Abstract
Among the mechanisms whereby sex is determined in animals, chromosomal sex determination is found in a wide variety of distant taxa. The widespread but not ubiquitous occurrence, not even within lineages, of chromosomal sex determination suggests that sex chromosomes have evolved independently several times during animal radiation, but firm evidence for this is lacking. The most favored model for this process is gradual differentiation of ancestral pairs of autosomes. As known for mammals, sex chromosomes may have a very ancient origin, and it has even been speculated that the sex chromosomes of mammals and birds would share a common chromosomal ancestry. In this study we showed that the two genes, ATP5A1 and CHD1, so far assigned to the female-specific W chromosome of birds both exist in a very closely related copy on the Z chromosome but are not pseudoautosomal. This indicates a common ancestry of the two sex chromosomes, consistent with the evolution from a pair of autosomes. Comparative mapping demonstrates, however, that ATP5A1 and CHD1 are not sex-linked among eutherian mammals; this is also not the case for the majority of other genes so far assigned to the avian Z chromosome. Our results suggest that the evolution of sex chromosomes has occurred independently in mammals and birds.
Although the concept of sexual reproduction is found among essentially all eukaryotes, the mechanisms whereby sex is determined are clearly diverse: chromosomal sex determination (CSD; with male or female heterogamety), mono- or polyfactorial sex determination not associated with heteromorphic sex chromosomes, environmental sex determination, cytoplasmic sex determination, and arrhenotoky (haplo-diploidy; ref. 1). The occurrence of these mechanisms is scattered across different animal groups. For instance, CSD can be found among as phylogenetically divergent taxa as Platyhelminthes, Nematoda, Crustacea, Insecta, Teleostomi, Amphibia, Reptilia, Aves, and Mammalia but is not necessarily the only mechanism present in the respective taxa. In turtles and lizards, for example, some species show temperature-dependent sex determination, whereas others possess CSD. In yet other taxa, however, such as birds and mammals, CSD is obligate. The taxonomic distribution of CSD throughout the animal kingdom strongly suggests that this type of sex-determining system has evolved independently in many different groups during animal radiation (1–6).
How do sex chromosomes arise? At the beginning of the century, Muller and Sturtevant (7) developed the theory that, from an initial state of similarity, sex chromosomes would evolve into one active and one degenerate copy. The ancestral state should hence generally have been that of a pair of autosomes. Despite being a commonly held view (1–6), the transition from homology to heteromorphism has only occasionally been supported by empirical data. The most prominent evidence comes from mammalian genome analysis: in primates and mice, genes or other DNA sequences similar to those on the non-recombining part of the X chromosome can be found on the Y chromosome, indicating the common ancestry of the two chromosomes (reviewed in refs. 8 and 9). Moreover, the existence of a pseudoautosomal (recombining) region further points to a common origin of the two chromosomes. Evidence from other taxa are only circumstantial: for instance, the gradual change from an undifferentiated homomorphic chromosome pair to highly differentiated Z and W chromosomes in different snake families (10).
Birds and reptiles are the closest relatives to mammals among extant taxa. Birds are characterized by female heterogamety: males have two copies of the Z chromosome (hence, denoted ZZ) and females have one copy of the Z chromosome and one of the W chromosome (ZW). The W chromosome is generally much smaller than the Z chromosome and also shows other typical signs of a degenerated sex chromosome, i.e., a low gene content that is rich in heterochromatic, repetitive DNA of the satellite type (11, 12). In this study we addressed the question of how the avian Z and W chromosomes have evolved. First, we asked whether the two chromosomes share a common ancestry, similar to the situation for mammalian sex chromosomes. Second, by comparative mapping we analyzed the genetic relationships between the sex chromosomes of birds and mammals to reveal the evolutionary history of sex chromosomes among higher animals. The latter issue should be seen in the perspective of the X chromosome being almost completely conserved among all eutherian mammals and also showing strong homology among eutherian mammals, monotremes, and marsupials (13, 14), indicating an ancient origin.
MATERIALS AND METHODS
Chicken Linkage Mapping.
Genetic mapping was done in one of the two internationally recognized chicken mapping populations, the East Lansing reference family. The family is built up by a cross between a Jungle Fowl sire and a White Leghorn dam, followed by backcross between one F1 male and four White Leghorn females (15). Fifty-two F2 progeny from this backcross were genotyped with markers described in this study, and linkage analysis was performed with map manager, version 2.6.5 (16), and mapmaker, version 3.0 (17), against a set of some 890 markers already typed in the pedigree (18). Restriction fragment length polymorphism (RFLP) analysis of the CHD1Z gene was done with a probe from Jungle Fowl DNA, amplified by PCR, by using primers 2895 (CGGCTAGTCACAAAAGGATC) and 3225 (TTGAACTGTGAAAGCAACTC) that were hybridized to HindIII-digested DNA. Length polymorphism in a poly(A) mononucleotide repeat present in intron 10 (GenBank accession no. AJ223297) of the chicken ATP5A1Z gene was scored by using exon-flanking primers 489 (TGCTGGGCCGTGTTGTAGAT) and 616 (GGTTCCCGCACAGAGATTC). One primer was fluorescently labeled, and the length variation was detected on an ABI377 sequencing instrument (Perkin–Elmer).
