Abstract
Loss of heterozygosity on chromosome 11q23 is observed at high frequency in human nonsmall cell lung carcinomas (NSCLCs), suggesting the presence of a tumor suppressor gene. Previous analysis of DNA from 79 patients identified a commonly deleted segment of 5 centimorgans. Complementation analysis was used to further localize a putative tumor suppressor gene. Three yeast artificial chromosome (YAC) clones spanning the minimal loss of heterozygosity region were modified, and spheroplast fusion was used to transfer them into human A549 NSCLC or murine Lewis lung carcinoma (LLC) cell lines. The resulting yeast × human hybrid cell lines containing an intact copy of a 1.6-Mb YAC, 939b12, showed reduced growth in vitro. Injection of parental A549 cells into athymic (nu/nu) mice resulted in tumor formation at 27 of 28 injection sites. In contrast, two independent 939b12-containing cell lines formed tumors at only 3 of 20 injection sites. 939b12 also suppressed tumor formation by LLC NSCLC cells in nude mice, but YACs 785e12 and 911f2, which flank 939b12, had no suppressor activity. Further localization of tumor suppression activity on 939b12 was accomplished by introduction of defined fragmentation derivatives into A549 cells and by analysis of YACs that were broken on transfer into LLC cells. This complementation approach localized tumor suppression activity to the central 700 kb of 939b12 and provides a functional assay for positional cloning of this tumor suppressor gene.
Lung cancer is one of the most common human malignancies. These cancers are divided by their biological features into two distinct categories: small cell lung carcinoma and nonsmall cell lung carcinoma (NSCLC) (1). The multiple genetic alterations resulting in NSCLC are not well understood, and this remains an aggressive cancer with a poor prognosis. Mutations of the KRAS and NRAS genes and inactivation of the p53 and RB genes have been demonstrated in a large number of NSCLCs (2–4). These changes seem likely to be secondary to the initiation of tumor formation. Loss of heterozygosity (LOH) on chromosome 3p (Chr 3p) and Chr 11 has been reported in NSCLC, suggesting the presence of additional tumor suppressor genes in these regions (5–7).
We reported previously that LOH in the chromosomal region 11q23 is observed frequently in human NSCLC. Analysis of 79 NSCLCs identified a 5-cM region that is commonly deleted (8). The presence of a tumor suppressor gene(s) on Chr 11q in NSCLCs was also demonstrated by using a functional assay. A549, a human NSCLC-derived cell line, shows complete loss of one copy of Chr 11. When a Chr 11 from a non-NSCLC cell line is introduced into A549 cells by microcell-mediated chromosome transfer, the transformed phenotype is lost as measured by several growth parameters and suppression of tumorigenicity in nude mice (9). Although this assay clearly implicates Chr 11, it does not provide for the precise subchromosomal localization of the tumor suppressor gene(s).
Procedures that introduce large segments of DNA into cells can be used to localize genes for which a functional assay is available (10). In particular, spheroplast fusion provides a method for the transfer of intact yeast artificial chromosomes (YACs) into cultured cells (11). Genes on transferred YACs are expressed under the control of their normal promoters. Because the minimal region of LOH on 11q23 in NSCLCs is completely spanned by a YAC contig (12), we transferred candidate YACs of up to 1.6 Mb and their defined fragmentation derivatives into A549 cells to further localize the region containing a gene(s) that will suppress A549 tumorigenicity in nude mice.
MATERIALS AND METHODS
Cells and Yeast Strains.
A human lung adenocarcinoma cell line, A549, and the murine Lewis lung carcinoma (LLC) line were obtained from Riken Cell Bank (Tsukuba, Japan) and grown in DMEM (A549) or Eagle’s minimal essential medium (LLC) supplemented with 10% fetal bovine serum, penicillin, and streptomycin (GIBCO/BRL) (13). Human-derived YACs from Chr 11 cloned in the pYAC4 vector and strain AB1380 were described (12). YPH925 (MATα, ura3, trp1, cyhr, leu2, HIS5, his3, kar1Δ15) was kindly provided by F. Spencer (Johns Hopkins School of Medicine, Baltimore). YAC manipulation was accomplished by using established procedures for Kar transfer (14), YAC fragmentation (15), and spheroplast fusion (16).
