis of the fragility of the FRA7H region, we completely sequenced a contig of 4 cosmids spanning the FRA7H region (Fig. 3). The final sequence of 161,155 bp (GenBank accession no. AF017104) was extensively analyzed for repetitive elements and regions with coding potential (for details see http://genome.imb-jena.de). The analysis indicated that the region is A+T-rich (58%). It is composed of 13.1% short interspersed elements, 13.8% long interspersed elements, 5% long terminal repeats, and 0.7% DNA transposons. No CGG or any other expended repeats were revealed. Thus, the molecular basis for the fragility of FRA7H is probably different from that of the rare fragile sites and does not require expanded repeats. Similarly, FRA3B also seems to lack expanded repeats (12–14, 16).
Numerous exons were predicted in FRA7H, but none by more than two different programs. It is likely that these predictions are false positives. blast similarity searches revealed a number of EST clones, none of which predicted exons. Resequencing of many of these EST clones revealed no ORFs. Similarity searches identified areas with 88% similarity to the zinc finger protein ZNF131 and 74% to histone H4. However, detailed analysis revealed that both are pseudogenes. The ZNF131 pseudogene most probably resulted from integration of a ZNF131 transcript into which subsequently several transposable elements integrated. The observation that several integration events occurred in FRA7H might be associated with the chromosomal instability of the region. These observations suggest that the region is gene poor. We could not exclude the possibility, however, that FRA7H constitutes a large gene, as reported for FRA3B (33).
Helix Flexibility and Stability.
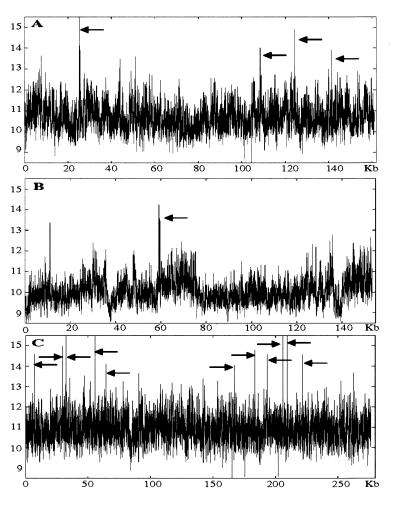
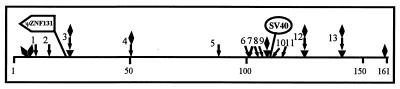
In the absence of any obvious DNA sequences that could account for the fragility we undertook a new approach. The FRA7H sequence was analyzed for structural characteristics of the DNA sequence that might be associated with the fragility. In rare fragile sites the expanded CGG repeats affect helix flexibility, leading to repression of nucleosome assembly, decondensation, and fragility (34, 35). Accordingly, we first searched the FRA7H sequence for potential local variations in the DNA flexibility, which appears to play an important role in protein–DNA interactions and hence might affect chromatin condensation (25). The analysis revealed four regions with potential high flexibility deviating significantly (>4.5 SD, based on the value of the lowest region among the four) from the average value of the entire FRA7H sequence (x̄ = 10.7°; SD = 0.65; P < 0.0001) (Fig. 4A). Three of these regions are clustered within a 33-kb region (Fig. 4A). The SV40 and the ZNF131 integration sites are located only 1–2 kb from high-flexibility regions (Fig. 5).
Figure 4.
Analysis of DNA flexibility. Arrows point to regions with significantly high flexibility. x axis, nucleotide position at the beginning of a 100-bp window. y axis, degrees of inclination in the twist angel. (A) FRA7H region. (B) Example of a control sequence, hsu52111. (C) FRA3B combined sequences.
Figure 5.
Map of FRA7H showing DNA regions of high flexibility (▾), low stability (⧫), and non-B-DNA structure ( ). The SV40 integration site and the ZNF131 pseudogene are marked. The numbers below the map represent the base location in FRA7H.