Murine Linkage Mapping.
Interspecific backcross progeny were generated by mating (C57BL/6J × Mus spretus) F1 females and C57BL/6J males as described (19). This interspecific backcross-mapping panel has been typed for more than 2500 loci that are well distributed among all of the autosomes as well as the X chromosome (19). A total of 205 F2 mice were used to map the Chd1 and Atp5a1 loci. DNAs were digested with several enzymes and analyzed by Southern blot hybridization (20) for informative RFLPs by using mouse cDNA probes. The Chd1 probe, a 1.7-kb EcoRI–HindIII fragment of mouse cDNA (21), revealed the presence or absence of 9.0- and 3.2-kb XbaI M. spretus-specific fragments, which cosegregated and were followed in backcrossed mice. The Atp5a1 probe, a 1.7-kb EcoR–XhoI fragment of mouse cDNA (22), detected 5.6-, 1.9-m and 1.5-kb BamHI fragments in C57BL/6J DNA and a major 6.8-kb BamHI fragment in M. spretus DNA. The presence or absence of the M. spretus-specific 6.8-kb BamHI fragment was followed in backcrossed mice. A description of the probes and RFLPs for the loci linked to Chd1, including Mas1 and Nkx2–5, has been reported (23); those linked to Atp5a1 include Dcc and Mbp (24). Recombination distances were calculated by using map manager. Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns.
Fluorescent in Situ Hybridization (FISH) in Chickens.
For physical assignments of chicken genes, the following probes were used: a 1.8-kb fragment prepared by PCR amplification of CHD1Z from male genomic chicken DNA by using primers 2895 and 3555 (AAAGGATTTAGCGATGCAGA); 2.3- and 1.8-kb fragments PCR amplified from ATP5A1Z of male genomic chicken DNA by using primers 141 (TTGCTGCAAGAAACATCCATGC) and 616 and primers 965 (GACAATGGAAAACATGCGTTG) and 1389 (CCACTTCACGGTACTGAGC), respectively. Probes were labeled with biotin-14–dATP or digoxigenin-11–dUTP via nick translation (BioNick labeling system, Life Technologies, Grand Island, NY). Chromosome preparations were made from chicken bone marrow by using standard methods (25). Slides were treated with RNase before hybridization, which was carried out overnight in a moist chamber at 37°C in a mixture of 50% formamide, 10% dextran sulfate, 1 μg chicken genomic DNA, and 100 ng labeled probe. The preparations were washed in 50% formamide/2× standard saline citrate (1× = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7.0) and 4× standard saline citrate at 40°C. Specific hybridization signals were detected with avidin–fluorescein isothiocyanate (biotin-labeled probes) and anti-digoxigenin–rhodamine (digoxigenin-11–dUTP-labeled probes) by using standard procedures. Finally, chromosomes were counterstained with 4′,6-diamidino-2-phenylindole, and metaphases were analyzed by using a BX60 fluorescence microscope (Olympus, New Hyde Park, NY) equipped with appropriate filters. Images were captured with an IMAC-CCD S30 video camera (Metasystems GmbH, Altlussheim, Germany) and analyzed by using isis, version 1.65 (Metasystems), software.
Radiation Hybrid Mapping.
Primers A1 (ATCACCCAGCCCAAGAATCAT) and A2 (GGCACTCCTCCCCATACACC) were selected to amplify a 297-bp PCR product from intron 3 of human ATP5A1 (GenBank accession no. D28126). No products were obtained from amplification of rodent DNA with the amplification conditions used. The PCR assay was used to score arrayed templates from the Genebridge4 radiation hybrid-screening panel in duplicate. Results were submitted to the server implemented at http://www-genome.wi.mit.edu/cgi-bin/contig/rhmapper.pl, for placement on the framework radiation hybrid map.
Comparative Mapping.
Map data for chicken and bovine genes were obtained from The Roslin Institute Online web pages (http://www.ri.bbsrc.ac.uk/genome_mapping.html). Map data for human genes were extracted from the Genome Data Base (http://www.hgmp.mrc.ac.uk/gdb/gdbtop.html), and data for mouse genes were extracted from the Mouse Genome Database (http://www.informatics.jax.org/mgd.html).
Sequence and Phylogenetic Analysis.
