Abstract
In the fission yeast Schizosaccharomyces pombe, S phase is limited to a single round per cell cycle through cyclin-dependent kinase phosphorylation of critical replication factors, including the Cdc18 replication initiator protein. Because defects in Cdc18 phosphorylation lead to a hyperstable and hyperactive form of Cdc18 that promotes high levels of overreplication in vivo, we wished to identify the components of the Cdc18 proteolysis pathway in fission yeast. In this paper we describe one such component, encoded by the sud1+ gene. sud1+ shares homology with the budding yeast CDC4 gene and is required to prevent spontaneous re-replication in fission yeast. Cells lacking sud1+ accumulate high levels of Cdc18 and the CDK inhibitor Rum1, because they cannot degrade these two key cell cycle regulators. Through genetic analysis we show that hyperaccumulation of Rum1 contributes to re-replication in Δsud1 cells, but is not the cause of the defect in Cdc18 proteolysis. Rather, Sud1 itself is associated with the ubiquitin pathway in fission yeast and binds to Cdc18 in vivo. Most importantly, Sud1-Cdc18 binding requires prior phosphorylation of the Cdc18 polypeptide at CDK consensus sites. These results provide a biochemical mechanism for the phosphorylation-dependent degradation of Cdc18 and other cell cycle regulators, including Rum1. Evolutionary conservation of the Sud1/CDC4 pathway suggests that phosphorylation-coupled proteolysis may be a general feature of nearly all eukaryotic cell cycles.
The timely and rapid destruction of regulatory proteins is one of the major themes in cell cycle control. In particular, the degradation of cyclins and cyclin-dependent kinase (CDK) inhibitors plays an important role in controlling progression through S phase and mitosis (reviewed in ref. 1). Studies in yeast, Xenopus oocytes, and mammalian cells indicate that two distinct proteolytic pathways are important for these cell-cycle events. Degradation of mitotic cyclins at the metaphase-anaphase transition depends on a large ubiquitin-protein ligase (E3) complex, termed the cyclosome or anaphase-promoting complex. Anaphase-promoting complex activity is cell cycle-regulated and uses as its substrate recognition signal a conserved “mitotic destruction box” that is present both in mitotic cyclins and in various noncyclin substrates (1). By contrast, ubiquitination and degradation of G1 cyclins and other G1/S substrates appears to regulated by the phosphorylation state of the substrate protein. For example, phosphorylation of the budding yeast G1 cyclin CLN2 and S phase CDK inhibitor SIC1 targets these proteins for rapid degradation by another putative E3 complex, containing the CDC4, CDC34, and CDC53 proteins (2–6).
In fission yeast, genetic analysis has provided insight into the cell cycle controls that prevent excess DNA replication (7, 8). To date, three broad classes of overreplication mutants have been described. The first group comprises mutants in which the activity of mitotic cyclin/CDK complexes (Cdc13/Cdc2 in fission yeast) has been quantitatively eliminated, for example through depletion of the mitotic cyclin Cdc13, or overproduction of the mitotic cyclin/CDK inhibitor Rum1 (9–13). The second group of overreplication mutants consists of those expressing high levels of Cdc18, a key replication initiator protein (6, 14, 15). Interestingly, both Cdc18 and Rum1 are highly unstable proteins that normally accumulate during the G1 phase of the cell cycle and then are rapidly degraded (13, 14). A third group of overreplication mutants includes the recently described pop1 mutant (16). Interestingly, fission yeast cells lacking pop1+ were found to express higher than normal levels of Rum1 and Cdc18, although the precise mechanism of this effect (e.g., increased transcription of cdc18+, increased translation of cdc18+ mRNAs, or reduced degradation of the Cdc18 polypeptide) remains unknown. Taken together, these data suggest that rapid proteolysis of Cdc18 and/or Rum1 might be important in limiting DNA replication to once per cell cycle.
Work in our laboratory has elucidated several genetic and biochemical connections between Cdc18 and CDK activity in fission yeast. (i) Previously we found that increased levels of the Rum1 CDK inhibitor reversed the DNA replication and cell cycle defects of a temperature-sensitive cdc18 mutant and promoted the stable accumulation of Cdc18 protein during Rum1-induced overreplication (12). (ii) We subsequently reported that fission yeast Cdc18 protein copurifies with S phase and mitotic cyclin/CDK complexes and is a substrate for these kinases in vitro (17). (iii) We have recently demonstrated that CDK-dependent phosphorylation of Cdc18 directly inhibits its replication activity and promotes its rapid destruction in vivo (6).