To assess the significance of the potential high-flexibility pattern identified in the FRA7H region, we analyzed 1.1 Mb composed of 14 genomic sequences mapped to chromosomal bands in which fragile sites were not described (GenBank locus names: hsablgr1, hsablgr2, hsl185e6a, hsl19h1, hsl247f6, hsl79f5a, hsq25, hsq27, hsu07000, hsu09822, hsu29953, hsu36341, hsu40455, hsu41384, humhprtb, hsu52111, hsu52112, and humretblas). The analysis revealed that regions with high flexibility appear every ≈100 kb (Fig. 4B). Thus, regions with high flexibility in FRA7H are more frequent than predicted by the control analysis.
We also analyzed the FRA7H sequence for helix stability. The analysis revealed seven sites with potential low stability that deviated significantly (>2.5 SD) from the average value of the entire FRA7H sequence (x̄ = 1.57 kcal/mol, SD = 0.124, P < 0.0062), based on the value for highest region among the seven sites. All the regions in FRA7H that were predicted to show high flexibility also showed low stability (Fig. 5).
Potential Non-B-DNA Sequences.
The replication of rare fragile sites with CGG expansion is delayed relative to that of normal alleles (36, 37). CGG repeats have a propensity to adopt a non-B-DNA structure (hairpin or tetraplex), leading to arrest of DNA replication in vitro (38–40) and in vivo (41). The replication of the common fragile site FRA3B is delayed by exposing cells to the fragile site inducer aphidicolin (42). Hence, we searched the FRA7H region for two types of potential non-B-DNA sequences under negative superhelical strain: triple helixes and MARs. Triple-helix-forming sequences play an important role in triggering gene expression and homologous recombination (43), block DNA replication in vitro (44), and decrease the efficiency of replication in vivo (45). The MARs are prone to unwinding by continuous base unpairing (46). Chromosomal attachments to the nuclear matrix create independent loop domains that affect DNA replication, transcription, and condensation (47). Our analysis revealed 13 sites with non-B-DNA-forming potential, 6 sites with potential to form a triple-helical DNA structure (sites 1, 2, 4, 7, 8, and 10), and 7 potential MARs (sites 3, 5, 6, 9, 11, 12, and 13; Fig. 5). Six of these (sites 6–11) were in a cluster that colocalized with a high flexibility and low stability and with the SV40 integration site (Fig. 5). Thus, the frequency and distribution of these sites might be an important feature of the FRA7H region.
In Vitro Analysis of Non-B-DNA Structures.
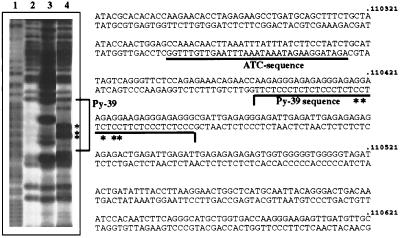
CAA has been used for non-B-DNA structure analysis (48). CAA reaction is ideal for detecting only unpaired bases (even only a single unpaired base) within double-stranded DNA (29). In vitro analysis of triple-helix formation was performed on a 4.4-kb EcoRI fragment, cloned from cosmid 137g12 (base position 106,853–111,288) (Fig. 4), which harbors the SV40 integration site and a potential triple helix site with a 39 homopyrimidine tract (site 10, Figs. 5 and 6). When a homopyrimidine/homopurine sequence forms triple helix structure (Py⋅Pu⋅Py triplex, H-DNA) the center of the homopyrimidine strand and the purine bases over the 5′ half of the homopurine strand are unpaired and are reactive with CAA. Thus, the H-DNA structure can be demonstrated by the cleavage of DNA at these CAA-modified residues. CAA-reacted cytosines were detected in the center of the Py-39 of site 10 in the presence of Mg2+ at pH 7, conditions that support the formation of H-DNA (Fig. 6, lane 4). Under the same conditions, CAA-reacted adenines were also detected over the 5′ half of the purine strand of the Py-39 sequence (data not shown). Thus, the predicted site of potential non-B-DNA structure was proved to form non-B-DNA structure under torsional stress in vitro. In the absence of metal ions the Py-39 sequence also showed a strong unpaired DNA structure (Fig. 6, lane 3 compared with the control lane 2) with a CAA-reactive pattern different from that of a normal H-DNA structure (Fig. 6, lane 4). The nature of this pattern is unknown, and it might be affected by the nearby ATC sequence (Fig. 6). In vitro analysis of an adjacent 2.2-kb EcoRI fragment (position 104,637–106,853), in which no sites were predicted, did not reveal any apparent non-B-DNA sequences (data not shown). Further studies are required to investigate the in vivo role of such unusual DNA structures in the fragility mechanism.