Ostrich (Struthio camelus) mRNA was prepared from blood of an adult female with a Quick Prep mRNA purification kit (Pharmacia). The Access RT-PCR system (Promega) was used together with different sets of primers to amplify parts of the ostrich CHD1 gene: 1105 (GTGGAATATTATAATTGCCAGCA) and 2128 (GACCAAAGCTCTTTGAGG), 1628 (ACTGAACTGGCTTGCTCA) and 2469 (CTGGTGGTTTAATGAGGTAA), 2895 and 3681 (GTAACTCTTGATAAATCGTCTA), and P3 (AGATATTCCGGATCTGATAGTGA) and 4104 (TCAGTAATTTAATGAGGTAGT). Amplification products were gel purified, cycle sequenced with dye terminator chemistry, and analyzed on an Applied Biosystems 377 instrument. In total, 1492 bp of the ostrich CHD1 gene sequence was obtained (GenBank accession nos. AF059276 and AF060700–2), and this was used for phylogenetic analysis together with human and murine CHD1 and chicken CHD1Z and CHD1W sequences. A phylogenetic tree was constructed with the maximum parsimony method by using paup, version 3.1.1.
RESULTS
Z and W Chromosome Homology.
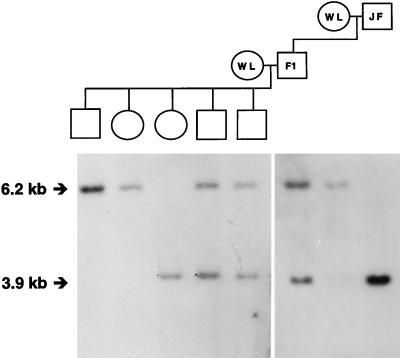
The first known gene shown conclusively to segregate with the female sex of birds, i.e., being located on the W chromosome, is a member of the chromo-helicase-DNA-binding protein family, CHD1W (W denoting its chromosomal location; refs. 26 and 27). Hybridization studies indicated that the gene is present on the W chromosome of probably all avian species, with the exception of ratites, and that the gene also exists in a second, very similar, but not W-linked, copy in the non-ratite bird genome (26, 27). We used a probe derived from this latter CHD1 copy to detect an HindIII RFLP in the East Lansing reference pedigree for chicken genome mapping (Fig. 1). Linkage analysis assigned the gene to the q arm of the Z chromosome, with a maximum logarithm of odds score of 15.7 at a distance of 0 centimorgan (cM) from the markers MSU0057, MSU0070, MSU0392, LEI0121, and LEI0144. We hence termed this gene CHD1Z, to distinguish it from CHD1W. It is important to point out that CHD1W and CHD1Z do not recombine and thus cannot be pseudoautosomal (28). Although they obviously originate from a common ancestral gene, sequence data confirm that they now evolve independently (28).
Figure 1.
An HindIII RFLP in the chicken CHD1Z gene demonstrating Z chromosome linkage JF, Jungle Fowl; WL, White Leghorn.
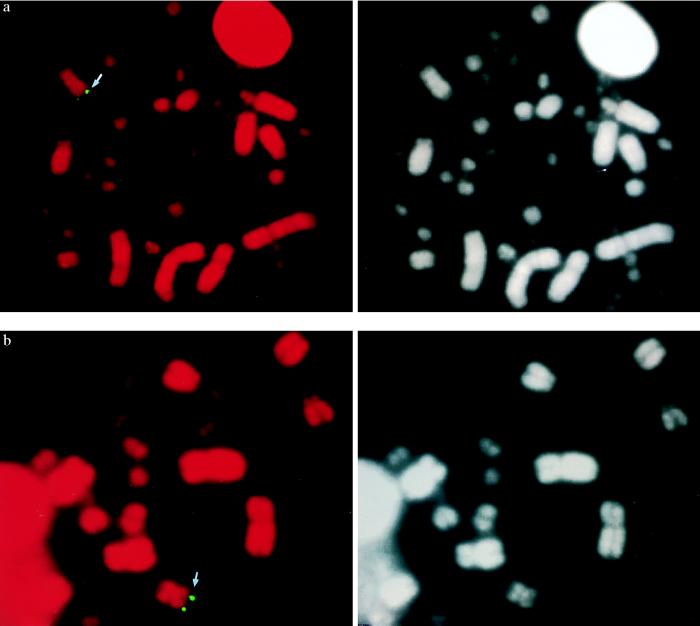
FISH with the CHD1Z probe showed a strong signal on the distal part of Zq (Fig. 2a), assigning the gene to bands Zq16-q21, i.e., at the border of the distal heterochromatic band of Zq (Fig. 3). This was somewhat surprising because linkage analysis had placed CHD1Z 60 cM away from the IREB1 gene (Fig. 3), which has been physically mapped to the same chromosomal region (29). Repeated FISH analysis with probes derived from other parts of CHD1Z, however, consistently gave the same localization (data not shown). Possibly, these observations could be explained by the region being a hot spot for recombination or one that is unusually contracted in metaphase. In some metaphases, the probes also hybridized to CHD1W. This signal was on the distal part of one of the arms of the W chromosome; the two arms are nearly indistinguishable with standard staining techniques.
Figure 2.