Tumorigenicity in Nude Mice.
For each inoculation, a suspension of 1 × 105 cells in 0.2 ml PBS was injected s.c. into one to four sites on the flanks of 5- to 6-week-old female BALB/c athymic nu/nu mice (Charles River Breeding Laboratories). Mice were monitored at least twice a week. Tumors were resected, and DNA was extracted from each tumor. Cell clones were judged nontumorigenic if no tumor was seen by 10 weeks after injection for A549 clones or 5 weeks for the more aggressive LLC clones. All animal experiments were performed in accordance with institutional guidelines.
DNA Marker Analysis.
DNA was extracted from ca. 107 cultured cells by proteinase K–phenol–chloroform extraction and from yeast containing YACs by using a glass bead–phenol procedure (17). PCR primer sequences for DNA markers on Chr 11 were obtained from the Genome Data Base (http://www.gdb.org). PCR with 32P-labeled primers was performed by using standard procedures (18). PCR products were separated on denaturing gels containing 7% acrylamide and 34% urea and autoradiographed at −80°C for 4–12 h on Hyperfilm-MP (Amersham) with an intensifying screen.
RESULTS
Cloning the Minimal Region of LOH.
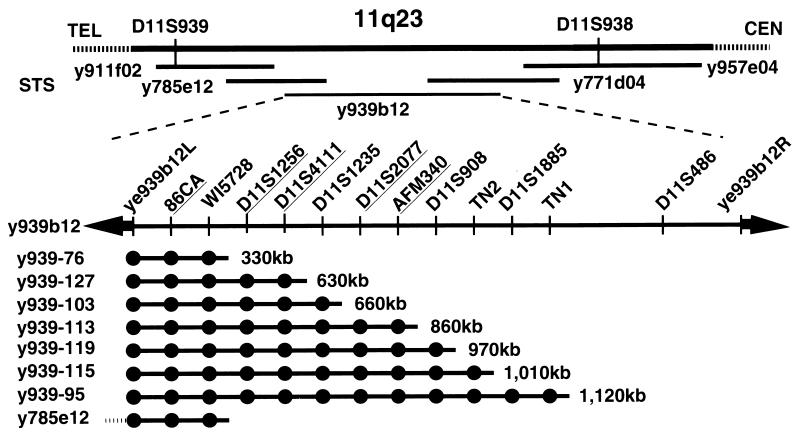
A region of LOH was identified originally in 25 of 79 patients with NSCLC at 11q23.2–23.3, across the 5-cM region between D11S939 and D11S938 (8) (Fig. 1). A YAC contig established by Arai et al. (12) includes five clones spanning this segment. Three YACs, 911f02 (780 kb), 785e12 (600 kb), and 939b12 (1.6 Mb) were selected initially for analysis.
Figure 1.
Physical mapping of the minimal NSCLC region of LOH on 11q23. Five YACs span the LOH region defined by D11S939 and D11S938 (12). Orientation relative to the Chr 11q centromere (CEN) and telomere (TEL) is indicated. The central 1,600-kb YAC, y939b12, was typed for 14 markers, including the vector-insert junction fragments, and subjected to fragmentation analysis; nine marker clusters (including the fixed positions of the vector-junction fragments) were identified. Three markers mapping within 330 kb of the centric end of YAC 939b12 were found on the YAC 785e12, which extends further toward the Chr 11q telomere; only the overlapping portion is diagrammed here. Five markers that are polymorphic between the insert of y939b12 and A549 are underlined. All deletion derivatives are products of acentric fragmentation and thus retain the centric YAC vector arm (adjacent to junction fragment ye939b12L).
YAC 939b12 was transferred from yeast strain AB1380 to strain YPH925 by kar transfer to provide a wider range of selectable markers for YAC manipulation (14). To order additional markers in the minimal LOH region, acentric YAC fragmentation with the pBCL1 vector (19) was used to create a series of nested deletion derivatives (15). pBCL1 targets recombination to Alu repeat elements spaced randomly along the YAC, introducing a LYS2-selectable marker and a telomere at the site of recombination. The telomere provides a new YAC end at the site of recombination, eliminating the URA3 marker in the acentric YAC vector arm. Deletion derivatives are selected for growth on media lacking lysine and screened for auxotrophy for uracil.