Figure 6.
Non-B-DNA structure adopted by plasmid DNA containing site 10. Plasmid DNA with the 4.4-kb EcoRI fragment was unmodified (lanes 1 and 2) or modified with CAA (lanes 3 and 4). Lane 1, hydrazine reaction of control DNA. Lanes 2–4, formic acid reaction of control DNA (lane 2), CAA-treated DNA in the absence of Mg2+, at pH 7 (lane 3), and CAA-treated DNA in the presence of Mg2+, at pH 7 (lane 4). The nucleotide sequence is from FRA7H sequence. The asterisks mark sites that reacted prominently with CAA.
FRA7H Shows Unusual Chromatin Organization.
Our extensive FISH studies identified nonoverlapping cosmid clones that showed FISH signals spanning FRA7H (Fig. 3, Table 2). These results suggest that the under-condensation of induced fragile sites can occur at different locations along the fragile region. Similar FISH patterns have been seen for other fragile sites.
Several YAC and cosmid clones, flanking the 161-kb sequenced region, showed FISH signals in opposite orientation relative to the physical map (Figs. 1 and 3, Tables 1 and 2). Similarly, clones spanning FRA3B yielded signals in opposite orientation on chromosomes from different individuals (13, 15, 17). We cannot exclude the possibility that genomic rearrangements associated with the region surrounding the fragile site might account for these FISH observations. We speculate, however, that the unexpected nonlinear FISH hybridization patterns at fragile sites might reflect regions of chromatin organization in which the linearity of the DNA relative to the chromosomal axis is altered.
Comparison of FRA7H, FRA3B, and the Putative Partial Sequence of FRA7G.
To identify shared sequence features between aphidicolin-induced common fragile sites, we first compared the sequences of FRA7H and FRA3B (GenBank accession no. AF017104 and U66722, respectively). Both regions are A+T-rich (58% and 61%, respectively) and are gene poor. In both fragile sites insertions of viral genomes and pseudogenes occurred, further indicating that fragile sites are unstable regions.
To assess the relevance of the high-flexibility and low-stability patterns to the aphidicolin-induced fragility, we analyzed the FRA3B combined sequences (GenBank accession nos. U66722 and AF020503). Eleven regions with potential high flexibility were identified, with values comparable to those of the high regions of FRA7H. These regions appeared in two clusters (Fig. 4C). One of the regions with high flexibility is at the same site as a reported aphidicolin-induced breakpoint cluster (position 39,597, GenBank accession no. U66722, position 205,959 in Fig. 4C). Interestingly, this site meets many of the criteria for being a MAR (49). We also analyzed the stability pattern of FRA3B and revealed that the high-flexibility and low-stability regions colocalized, as in the FRA7H region (data not shown). The impressive cluster of high-flexibility and low-stability regions in FRA3B might reflect the fact that it is the most inducible common fragile site in the human genome. Thus, regions with high flexibility and low stability, in FRA7H and FRA3B, might contribute to the overall chromatin structure of these fragile sites and hence to their breakage and rearrangement.
We also searched for similarity in the repeat free sequences between FRA7H and FRA3B. The comparison revealed similarities (>95%) among short sequences within the regions with high flexibility. These sequences are A+T-rich, as expected from the flexibility value for this dinucleotide (25). Thus, these short sequence elements might be associated with the fragility of both FRA7H and FRA3B. Sequence similarities (>95%) were also found in two of the regions predicted to form non-B-DNA structure in FRA7H (sites 3 and 7). Together these observations indicate that FRA7H and FRA3B share sequence similarities in regions with potential to form unusual DNA structure, and they support our contention that the unusual DNA structure may be associated with the mechanism of fragility in FRA7H and FRA3B. Nine additional sequences of 17–22 bp showed sequence similarity of >95% between the two fragile sites, the significance of which needs further investigation.