Fig. 2. FISH localization of the chicken CHD1Z (a) and ATP5A1Z (b) genes. (Left) Hybridization signal. (Right) 4′,6-Diamidino-2-phenylindole-banding pattern (in black and white) of the corresponding metaphases.
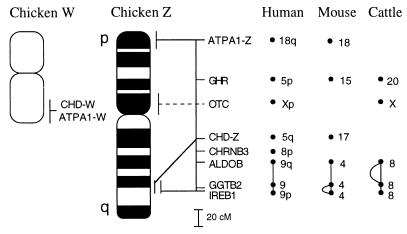
Figure 3.
A summary of genetic, physical, and comparative map data for the chicken Z and W chromosomes. Note that the orientation of the W chromosome is not known; the signals of CHD1W and ATP5A1W are both from one of the chromosome ends. The location of Z chromosome genes have been drawn to scale according to the recombination distances separating them in the linkage map derived from the East Lansing reference population. A large number of anonymous markers placed between the genes are not included in the figure. Also note that the OTC gene has not been genetically mapped in chickens. The curved lines next to the ALDOB–GGTB2–IREB1 linkage groups in mice and cattle indicate rearranged gene orders.
Dvorak et al. (30) reported an anonymous cDNA clone derived from turkeys that hybridized to the W chromosome of a wide variety of bird species (but not ratites). As for CHD1, this clone also hybridized to a second, non-W-linked copy in all non-ratite birds investigated. The two gene copies have been sequenced and identified as avian homologs of the ATP synthase α-subunit, ATP5A1 (International Patent Publication no. WO 94/07907). By using primers derived from the turkey sequence, we amplified the two gene copies in chickens and identified a length polymorphism in an intronic (A)n repeat in the non-W copy of the gene, providing a means for linkage mapping. An assignment was made to the distal part of the p arm of chromosome Z, with MSU0342 as the closest marker at 0 cM (logarithm of odds score of 15.7). Hence, the two avian ATP5A1 gene copies are hereafter denoted ATP5A1Z and ATP5A1W. Physical mapping with FISH placed the ATP5A1Z gene close to the telomere of Zp (Fig. 2b). As for CHD1, in some metaphases the ATP5A1 probe also gave a signal from the end of one of the arms of the W chromosome. We used two-color FISH to address whether ATP5A1W and CHD1W were located on the same arm of the W chromosome, but we failed to obtain any clear signals when both probes were applied together.
Comparative Mapping.
To comparatively address the chromosomal locations of the sex-linked avian genes in mammals, we sought map data for the genes in humans and mice. In humans, there are at least four related CHD genes (CHD1–4); phylogenetic analysis shows clearly that CHD1 corresponds to avian CHD1Z and CHD1W (31). In humans, CHD1 has been mapped to HSA5q15–21 (31). The genomic localization of the cloned, but previously unmapped, human ATP5A1 gene was determined by placing an intronic sequence-tagged site corresponding to ATP5A1 on the Genebridge4 radiation hybrid-mapping panel. By using this assay we assigned the human ATP5A1 gene to a region between markers WI-2986 and D18S72 on HSA18q11–12.
In mice only one full-length CHD gene, Chd1, has been cloned (20); however, murine homologs of human CHD1–4 are present in dbEST, a database of expressed sequence tagged sites. Phylogenetic analysis shows that mouse Chd1, human CHD1, and avian CHD1Z and CHD1W are more closely related to each other than to any other known CHD gene and that they are likely to be derived from a single ancestral gene (31). The mouse chromosomal location of Chd1 was determined by interspecific backcross analysis by using progeny derived from matings of [(C57BL/6J × M. spretus)F1 × C57BL/6J] mice. The mapping results indicated that Chd1 is located in the proximal region of the mouse chromosome 17 linked to Mas1 and Nkx2–5. The ratios of the total number of mice exhibiting recombinant chromosomes to the total number of mice analyzed for each pair of loci and the most likely gene order are: centromere–Mas1–0/164–Chd1–3/129–Nkx2–5. The genetic distances (in cM ± SEM) are: [Mas1, Chd1]–2.3 ± 1.3–Nkx2–5. No recombinants were detected between Mas1 and Chd1 in 164 animals typed in common, suggesting thar the two loci are within 1.8 cM of each other (upper 95% confidence interval). A 6.8-kb BamHI M. spretus RFLP (see Materials and Methods) was used to follow the segregation of the Atp5a1 locus in backcrossed mice. Atp5a1 mapped to the distal region of mouse chromosome 18, linked to Dcc and Mbp. The ratios of the total number of mice exhibiting recombinant chromosomes to the total number of mice analyzed for each pair of loci and the most likely gene order are: centromere–Dcc–8/169–Atp5a1–4/133–Mbp. The corresponding distances (cM ± SEM) are: –Dcc–4.7 ± 1.6–Atp5a1–3.0 ± 1.5–Mbp.