Twenty-four derivatives ranging in size from 200 to 1,600 kb were typed for the presence or absence of 12 markers. Including the vector-insert junction clones, ye939b12L and ye939b12R, this defines nine marker groups on YAC 939b12 and accurately measures the physical size of each segment (Fig. 1). Five of the 14 markers were found to be polymorphic between the DNA in the 939b12 YAC insert and A549 and, thus, were useful for assessing YAC integrity in yeast ×human hybrid (ybrid) cells.
YAC Modification and Introduction into A549 Cells.
The integrating plasmid, pDC47 (16), was used to introduce a mammalian selectable marker conferring resistance to G418 and a yeast HIS3 gene into YACs 939b12, 785e12, and 911f02. The neo-containing 939b12 YAC was introduced into A549 cells by spheroplast fusion. Spheroplast fusion is the only procedure that can be used to routinely introduce an intact segment of this size (1.6 Mb) into cultured cells. Furthermore, when this procedure is performed so as to preclude the lysis of spheroplasts during sequential washing, it results in far less breakage of the YAC than other methods of YAC transfer (see Discussion). This was an important consideration in these experiments because only five markers (plus the vector arm sequences) were available that would indicate the presence of the 1.6 Mb of human DNA from 939b12 on the A549 (human) background.
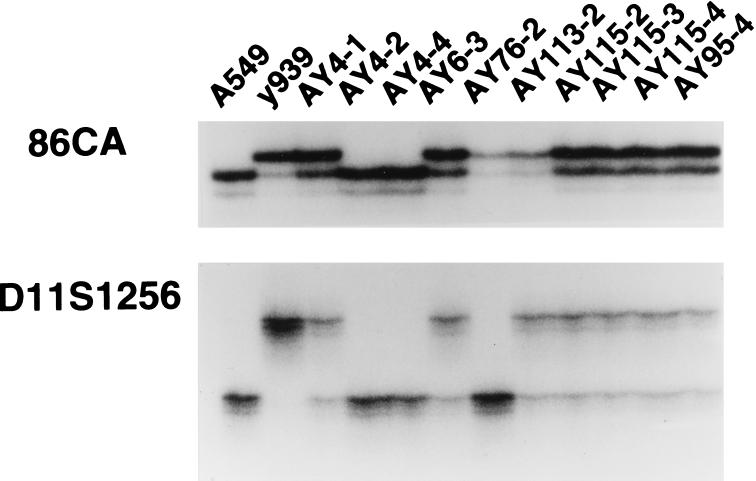
Four G418-resistant ybrids were selected from spheroplast fusions between y939b12 and A549. The frequency of ybrid formation was lower than we have observed previously in fusions between yeast spheroplasts and mouse cells (11). To provide additional cell lines for biological analysis, fusions were also performed to introduce 939b12, 785e12, and 911f02 into mouse LLC cells. All ybrids were typed for the presence or absence of YAC vector arms. In addition, A549 ybrid lines were analyzed for five polymorphic markers and the LLC cell ybrids were analyzed for 12 human DNA markers (Fig. 2).
Figure 2.
Assessment of YACs in ybrid cell lines carrying full-length or fragmented YACs. PCR reactions were for 86CA and D11S1256, two of five markers that were found to be polymorphic between the A549 (human) cell line and human DNA cloned in YAC y939b12 (y939).
Introduction of 939b12 Reduces Growth of A549 Cells.
Ybrid cells from line AY4-1 (A549 containing 939b12) and control A549neor cells were trypsinized, washed, and counted. Duplicate 6 × 35 mm2 well plates were made with 104, 5 × 104, or 105 cells per well (Table 1). Twenty-four hours after plating, all wells in one plate were trypsinized and counted. Similar numbers of cells were seen in both the ybrid and control lines at 24 h, indicating that the plating efficiency was similar for each line. Each well from the second plate was trypsinized and counted after 5 days to determine whether the two lines grew at the same rate. AY4-1 ybrid cells demonstrated little or no increase in cell number, whereas control cells showed a 5- to 10-fold expansion. The same pattern was seen in two additional experiments, in which the reduced rate of AY4-1 growth was evident (by 2-fold or more) as early as 48 h after plating (data not shown).