In this paper we provide direct evidence that proteolysis of Cdc18 requires the ubiquitin-proteasome pathway. We also describe one of the cellular factors responsible for the phosphorylation-dependent degradation of Cdc18, which we have named sud1+ (stops unwanted diploidization). sud1+ is phylogenetically related to the budding yeast CDC4 gene and the fission yeast pop1+ gene (16). As its name suggests, cells lacking sud1+ undergo spontaneous re-replication of the genome, resulting in the accumulation of diploid and tetraploid cells. At a molecular level, Δsud1 mutants are unable to degrade Cdc18 and Rum1 rapidly and accumulate high levels of both cell-cycle proteins. Genetic analysis indicates that these proteolysis defects are independent of one another and suggests that excessive accumulation of Rum1 in particular plays a major role in spontaneous diploidization. Coimmunoprecipitation experiments demonstrate that Sud1 physically associates both with the ubiquitin pathway and with Cdc18, one of its physiologic substrates. Most strikingly, Sud1 binding requires CDK phosphorylation of Cdc18, thus explaining the phosphorylation dependence of Cdc18 degradation.
MATERIALS AND METHODS
Genetic Methods.
Protocols for growth and genetic manipulation of fission yeast have been described (18). The sud1+ gene was identified in a tblastn search with the Saccharomyces cerevisiae CDC4 protein sequence against the S. pombe genome sequence database (http://www.sanger.ac.uk/Projects/S_pombe/blast_server.shtml). The sud1+ gene physically maps to chromosome I, cosmid c4D7. To delete sud1+ by homologous recombination, sequences 5′ and 3′ to the sud1+ ORF were separately amplified by PCR, digested with XhoI/BglII and BglII/NotI, respectively, and ligated to XhoI/NotI-digested pBluescript. This plasmid was linearized with BglII, treated with calf intestinal alkaline phosphatase, and ligated with a BglII fragment containing the his3+ gene (19, 20). An ApaI/NotI fragment of the final disruption construct was transformed into an appropriate diploid strain (h+/h− his3-D1/his3-D1 leu1–32/leu1–32 ura4-D18/ura4-D18 ade6-M210/ade6-M216). Stable His+ Ade+ transformants were isolated and sporulated, and tetrads dissected. Tetrads yielding a monogenic segregation pattern of 2 His+: 2 His− colonies were then analyzed by Southern blotting to verify proper disruption of sud1+. Because Δsud1 mutants were sterile, genetic crosses were performed by using a Δsud1 strain that harbors a sud1+/ura4+ plasmid. The resulting progeny strains were then tested on Edinburgh minimal medium minus uracil plates to confirm meiotic loss of the plasmid. Cellular DNA content was measured by propidium iodide staining of ethanol-fixed cells and flow cytometry (6).
nmt1 Promoter Shutoff Experiments.
Acute repression of genes under the control of the thiamine-repressible nmt1 promoter was achieved by adding thiamine (10 μg/ml final) to cells grown overnight in thiamine-free Edinburgh minimal medium. In some experiments, cycloheximide (100 μg/ml final) was added at the same time to inhibit ongoing protein synthesis (6). Samples were harvested at the indicated intervals and quick-frozen at −70°C.
RNA and Protein Analysis.
Procedures for RNA isolation and Northern blot analysis have been described (12). To extract protein from S. pombe, two methods were used. For samples to be subjected only to SDS/PAGE, 108 cells were lysed with glass beads directly in SDS loading buffer. For soluble native extracts, about 109 cells were lysed in ice-cold HB buffer (18), and the supernatant harvested after centrifugation at 15,000 × g for 30 minutes at 4°C. Subsequent immunoprecipitation and immunoblotting procedures were identical to those already described (6, 12). Anti-tubulin mAb B512 and polyclonal anti-ubiquitin antibodies were obtained from Sigma. Anti-HA mAb 12CA5 and anti-MYC mAb 9E10 were purchased from Babco. 9E10 mAb-horseradish peroxidase conjugate was purchased from Boehringer Mannheim. Anti-Rum1 antiserum was a generous gift of Sergio Moreno (Universidad de Salamanca) and was affinity purified before use.