Human and murine map data for CHD1 and ATP5A1 thus show that none of these genes are sex-linked in mammals. In Fig. 3 we have summarized mammalian map locations for other genes assigned to the chicken Z chromosome. Presently, there are six such genes, all of which have been mapped in humans, six in cattle, and five in mice. As yet, there is only one example of a chicken Z chromosome gene, ornithine transcarbamylase (OTC), that is sex-linked in mammals (humans, cattle). Other Z chromosome genes are spread over different mammalian autosomes, e.g., in the case of humans, on five autosomes (CHD1 and ATP5A1 included). The largest conserved syntenic group contains three genes, ALDOB, GGTB2, and IREB1, which are all on HSA9, MMU4, and BTA8, respectively. The genetic distance of this conserved group in chickens is 36 cM.
Dating Avian Sex Chromosome Evolution.
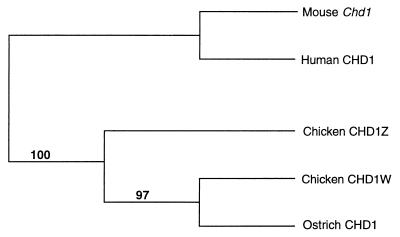
As mentioned before, neither the CHD1 or the ATP5A1 gene shows any evidence of sex linkage in ratites, i.e., the ostrich and its allies (26–27, 30). Given that the divergence of ratites may represent the deepest branch among extant bird lineages, a possible explanation for the failure to detect sex linkage of the CHD1 and ATP5A1 genes in ratites is that the sex chromosomes of non-ratite birds began to differentiate after the split of the ratites. To address this, we sequenced part of the single CHD1 gene found in ostriches and constructed a phylogenetic gene tree based on this sequence, chicken CHD1W and CHD1Z sequences, with human and murine CHD1 as an outgroup. Surprisingly, the ostrich CHD1 gene clustered with chicken CHD1W, suggesting that the chicken CHD1Z branched off before the split of the former two genes (Fig. 4). The topology of the tree has strong bootstrap support.
Figure 4.
A tree depicting the phylogenetic relationship among chicken CHD1Z, chicken CHD1W, ostrich, human, and murine CHD1 gene. Figures indicate bootstrap support for branches (1,000 replicates). The total tree length was 242 steps, the consistency index = 0.781, the retention index = 0.701, and the homeoplasy index = 0.219.
DISCUSSION
The mammalian X and Y chromosomes share several homologous regions. There are two pseudoautosomal segments, the largest at the ends of Xp and Yp (32) and a second and smaller region at the Xq and Yq telomeres (33). In addition, there are at least ten different regions spread over both arms of the X and Y chromosomes that are homologous, but not involved, in chromosome pairing at meiosis, thus being truly sex-linked (9). It is reasonable to see these regions of homology as vestigial remnants of an ancestral pair of autosomes, and this is indeed a commonly held view. Our data show now that a similar relationship between the sex chromosomes of birds might exist. Two genes (ATP5A1W and CHD1W) from the chicken W chromosome, so far the only genes that have been mapped to this gene-poor chromosome, are both present in copies also on the Z chromosome. The regions of homology are two parts of the Z chromosome, one close to the Zp telomere and one near the border of distal heterochromatin on Zq, and either one or both ends of the small W chromosome (Fig. 3). Because both ATP5A1 and CHD1 have been found to be W-linked as well as being present in another genomic copy in numerous species throughout avian phylogeny (26, 27, 30), it appears likely that the observed Z and W chromosome homology in chickens is characteristic of non-ratite birds in general. This is corroborated by the fact that all other chicken Z chromosome genes, which also have been mapped in other bird species, are consistently Z-linked (34–36).
Several other observations support a homology of the avian Z and W chromosomes. First, and importantly, the terminal, nonrepetitive part of one of the W chromosome arms pairs with the terminal part of chromosome Zp during pachytene and diplotene of female meiosis (37–40). Because pairing is associated with an obligate chiasmata, the region should be regarded as pseudoautosomal and must contain homologous, but as yet uncharacterized, sequences. Second, anonymous but sex-linked genomic clones from both chickens and geese have been found to cross-hybridize between the Z and W chromosome (41, 42). Third, by using starch gel electrophoresis of muscular creatine kinase (CKMM) in a hawk species, Morizot et al. (43) found a preliminary association of enzyme phenotypes and sex, interpreted as one gene copy on the W chromosome and one on the Z chromosome. Although this observation has to be confirmed by more detailed genetic studies, if correct, it may represent yet another homology between the region common to the avian Z and W chromosomes and mammalian autosomes; CKMM is on HSA19q and on MMU7, respectively.