Table 1.
Growth of control A549neo cells relative to ybrid cell line AY4-1, which contains the full length YAC 939b12
| Cell line | Time after plating, h | No. of cells plated (based on initial cell counts)*
|
||
|---|---|---|---|---|
| 104 | 5 × 104 | 105 | ||
| AY4-1 | 24 | 0.75 × 104 | 3.25 × 104 | 7.75 × 104 |
| A549neo | 24 | 1 × 104 | 4.75 × 104 | 5.75 × 104 |
| AY4-1 | 120 | 1.25 × 104 | 3 × 104 | 8 × 104 |
| A549neo | 120 | 1.73 × 105 | 2.35 × 105 | 3.45 × 105 |
*Average numger of cells per well is given for the identical time.
Introduction of 939b12 Reduces Tumorigenicity of A549 and LLC Cells.
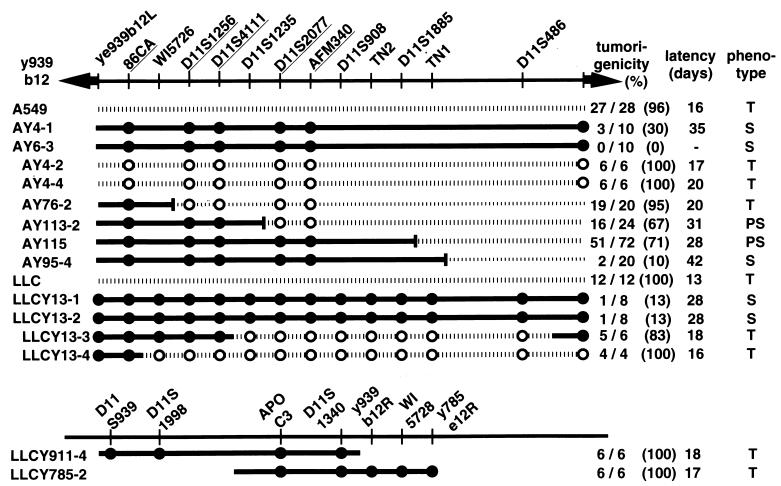
Two independently derived 939b12 ybrid lines made with A549 and two lines made with LLC were tested for tumorigenicity relative to the parental cell lines. Tumorigenicity was determined as formation of a tumor ≤35 days after injection of 105 cells into athymic, nude mice. A549 cells produced tumors at 27 of 28 sites in recipient mice with an average latency of 16 days (Fig. 3). In contrast, ybrid AY4-1 containing the full-length YAC 939b12, formed tumors in only three of 10 mice with extended latency (latency = 21, 35, and 50 days, average = 35 days), and a second A549 ybrid, AY6-3, formed no tumors at 10 injection sites. These lines contained the centric and acentric YAC vector arms and all five y939-specific alleles, suggesting that the entire YAC was present in each case.
Figure 3.
Localization of tumor suppressor activity on YAC 939b12 demonstrates that the distal 1.1 Mb is sufficient to suppress tumorigenesis of A549 or LLC cells injected in nu/nu mice. Number and percentage of sites forming tumors and the average latency are shown for parental lines A549 and LLC, for two independent ybrids with each parental line containing full-length YAC 939b12 (i.e., AY4-1, AY6-3, LLCY13-1 and LLCY13-2), and for ybrids containing fragmented or broken YACs retained in either parental cell line. Data for AY115 represents the aggregate tumor incidence from three independently derived ybrids containing the 1,010-kb fragmentation derivative, all of which showed ca. 70% tumor incidence with prolonged latency. The name of the ybrid line includes the name of ybrids made with fragmented YACs includes the name of the fragmentation derivative, e.g., AY115 results from a fusion with fragmentation derivative y939-115 (Fig. 1). Ybrids containing the overlapping YACs 911f02 or y785e12 showed no suppression, forming tumors at all injection sites. Phenotype designations are: T, tumorigenic; S, suppressed; PS, partial suppression of tumorigenesis. •, Markers present in hybrid cells; ○, markers absent in hybrid cells.