RESULTS
Destruction of the Cdc18 Replication Initiator Protein is Mediated by the Ubiquitin-Proteasome Pathway.
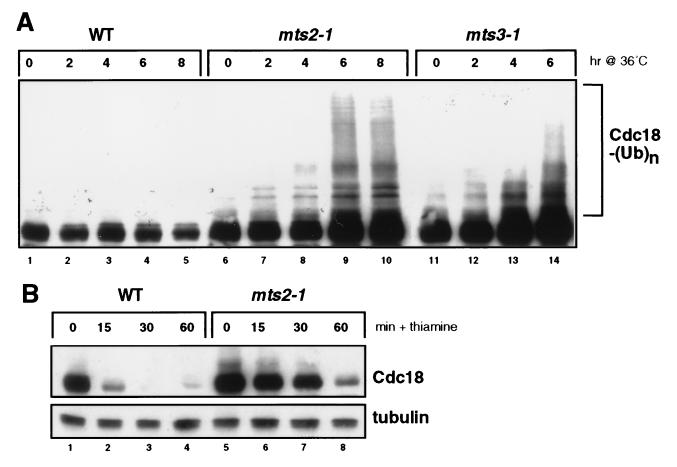
To address the mechanism of Cdc18 proteolysis, we examined the role of proteasome function in this process. In fission yeast, the mts2+ and mts3+ gene products have been identified as subunits of the 26S proteasome, which is responsible for the ubiquitin-dependent degradation of a variety of cellular proteins in eukaryotic cells (21, 22). We used temperature-sensitive mutations in mts2+ and mts3+ to inactivate proteasome function conditionally. Increased expression of Cdc18 in mts3–1 (but not mts2–1) mutants has been reported (16), although it is not clear whether this reflects an increased half-life of the Cdc18 polypeptide, or merely accumulation of cells in a portion of the cell cycle where cdc18+ is highly transcribed. To circumvent these limitations in our experiments, Cdc18 was expressed constitutively from an attenuated version of the thiamine-repressible nmt1 promoter. We found that Cdc18 readily accumulated in both mts2–1 and mts3–1 strains at 36°C (Fig. 1A). We also observed a ladder of higher molecular weight species, consistent with the accumulation of Cdc18-multiubiquitin adducts in the absence of proteasome function. To demonstrate directly that Cdc18 proteolysis requires the proteasome, we acutely repressed Cdc18 synthesis from the nmt1 promoter with thiamine and monitored protein turnover by immunoblotting (Fig. 1B). As expected, Cdc18 is rapidly destroyed in wild-type cells, with a half-life of about 5 min (6, 14). By contrast, Cdc18 was degraded very slowly in mts2–1 and mts3–1 mutant cells, persisting for up to 1 hr after shutoff (Fig. 1B). Taken together, our data indicate that Cdc18 proteolysis requires the ubiquitin-proteasome pathway.
Figure 1.
Degradation of the Cdc18 replication initiator protein is mediated by the ubiquitin-proteasome pathway in fission yeast. (A) A hemagglutinin (HA) epitope-tagged form of Cdc18 was expressed from the weak thiamine-repressible nmt1 promoter (REP41X) in a wild-type strain (lanes 1–5) or in two temperature-sensitive (ts) proteasome mutants (mts2–1, lanes 6–10; mts3–1, lanes 11–14). Logarithmically growing cells were shifted from 25°C to 36°C, and the abundance and electrophoretic mobility of Cdc18 assessed by immunoblotting with anti-HA mAb 12CA5. The position of the Cdc18-multiubiquitin adducts is indicated. (B) The half-life of Cdc18 in wild-type and mts2–1 strains was measured by shifting cells to 36°C for 3 hr (to inactivate the mutant proteasomes) and then adding thiamine to acutely repress de novo expression from the nmt1 promoter. Cdc18 degradation was monitored by immunoblotting with 12CA5 mAb. The same membrane was blotted with anti-tubulin mAb B512 to confirm equal loading.
Identification of sud1+, a CDC4-Related Gene in Fission Yeast.