It was of interest to analyze the FRA7G sequence; however, the region spanned by FRA7G is not completely defined. Nevertheless, we performed analysis of helix flexibility and stability on the available 150-kb sequence harboring the DNA marker D7S522 reported to be part of FRA7G (accession. no. AC002066; ref. 18). Eight regions with potential high flexibility were identified. Three of these sites were clustered within 18 kb harboring D7S522 (data not shown). Similarly to FRA7H and FRA3B, the stability pattern of FRA7G revealed that the high-flexibility and low-stability regions colocalized (data not shown).
We also searched for similarity in the repeat free sequences between FRA7H and the partial sequence of FRA7G. The comparison revealed similarities (>95%) among short sequences, the significance of which needs further investigation.
In summary, we identified a common fragile site, FRA7H, by cloning a viral integration site. This approach could be of general value for the isolation of common fragile sites in the human genome. Sequence analysis of FRA7H and FRA3B revealed several sequence-based features that are suggestive of unusual DNA structures. The sequence analysis of the putative partial FRA7G also revealed similar features. These are possibly intrinsic properties of common fragile sites that may affect their replication and condensation as well as organization and may lead to fragility. Identification and characterization of other aphidicolin-induced common fragile sites and studies of specific regions within these sites are required to understand the significance of the unusual DNA structures found in FRA7H, FRA3B, and FRA7G for the general mechanism of fragility.
Acknowledgments
We thank Raveh Gill-More, Compugen Ltd., for assistance with the computerized sequence analysis and Dr. Irit Baram for assistance in FISH. This research was supported by grants from the Israel Academy of Sciences and Humanities to B.K.; by grants from the Canadian Genome Analysis and Technology Program and the Canadian Genetic Disease Network and a Howard Hughes International Scholar Award to L.-C.T.; by Grant CA51377 from the National Institutes of Health to Y.K.; and by grants from the German Federal Ministry of Education, Science and Technology, Projekttraeger BEO. to B.H. and B.D.
ABBREVIATIONS
- YAC
yeast artificial chromosome
- SV40
simian virus 40
- FISH
fluorescent in situ hybridization
- EST
expressed sequence tag
- MAR
matrix attachment region
- CAA
chloroacetaldehyde
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF017104).
References
- 1.Sutherland G R, Richards R I. Curr Opin Genet Dev. 1995;5:323–327. doi: 10.1016/0959-437x(95)80046-8. [DOI] [PubMed] [Google Scholar]
- 2.Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas M F, Mandel J L. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 3.Gecz J, Gedeon A K, Sutherland G R, Mulley J C. Nat Genet. 1996;13:105–108. doi: 10.1038/ng0596-105. [DOI] [PubMed] [Google Scholar]
- 4.Jones C, Penny L, Mattina T, Yu S, Baker E, Voullaire L, Langdon W, Sutherland G, Richards R, Tunnacliffe A. Nature (London) 1995;376:145–149. doi: 10.1038/376145a0. [DOI] [PubMed] [Google Scholar]