LLC cell-derived ybrids provided a more thorough assessment of the incorporated YAC because 14 markers could be used. As in the A549 tumorigenesis experiments, the parental LLC cell line showed no suppression, with tumors forming at an aggregate 12 of 12 injection sites after only 13 d (Fig. 3). The presence of the 939b12 YAC in ybrids LLCY13-1 and 13-2 resulted in suppression, with only two tumors appearing from 16 injection sites by 35 da after injection. Tumorigenicity was also assessed for LLC cell ybrids containing intact copies of the flanking YACs, 911f02 and 785e12. No suppression was seen in either case, with tumors forming at 6 of 6 injection sites for each line. Because YAC 785e12 overlaps the centric end of 939b12 by ca. 200 kb, all or part of the tumor suppressor gene must be contained in the remaining 1.4 Mb.
Fragmentation Further Localizes the Tumor Suppressor Gene.
Six ybrid lines were constructed between A549 and four 939b12 fragmentation derivatives ranging from 330 to 1,120 kb. Ybrid AY76-2, containing the 330-kb fragmentation derivative, y939-76, showed no suppression, forming tumors at 19 of 20 sites (Fig. 3). In contrast, ybrid AY95-4, containing the 1,120-kb derivative y939-95, showed strong suppression with only two tumors at 20 sites 42 d after injection.
One ybrid line containing an 860-kb derivative (y939-113) and three with a 1,010-kb fragmented YAC (y939-115) showed partial suppression, with tumors appearing at ca. 70% of injection sites with a longer latency than the parental cell line. Partial suppression could arise if the fragmentation breakpoint interferes with expression of the tumor suppressor gene, if expression of genes on a YAC is subject to a position effect because of the integration site of the YAC, or both factors could be important. In this regard, it is worth noting that the integration site had no inhibitory effect on tumor suppression in the five independent ybrid lines containing either the 1.1- or 1.6-Mb (full length) versions of YAC 939b12. Three independent ybrid lines (i.e., three integration sites) containing the 1,010-kb deletion derivative y939-115 all had the ca. 30% reduction in tumor formation reflected in the aggregate 51 of 72 sites (Fig. 3).
Ybrid LLCY13-3 formed between full-length 939b12 and LLC retained both vector arms and markers occurring within ca. 400 kb of the centric YAC arm but lost the eight remaining markers. This ybrid formed tumors at five of six sites with short latency. Taken together, the A549 and LLC ybrid data exclude all or part of the gene responsible for tumor suppression from the centric 400 kb of the YAC and completely exclude the gene from the distal (acentric) 500 kb of the 1.6-Mb YAC. The 1.1-Mb fragmentation derivative in ybrid AY95-4 gives full suppression; this suppression is substantially reduced but not eliminated by deletion of an additional 90 kb, as shown by three independent AY115 ybrids containing a 1,010-kb derivative of y939b12. Thus, sequences that contribute significantly to the tumor suppressor activity fall into this small region, but additional information that contributes to this phenotype cannot be excluded from the central 700 kb of the 1.6-Mb parental YAC.
DISCUSSION
A functional cloning/complementation approach was used to localize tumor suppressor activity to a 700-kb cloned region within a 5-cM interval of LOH on Chr 11q23. Because heritable forms of this disease have not been reported, conventional linkage analysis to identify relevant genes is not possible. However, assessment of LOH in sporadic tumors provides a useful approach to the identification of genes that are important in the initiation and progression of cancers that are affected by tumor suppressor genes.