The gene content of the eutherian X chromosome is conserved essentially in toto, as predicted by Ohno (2). The q arm of the eutherian X is also conserved in marsupials (dasyurids and kangaroos) and monotremes (platypus and echidna), and comparative gene-mapping data suggest that the marsupial X chromosome represents that of an early mammalian ancestor, which in the monotreme and eutherian lineages subsequently gained different chromosomal segments through translocations with autosomes (44–47). Hence, large parts of the mammalian sex chromosomes have an ancient origin (monotremes diverged from other mammals approximately 150 million years ago). Independently of these observations, Ohno (2) raised the provocative idea that the X–Y and Z–W sex chromosomes of mammals and birds would in fact be derived from the same pair of autosomes in an ancestral vertebrate. However, our data (Fig. 3) suggest that the evolution of sex chromosomes in mammals and birds represent independent events. Importantly, the autosomal location in mammals of the genes found to be contained within the region of Z and W chromosome homology indicates that the ancestral, protoavian autosome pair harboring these genes was not homologous to the protomammalian pair giving rise to X and Y chromosomes. Moreover, the eight genes now assigned to the chicken Z chromosome (Fig. 3) are found on six different human autosomal arms, clearly demonstrating the absence of significant homology to the X chromosome. One of these genes, OTC, is, however, X-linked in eutherian mammals. Although it cannot be formally excluded that its chromosomal location in the two taxa is indicative of common ancestry, the most plausible interpretation of this single observation would be that one or more chromosomal rearrangements during vertebrate evolution placed the gene on the chromosomes that subsequently evolved into the sex chromosomes of the respective taxa. This is further suggested by the fact that OTC is autosomal in marsupials and monotremes (14).
The conservative nature of the avian Z chromosome resembles that of the mammalian X chromosome. As mentioned above, however, ratites probably constitute an exception. The divergence of ratites from other birds constitute one of the deepest branches among extant bird lineages, possibly the very deepest (48, 49). Most ratite species, such as ostriches, emus, and kiwis, do not possess clearly heteromorphic sex chromosomes: the size of the Z and W chromosome differs only slightly, and they show strong banding homology and are both euchromatic, in contrast to the situation for most non-ratite birds (50). Hybridization with CHD1 (26, 27) and ATP5A1 (30) probes does not reveal sex-specific RFLP patterns in ostriches. There are at least two possible explanations for this situation. One is that the sex chromosomes of ratites and non-ratite birds originate from different pairs of autosomes. This would be compatible with ratites branching off before all other extant avian orders diverged and would imply that the sex chromosomes of non-ratite birds started to differentiate after the split of ratites, i.e., about 60–100 million years ago (48, 51, 52). Alternatively, the sex chromosomes of all birds could be derived from the same ancestral pair of protoavian autosomes. If this were the case, the full sex chromosomal differentiation leading to independent evolution of the two Z- and W-linked genes analyzed in this study must still have occurred after the split of ratites from other birds. An important consequence of these two scenarios is that we would expect all CHD1W genes to be more closely related to all CHD1Z genes than to any ratite CHD1 gene. However, a phylogenetic analysis placed the ostrich CHD1 gene together with chicken CHD1W, with chicken CHD1Z on a more distant node (Fig. 4). How can this be explained? One possibility is that the avian sex chromosomes started to differentiate close in time to the basal radiation of major extant bird lineages. If the sequence, ratite split–sex chromosome differentiation–basal radiation of other lineages, occurred within a limited evolutionary period, a phylogenetic analysis may fail to derive the correct topology of the CHD1 gene tree. This would be because CHD1W evolves at a much slower tempo than CHD1Z, an effect of the male-biased mutation rate demonstrated for birds (28). Indeed, simulations show that topological errors in parsimony trees may be hard to avoid when branch lengths are short and the rate of evolution differs significantly between lineages (P. Pamilo, personal communication). Also, results of several genetic studies have suggested that the basal radiation of major extant avian lineages has the character of a star phylogeny (48, 49). The conclusion from this would be that the avian sex chromosomes evolved at approximately the Cretaceous-Tertiary boundary, approximately 60–100 million years ago. Chromosomal sex determination would in such a case have a more recent origin among birds than among mammals (46–47).
In summary, we asked in this study whether the avian sex chromosomes share a common ancestry and whether this ancestry is also common to the mammalian sex chromosomes. The answers appear to be yes and no, respectively. Genes and DNA sequences present on the W chromosome of chickens and other birds can also be found on the Z chromosome, although not in a pseudoautosomal fashion. The homology involves two segments at the terminal part of Zp and at the border of the distal heterochromatin of Zq, respectively. Genes from these regions are not sex-linked in mammals, as is also not the case for the majority of genes so far mapped to other parts of the Z chromosome. The evolution of the avian sex chromosomes from a pair of autosomes, possibly dating from 60–100 million years ago, would thus seem to represent an independent event in vertebrate genome evolution.
Acknowledgments
We thank Debra J. Gilbert for technical assistance, Robert Perry and René St.-Arnaud for providing clones, and Pekka Pamilo for helpful discussion. This research was supported, in part, by the National Cancer Institute, Department of Health and Human Services, under contract with Advanced BioScience Laboratories. H.E. is sponsored by the Swedish Research Councils for Natural Sciences and for Agriculture and Forestry.