- 5.Kremer E J, Pritchard M, Lynch M, Yu S, Holman K, Baker E, Warren S T, Schlessinger D, Sutherland G R, Richards R I. Science. 1991;252:1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- 6.Knight S, J, Flannery A V, Hirst M C, Campbell L, Christodoulou Z, Phelps S R, Pointon J, Middleton-Price H R, Barnicoat A, Pembrey M E, et al. Cell. 1993;74:127–134. doi: 10.1016/0092-8674(93)90300-f. [DOI] [PubMed] [Google Scholar]
- 7.Nancarrow J, Kremer E, Holman K, Eyre H, Doggett N, Le Paslier D, Callen D, Sutherland G, Richards R. Science. 1994;264:1938–1941. doi: 10.1126/science.8009225. [DOI] [PubMed] [Google Scholar]
- 8.Parrish J E, Oostra B A, Verkerk A J, Richards C S, Reynolds J, Spikes A S, Shaffer L G, Nelson D L. Nat Genet. 1994;8:229–235. doi: 10.1038/ng1194-229. [DOI] [PubMed] [Google Scholar]
- 9.Yu S, Mangelsdorf M, Hewett D, Hobson L, Baker E, Eyre H J, Lapsys N, Le Paslier D, Doggett N A, Sutherland G R, Richards R I. Cell. 1997;88:367–374. doi: 10.1016/s0092-8674(00)81875-9. [DOI] [PubMed] [Google Scholar]
- 10.Kornberg A, Baker T A. DNA Replication. New York: Freeman; 1992. [Google Scholar]
- 11.Wilke C M, Guo S W, Hall B K, Boldog F, Gemmill R M, Chandrasekharappa S C, Barcroft C L, Drabkin H A, Glover T W. Genomics. 1994;22:319–326. doi: 10.1006/geno.1994.1390. [DOI] [PubMed] [Google Scholar]
- 12.Paradee W, Wilke C M, Wang L, Shridhar R, Mullins C M, Hoge A, Glover T W, Smith D I. Genomics. 1996;35:87–93. doi: 10.1006/geno.1996.0326. [DOI] [PubMed] [Google Scholar]
- 13.Wilke C M, Hall B K, Hoge A, Paradee W, Smith D L, Glover T W. Hum Mol Genet. 1996;5:187–195. doi: 10.1093/hmg/5.2.187. [DOI] [PubMed] [Google Scholar]
- 14.Boldog F, Gemmill R M, West J, Robinson M, Robinson L, Li E, Roche J, Todd S, Waggoner B, Lundstrom R, Jacobson J, Mullokandov M R, Klinger H, Drabkin H A. Hum Mol Genet. 1997;6:193–203. doi: 10.1093/hmg/6.2.193. [DOI] [PubMed] [Google Scholar]
- 15.Zimonjic D B, Druck T, Ohta M, Kastury K, Croce C M, Popescu N C, Huebner K. Cancer Res. 1997;57:1166–1170. [PubMed] [Google Scholar]
- 16.Inoue H, Ishii H, Alder H, Snyder E, Druck T, Huebner K, Croce C M. Proc Natl Acad Sci USA. 1997;94:14584–14589. doi: 10.1073/pnas.94.26.14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rassool F V, Le Beau M M, Shen M-L, Neilly M, E, Espinosa R, III, Ong S T, Boldog F, Drabkin H, McCarroll R, McKeithan T W. Genomics. 1996;35:109–117. doi: 10.1006/geno.1996.0329. [DOI] [PubMed] [Google Scholar]
- 18.Huang H, Qian C, Jenkins R B, Smith D I. Genes Chromosomes Cancer. 1998;21:152–159. doi: 10.1002/(sici)1098-2264(199802)21:2<152::aid-gcc11>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 19.Popescu N, Zimonjic D, DiPaolo J. Hum Genet. 1990;84:383–386. doi: 10.1007/BF00195804. [DOI] [PubMed] [Google Scholar]
- 20.Smith P P, Friedman C L, Bryant E M, McDougall J K. Genes Chromosomes Cancer. 1992;5:150–157. doi: 10.1002/gcc.2870050209. [DOI] [PubMed] [Google Scholar]
- 21.Scherer S W, Tompkins B J, Tsui L C. Mamm Genome. 1992;3:179–181. doi: 10.1007/BF00352464. [DOI] [PubMed] [Google Scholar]
- 22.Ioannou P A, Amemiya C, Garnes J, Kroisel P M, Shizuya H, Batzer M A, de Jong P J. Nat Genet. 1994;6:84–88. doi: 10.1038/ng0194-84. [DOI] [PubMed] [Google Scholar]
- 23.Lichter P, Cremer T, Borden J, Manuelidis L, Ward D C. Hum Genet. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- 24.Craxton M. Methods Mol Biol. 1993;23:149–167. doi: 10.1385/0-89603-248-5:149. [DOI] [PubMed] [Google Scholar]
- 25.Sarai A, Mazur J, Nussinov R, Jernigan R L. Biochemistry. 1989;28:7842–7849. doi: 10.1021/bi00445a046. [DOI] [PubMed] [Google Scholar]