Chr 11 in particular was a target for LOH studies, because in vitro experiments demonstrated reversal of transformed phenotypes when a normal Chr 11 was introduced into A549 cells through microcell-mediated chromosome transfer (9). Chr 11 also demonstrates tumor suppressor activity when introduced into cell lines derived from cervical, prostate, or breast cancers or rhabdomyosarcomas (20–23). Chromosome transfer methods can be combined with this type of biological assay to further refine the positions of candidate genes when donor cells are irradiated to create fragments of chromosomes before fusion. This approach was utilized to localize regions harboring prostate tumor suppressor genes to segments of 28 cM on Chr 17q and 17 cM on Chr 10p (18, 24). For NSCLC, the combined analysis by using microcell-mediated hybrids and LOH (8) identified a candidate region of <5 cM on Chr 11. Further assessment of these patients has reduced the minimal LOH region to 2 cM (Y.S.M., unpublished data), which is completely concordant with the tumor suppression results reported here. This resolution is quite good for LOH studies but presents an area that is very large for conventional positional cloning approaches.
The availability of an in vitro complementation assay by using A549 cells suggested that a functional cloning strategy could be utilized for positional cloning (10). YACs spanning the region of LOH provided the substrate for this approach (12). Spheroplast fusion was used to introduce candidate human-derived YACs into (human) A549 cells because it is the only procedure that can routinely transfer YACs larger than approximately 600 kb without breakage. When spheroplasting is performed so as to preclude the lysis of yeast, this procedure results in a very low rate of YAC breakage, presumably because fusion between the yeast and mammalian cell membranes results in the introduction of the intact yeast nucleus to the cytoplasm. When vigorous conditions for spheroplasting are used in which many lysed cells (“ghosts”) are visible, the number of partial YACs recovered increases. This may be because of liberation of yeast nuclei that fuse to mammalian cells, introducing genomic DNA directly into the cytoplasm without the protection of nuclear packaging. In this study, fewer hybrids were observed after fusion with A549 human cells than with mouse cell lines that we have used previously, but spheroplast fusion clearly can be performed successfully with human cell lines. YAC sizes from 200 to 1,600 kb had no apparent effect on fusion frequency.
Chr 11q contains multiple genes that are implicated in a variety of neoplastic diseases. In addition to the common minimal region of deletion in NSCLC, a number of independent LOH studies have shown deletions, including 11q22–23 in breast, cervical, bladder, and ovarian cancers and malignant melanomas and neuroblastomas; individual reports exist for testicular germ cell tumors, squamous cell carcinomas of head and neck, and pancreatic carcinomas (for example, see refs. 25–34). The observation of Chr 11q deletions in multiple types of tumors raises the question of how many tumor suppressor genes exist in this region. LOH at the MEN1 locus on Chr 11q13, 20–30 cM proximal to the NSCLC locus characterized here, is seen in parathyroid tumors (35). Koreth et al. (26) reported two distinct regions, at 11q22-q23.1 and 11q25-qter, that are frequently lost in sporadic breast cancers but not colorectal cancers, and two regions of LOH, at 11q13.3–22 and 11q22–24, are reported for a group of 52 nasopharyngeal carcinomas (36). Although different studies vary in the degree to which commonly deleted regions are characterized and reported, the occurrence of unidentified tumor suppressor genes in addition to the NSCLC gene characterized here is quite feasible. The frequent occurrence of LOH for 11q22–23 in various tumors also raises the possibility that one or more genes in this region may be involved in a pathway of tumor progression common to a variety of neoplasias with different initiating events. Cloning of the gene responsible for the activity described here will provide information crucial to the determination of the types and frequency of predisposing alleles that may contribute to this range of diseases.
We used YAC complementation to identify a region of no more than 700 kb on Chr 11q23 that harbors all or part of a tumor suppressor gene important in NSCLC. Results of a functional assay by using YAC complementation were completely concordant with fine-scale LOH analysis. The coincident results of two different approaches strongly suggest that a tumor suppressor gene lies within the region. Ongoing conversion of the 939b12 YAC to bacterial artificial chromosomes carrying a mammalian selectable marker, which can be used directly in further complementation assays, will localize the gene to a segment amenable to genomic sequencing, providing substrate for a comprehensive search of candidate genes in the region.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Cancer Research (7D-1) and for the 2nd Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health and Welfare of Japan; a grant for Research on Human Genome and Gene Therapy from the Ministry of Health and Welfare of Japan; a grant-in-aid for Special Project for Cancer Research from the Ministry of Education, Science, Sports and Culture of Japan; the Program for Promotion of Fundamental Studies in Health and Sciences of the Organization for Drug ADR Relief, R & D Promotion and Product Review of Japan; and Public Health Service awards HD24605 and HG00405 (to R.H.R.). T.N. and K.T. are the recipients of a Research Resident Fellowship from the Foundation for Promotion of Cancer Research, Japan. P.F. and P.H. are students in the Predoctoral Training Program in Human Genetics (GM07814).