ABBREVIATIONS
- CSD
chromosomal sex determination
- RFLP
restriction fragment length polymorphism
- FISH
fluorescent in situ hybridization
- cM
centimorgan
Footnotes
References
- 1.Bull J. Evolution of Sex Determining Mechanisms. Menlo Park, CA: Benjamin/Cummings; 1983. [Google Scholar]
- 2.Ohno S. In: Monographs on Endocrinology. Labhart A, Mann T, Samuels L T, editors. Vol. 1. New York: Springer; 1967. [Google Scholar]
- 3.Charlesworth B. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- 4.Charlesworth B. Curr Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- 5.Jablonka E, Lamb M J. Biol Rev. 1990;65:249–276. doi: 10.1111/j.1469-185x.1990.tb01426.x. [DOI] [PubMed] [Google Scholar]
- 6.Rice W R. Bioscience. 1996;46:331–343. [Google Scholar]
- 7.Muller H J. J Exp Zool. 1914;17:325–336. [Google Scholar]
- 8.Watson J M, Wilcox S A, Graves J A M. In: Sex Chromosomes and Sex-Determining Genes. Reed K C, Graves J A M, editors. Bern, Switzerland: Harwood Academic; 1993. [Google Scholar]
- 9.Affara N A, Ferguson-Smith M A. In: Molecular Genetics of Sex Determination. Wachtel S S, editor. New York: Academic; 1994. [Google Scholar]
- 10.Jones K W, Singh L. Trends Genet. 1985;1:55–61. [Google Scholar]
- 11.Tone M, Nakano N, Takao E, Narisawa S, Mizuno S. Chromosoma. 1982;86:551–569. doi: 10.1007/BF00330126. [DOI] [PubMed] [Google Scholar]
- 12.Saitoh Y, Saitoh H, Ohtomo K, Mizuno S. Chromosoma. 1991;202:32–40. doi: 10.1007/BF00360684. [DOI] [PubMed] [Google Scholar]
- 13.Graves J A M, Watson J M. Chromosoma. 1991;101:63–68. doi: 10.1007/BF00357055. [DOI] [PubMed] [Google Scholar]
- 14.Graves J A M, Foster J W. Int Rev Cytol. 1994;154:191–259. doi: 10.1016/s0074-7696(08)62200-7. [DOI] [PubMed] [Google Scholar]
- 15.Crittenden L B, Provencher L, Santangelo I, Levin H, Abplanalp R W, Briles W E, Dodgson J B. Poult Sci. 1993;72:334–348. [Google Scholar]
- 16.Manly K F. Mamm Genome. 1993;4:303–313. doi: 10.1007/BF00357089. [DOI] [PubMed] [Google Scholar]
- 17.Lander E S, Green P, Abrahamson J, Barlow A, Daly M J, Lincoln S E, Newbury L. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 18.Cheng H H, Levin I, Vallejo R L, Khatib H, Dodgson J B, Crittenden B, Hillel J. Poult Sci. 1995;74:1855–1874. doi: 10.3382/ps.0741855. [DOI] [PubMed] [Google Scholar]
- 19.Copeland N G, Jenkins N A. Trends Genet. 1992;7:113–118. doi: 10.1016/0168-9525(91)90455-y. [DOI] [PubMed] [Google Scholar]
- 20.Jenkins N A, Copeland N G, Taylor B A, Lee B K. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delmas V, Stokes D G, Perry R P. Proc Natl Acad Sci USA. 1993;90:2414–2418. doi: 10.1073/pnas.90.6.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yotov W V, St-Arnaud R. Biochem Biophys Res Commun. 1993;191:142–148. doi: 10.1006/bbrc.1993.1195. [DOI] [PubMed] [Google Scholar]
- 23.Himmelbauer H, Harvey R P, Copeland N G, Jenkins N A, Silver L M. Mamm Genome. 1994;5:814–816. doi: 10.1007/BF00292022. [DOI] [PubMed] [Google Scholar]
- 24.Justice M J, Gilbert D J, Kinzler K W, Vogelstein B, Buchberg A M, Ceci J D, Matsuda Y, Chapman V M, Patriotis C, Makris A, et al. Genomics. 1992;13:1281–1288. doi: 10.1016/0888-7543(92)90047-v. [DOI] [PubMed] [Google Scholar]
- 25.Christidis L. Cytogenet Cell Genet. 1983;36:641–648. doi: 10.1159/000131988. [DOI] [PubMed] [Google Scholar]
- 26.Griffiths R, Daan S, Dijkstra C. Proc R Soc Lond Ser B. 1996;263:1251–1256. doi: 10.1098/rspb.1996.0184. [DOI] [PubMed] [Google Scholar]
- 27.Ellegren H. Proc R Soc Lond Ser B. 1996;263:1635–1641. doi: 10.1098/rspb.1996.0239. [DOI] [PubMed] [Google Scholar]