- 26.Breslauer K J, Frank R, Blocker H, Marky L A. Proc Natl Acad Sci USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank-Kamenetskii M D, Mirkin S M. Annu Rev Biochem. 1995;64:65–95. doi: 10.1146/annurev.bi.64.070195.000433. [DOI] [PubMed] [Google Scholar]
- 28.Dickinson L A, Joh T, Kohwi Y, Kohwi-Shigematsu T. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- 29.Kohwi Y, Kohwi-Shigematsu T. Proc Natl Acad Sci USA. 1988;85:3781–3785. doi: 10.1073/pnas.85.11.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell A R, Ambros P, Gosden J R, Morten J E, Porteous D J. Somat Cell Mol Genet. 1986;12:313–324. doi: 10.1007/BF01570725. [DOI] [PubMed] [Google Scholar]
- 31.Campo M S, Cameron I R, Rogers M E. Cell. 1978;15:1411–1426. doi: 10.1016/0092-8674(78)90065-x. [DOI] [PubMed] [Google Scholar]
- 32.Kunz J, Scherer S W, Klawitz I, Soder S, Du Y Z, Speich N, Kalff-Suske M, Heng H H, Tsui L C, Grzeschik K H. Genomics. 1994;22:439–448. doi: 10.1006/geno.1994.1407. [DOI] [PubMed] [Google Scholar]
- 33.Ohta M, Inoue H, Cotticelli M G, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, et al. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 34.Metzenberg S. Am J Hum Genet. 1996;59:252–253. [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y-H, Gellibolian R, Shimizu M, Wells R D, Griffith J. J Mol Biol. 1996;263:511–516. doi: 10.1006/jmbi.1996.0593. [DOI] [PubMed] [Google Scholar]
- 36.Hansen R S, Canfield T K, Lamb M M, Gartler S M, Laird C D. Cell. 1993;73:1403–1409. doi: 10.1016/0092-8674(93)90365-w. [DOI] [PubMed] [Google Scholar]
- 37.Subramanian P S, Nelson D L, Chinault A C. Am J Hum Genet. 1996;59:407–416. [PMC free article] [PubMed] [Google Scholar]
- 38.Fry M, Loeb L A. Proc Natl Acad Sci USA. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Usdin K, Woodford K J. Nucleic Acids Res. 1995;23:4202–4209. doi: 10.1093/nar/23.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weitzmann M N, Woodford K J, Usdin K. J Biol Chem. 1997;272:9517–9523. doi: 10.1074/jbc.272.14.9517. [DOI] [PubMed] [Google Scholar]
- 41.Samadashwily G M, Raca R, Mirkin S M. Nat Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 42.Le Beau M M, Rassool F V, Neilly M E, Espinosa R, III, Glover T W, Smith D I, McKeithan T W. Hum Mol Genet. 1998;7:755–761. doi: 10.1093/hmg/7.4.755. [DOI] [PubMed] [Google Scholar]
- 43.Kohwi Y, Kohwi-Shigematsu T. Genes Dev. 1991;5:2547–2554. doi: 10.1101/gad.5.12b.2547. [DOI] [PubMed] [Google Scholar]
- 44.Baran N, Lapidot A, Manor H. Proc Natl Acad Sci USA. 1991;88:507–511. doi: 10.1073/pnas.88.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao B S, Manor H, Martin R G. Nucleic Acids Res. 1988;16:8077–8094. doi: 10.1093/nar/16.16.8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kohwi-Shigematsu T, Kohwi Y. Biochemistry. 1990;29:9551–9560. doi: 10.1021/bi00493a009. [DOI] [PubMed] [Google Scholar]
- 47.Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, Kohwi-Shigematsu T. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- 48.Kohwi-Shigematsu T, Kohwi Y. Methods Enzymol. 1992;212:155–180. doi: 10.1016/0076-6879(92)12011-e. [DOI] [PubMed] [Google Scholar]
- 49.Wang L, Paradee W, Mullins C, Shridhar R, Rosati R, Wilke C M, Glover T W, Smith D I. Genomics. 1997;41:485–488. doi: 10.1006/geno.1997.4690. [DOI] [PubMed] [Google Scholar]