ABBREVIATIONS
- NSCLC
nonsmall cell lung carcinoma
- LOH
loss of heterozygosity
- Chr
chromosome
- YAC
yeast artificial chromosome
- LLC
Lewis lung carcinoma line
- hybrid
yeast ×human hybrid
References
- 1.Ihde D C, Minna J D. Curr Probl Cancer. 1991;15:105–154. doi: 10.1016/0147-0272(91)90012-y. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki Y, Orita M, Shiraishi M, Hayashi K, Sekiya T. Oncogene. 1990;5:1037–1043. [PubMed] [Google Scholar]
- 3.Kishimoto Y, Murakami Y, Shiraishi M, Hayashi K, Sekiya T. Cancer Res. 1992;52:4799–4804. [PubMed] [Google Scholar]
- 4.Sachse R, Murakami Y, Shiraishi M, Hayashi K, Sekiya T. Oncogene. 1994;9:39–47. [PubMed] [Google Scholar]
- 5.Kok K, Osinga J, Carritt B, Davis M B, van der Hout A H, van der Veen A Y, Landsvater R M, de Leij L F, Berendsen H H, Postmus P E, et al. Nature (London) 1987;330:578–581. doi: 10.1038/330578a0. [DOI] [PubMed] [Google Scholar]
- 6.Weston A, Willey J C, Modali R, Sugimura H, McDowell E M, Resau J, Light B, Haugen A, Mann D L, Trump B F, et al. Proc Natl Acad Sci USA. 1989;86:5099–6103. doi: 10.1073/pnas.86.13.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiraishi M, Morinaga S, Noguchi M, Shimosato Y, Sekiya T. Jpn J Cancer Res. 1987;78:1302–1308. [PubMed] [Google Scholar]
- 8.Iizuka M, Sugiyama Y, Shiraishi M, Jones C, Sekiya T. Genes Chromosomes Cancer. 1995;13:40–46. doi: 10.1002/gcc.2870130107. [DOI] [PubMed] [Google Scholar]
- 9.Satoh H, Lamb P, Dong J-T, Everitt J, Boreiko C, Oshimura M, Barrett J. Mol Carcinogen. 1993;7:157–164. doi: 10.1002/mc.2940070306. [DOI] [PubMed] [Google Scholar]
- 10.Gu J Z, Carstea E D, Cummings C, Morris J A, Loftus S K, Zhang D, Coleman K G, Cooney A M, Comly M E, Fandino L, et al. Proc Natl Acad Sci USA. 1997;94:7378–7383. doi: 10.1073/pnas.94.14.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavan W J, Hieter P, Reeves R H. Mol Cell Biol. 1990;10:4163–4169. doi: 10.1128/mcb.10.8.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arai Y, Hosoda F, Nakayama K, Ohki M. Genomics. 1996;35:196–206. doi: 10.1006/geno.1996.0339. [DOI] [PubMed] [Google Scholar]
- 13.Giard D J, Aaronson S A, Todaro G J, Arnstein P, Kersey J H, Dosik H, Parks W P. J Natl Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 14.Spencer F, Hugerat Y, Simchen G, Hurko O, Connelly C, Hieter P. Genomics. 1994;22:118–126. doi: 10.1006/geno.1994.1352. [DOI] [PubMed] [Google Scholar]
- 15.Pavan W J, Hieter P, Reeves R H. Proc Natl Acad Sci USA. 1990;87:1300–1304. doi: 10.1073/pnas.87.4.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reeves R H, Cabin D E, Lamb B. In: Current Protocols in Human Genetics. Dracopoli N C, Haines J L, Korf B R, Moir D T, Morton C C, Seidman C E, Seidman J G, Smith D R, editors. Unit 5.12. Brooklyn, NY: Current Protocols; 1995. [Google Scholar]