- 28.Ellegren H, Fridolfsson A-K. Nat Genet. 1997;17:182–184. doi: 10.1038/ng1097-182. [DOI] [PubMed] [Google Scholar]
- 29.Saitoh Y, Ogawa A, Hori T, Kunita R, Mizuno S. Chromosome Res. 1993;1:293–251. doi: 10.1007/BF00710129. [DOI] [PubMed] [Google Scholar]
- 30.Dvorak J, Halverson J L, Gulick P, Rauen K A, Abbott U K, Kelly B J, Schultz F T. J Hered. 1992;83:22–25. [Google Scholar]
- 31.Woodage T, Basrai M A, Baxevanis A D, Hieter P, Collins F S. Proc Natl Acad Sci USA. 1997;94:11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellis N A, Goodfellow P N. Trends Genet. 1989;5:406–410. doi: 10.1016/0168-9525(89)90199-6. [DOI] [PubMed] [Google Scholar]
- 33.Freije D, Helms C, Watson M S, Donis-Keller H. Science. 1992;258:1784–1787. doi: 10.1126/science.1465614. [DOI] [PubMed] [Google Scholar]
- 34.Baverstock P R, Adams M, Polkinghorne R W, Gelder M. Nature (London) 1982;296:763–766. doi: 10.1038/296763a0. [DOI] [PubMed] [Google Scholar]
- 35.Lacson J M, Morizot D C. Cytogenet Cell Genet. 1988;48:244–245. doi: 10.1159/000132638. [DOI] [PubMed] [Google Scholar]
- 36.Saitoh Y, Ogawa A, Hori T, Kunita R, Mizuno S. Chromosome Res. 1993;1:293–251. doi: 10.1007/BF00710129. [DOI] [PubMed] [Google Scholar]
- 37.Solari A J, Fechheimer N S, Bitgood J J. Cytogenet Cell Genet. 1988;48:130–136. doi: 10.1159/000132609. [DOI] [PubMed] [Google Scholar]
- 38.Solovei I, Gaginskaya E, Hutchison N, Macgregor H. Chromosome Res. 1993;1:153–166. doi: 10.1007/BF00710769. [DOI] [PubMed] [Google Scholar]
- 39.Solari A J, Dresser M E. Chromosome Res. 1995;3:87–93. doi: 10.1007/BF00710668. [DOI] [PubMed] [Google Scholar]
- 40.Hori T, Suzuki Y, Solovei I, Saitho Y, Hutchison N, Ikeda J-E, Macgregor H, Mizuno S. Chromosome Res. 1996;4:411–426. doi: 10.1007/BF02265048. [DOI] [PubMed] [Google Scholar]
- 41.Ogawa A, Solovei I, Hutchison N, Saitoh Y, Ikeda J-E, Macgregor H, Mizuno S. Chromosome Res. 1997;5:93–101. doi: 10.1023/a:1018461906913. [DOI] [PubMed] [Google Scholar]
- 42.Quinn T W, Cooke F, White B N. Auk. 1990;107:199–202. [Google Scholar]
- 43.Morizot D C, Bednarz J C, Ferrell R E. Cytogenet Cell Genet. 1987;44:89–91. [Google Scholar]
- 44.Spencer J A, Watson J M, Graves J A M. Genomics. 1991;9:598–604. doi: 10.1016/0888-7543(91)90352-f. [DOI] [PubMed] [Google Scholar]
- 45.Spencer J A, Sinclair A H, Watson J M, Graves J A M. Genomics. 1991;11:339–345. doi: 10.1016/0888-7543(91)90141-z. [DOI] [PubMed] [Google Scholar]
- 46.Watson J M, Spencer J A, Riggs A D, Graves J A M. Proc Natl Acad Sci USA. 1990;87:7125–7129. doi: 10.1073/pnas.87.18.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watson J M, Spencer J A, Riggs A D, Graves J A M. Proc Natl Acad Sci USA. 1991;88:11256–11260. doi: 10.1073/pnas.88.24.11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sibley C G, Ahlquist J E. Phylogeny and Classification of Birds. A Study in Molecular Evolution. New Haven, CT: Yale Univ. Press; 1990. [Google Scholar]
- 49.Mindell D P, Sorenson M D, Huddleston C J, Miranda H, Knight A, Sawchuk S J, Yuri T. In: Avian Molecular Evolution and Systematics. Mindell D P, editor. New York: Academic; 1997. [Google Scholar]
- 50.Ansari H A, Takagi N, Sasaki M. Cytogenet Cell Genet. 1988;47:185–188. doi: 10.1159/000132290. [DOI] [PubMed] [Google Scholar]
- 51.Hedges S B, Parker P H, Sibley C G, Kumar S. Nature (London) 1996;381:226–229. doi: 10.1038/381226a0. [DOI] [PubMed] [Google Scholar]
- 52.Harild A, Janke A, Arnason U. Mol Biol Evol. 1997;14:754–761. doi: 10.1093/oxfordjournals.molbev.a025815. [DOI] [PubMed] [Google Scholar]