- 17.Hoffman C S, Winston F. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 18.Murakami Y S, Brothman A R, Leach R J, White R L. Cancer Res. 1995;55:3389–3394. [PubMed] [Google Scholar]
- 19.Lewis B, Shah N, Braun B, Denny C. Genome Analysis Trends and Applications. 1992;9:86–90. doi: 10.1016/1050-3862(92)90003-n. [DOI] [PubMed] [Google Scholar]
- 20.Phillips K K, Welch D R, Miele M E, Lee J H, Wei L L, Weissman B E. Cancer Res. 1996;56:1222–1227. [PubMed] [Google Scholar]
- 21.Rinker-Schaeffer C W, Hawkins A L, Ru N, Dong J, Stoica G, Griffin C A, Ichikawa T, Barrett J C, Isaacs J T. Cancer Res. 1994;54:6249–6256. [PubMed] [Google Scholar]
- 22.Negrini M, Castagnoli A, Sabbioni S, Recanatini E, Giovannini G, Possati L, Stanbridge E J, Nenci I, Barbanti-Brodano G. Oncogene. 1992;7:2013–2018. [PubMed] [Google Scholar]
- 23.Oshimura M, Kugoh H, Koi M, Shimizu M, Yamada H, Satoh H, Barrett J C. J Cell Biochem. 1990;42:135–142. doi: 10.1002/jcb.240420304. [DOI] [PubMed] [Google Scholar]
- 24.Murakami Y S, Albertsen H, Brothman A R, Leach R J, White R L. Cancer Res. 1996;56:2157–2160. [PubMed] [Google Scholar]
- 25.Kerangueven F, Noguchi T, Coulier F, Allione F, Wargniez V, Simony-Lafontaine J, Longy M, Jacquemier J, Sobol H, Eisinger F, et al. Cancer Res. 1997;57:5469–5474. [PubMed] [Google Scholar]
- 26.Koreth J, Bakkenist C J, McGee J O. Oncogene. 1997;14:431–437. doi: 10.1038/sj.onc.1200847. [DOI] [PubMed] [Google Scholar]
- 27.Kersemaekers A M, Hermans J, Fleuren G J, van de Vijver M J. Br J Cancer. 1998;77:192–200. doi: 10.1038/bjc.1998.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomlinson I P, Bodmer W F. J Clin Pathol. 1996;49:386–390. doi: 10.1136/jcp.49.5.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosin M P, Cairns P, Epstein J I, Schoenberg M P, Sidransky D. Cancer Res. 1995;55:5213–5216. [PubMed] [Google Scholar]
- 30.Shaw M E, Knowles M A. Genes Chromosomes Cancer. 1995;13:1–8. doi: 10.1002/gcc.2870130102. [DOI] [PubMed] [Google Scholar]
- 31.Bethwaite P B, Koreth J, Herrington C S, McGee J O. Br J Cancer. 1995;71:814–818. doi: 10.1038/bjc.1995.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foulkes W D, Campbell I G, Stamp G W, Trowsdale J. Br J Cancer. 1993;67:268–273. doi: 10.1038/bjc.1993.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomlinson I P, Beck N E, Bodmer W F. Eur J Cancer. 1996;32A:1797–1802. doi: 10.1016/0959-8049(96)00198-0. [DOI] [PubMed] [Google Scholar]
- 34.Takita J, Hayashi Y, Kohno T, Shiseki M, Yamaguchi N, Hanada R, Yamamoto K, Yokota J. Oncogene. 1995;11:1829–1834. [PubMed] [Google Scholar]
- 35.Farnebo F, Teh B T, Dotzenrath C, Wassif W S, Svensson A, White I, Betz R, Goretzki P, Sandelin K, Farnebo L O, et al. Hum Genet. 1997;99:342–349. doi: 10.1007/s004390050369. [DOI] [PubMed] [Google Scholar]
- 36.Hui A B, Lo K W, Leung S F, Choi P H, Fong Y, Lee J C, Huang D P. Cancer Res. 1996;56:3225–3229. [PubMed] [Google Scholar]