We next examined a number of S. pombe mutants for their effects on Cdc18 degradation. We observed that the anaphase-promoting complex was probably not responsible for the bulk of Cdc18 proteolysis, as the half-life of Cdc18 in nuc2–663 and cut9–665 mutants was similar to that in wild-type (data not shown). This result confirms that the stabilization of Cdc18 in proteasome-defective cells (Fig. 1) was not an indirect effect of cell cycle synchronization, as mts2–1, mts3–1, nuc2–663, and cut9–669 mutants all arrest at the metaphase-anaphase transition with high levels of mitotic CDK activity (21–24). To identify cellular factors participating in Cdc18 proteolysis, we considered the fact that CDK phosphorylation of Cdc18 plays a major role in targeting the polypeptide for destruction (6). Because degradation of the cell-cycle phosphoproteins CLN2 and SIC1 is mediated by the CDC4, CDC34, and CDC53 gene products in budding yeast, we searched for fission yeast homologs as potential Cdc18 proteolysis factors. Systematic sequencing of the fission yeast genome has uncovered a gene related to CDC4, which we have named sud1+. The sud1+ gene encodes a polypeptide of 704 amino acids that is 32% identical to the budding yeast CDC4 protein and 35% identical to the pop1+ gene product. The maximum homology between these proteins is located within the C-terminal WD-40 repeats, while the N-terminal regions are much less well conserved. The N terminus of Sud1 contains a recognizable F-box sequence motif (LPFSIVQSILLNLDIHSFLSCRLVSPTWNRILDVHTSYWKHM; amino acids 242–283). This motif appears to mediate physical interaction of CDC4 and other F-box proteins with SKP1 (25).
sud1+ Stops Unwanted Diploidization.
To analyze the function of sud1+, we constructed a fission yeast strain in which the sud1+ gene was deleted, having been replaced by the his3+ prototrophic marker. After sporulation of a heterozygous Δsud1/sud1+ diploid strain, we readily recovered tetrads in which two of the colonies were His+ and two were His−, indicating that sud1+ is not essential for viability. However, the Δsud1 cells were frequently larger than wild-type cells and gave rise to darkly staining colonies on media containing the vital dye phloxin B. Because similar attributes are observed with diploid fission yeast cells, we examined the genomic DNA content of Δsud1 cells by flow cytometry. Interestingly, the Δsud1 mutant frequently carried out an extra round of DNA replication without mitosis, resulting in the accumulation of cells with diploid (4C) DNA content (Fig. 2A). We conclude that the normal function of sud1+ is to suppress unwanted diploidization, thus ensuring stable maintenance of the haploid state during vegetative growth. This may account in part for the fact that deletion of sud1+ also results in sterility (data not shown).
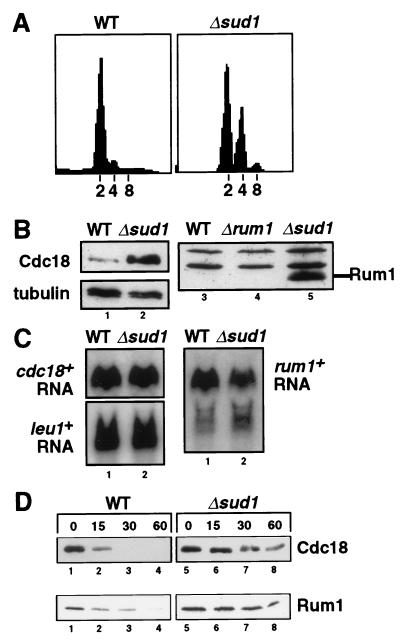
Figure 2.
sud1+ stops unwanted diploidization and is required for rapid degradation of both Cdc18 and the fission yeast CDK inhibitor Rum1. (A) DNA content of wild-type and Δsud1 cells was analyzed by propidium iodide staining and flow cytometry. The positions of normal haploid (2C), diploid (4C), and tetraploid (8C) DNA contents are indicated. (B) The abundance of Cdc18 and Rum1 proteins in wild-type, Δsud1, and Δrum1 strains was measured by immunoblotting with specific antibodies. (C) Northern blot analysis of cdc18+ and rum1+ mRNA transcripts in wild-type (lane 1) and Δsud1 cells (lane 2). The leu1+ mRNA was used as a loading control. PhosphorImager quantitation demonstrated that cdc18+ and rum1+ mRNAs were equally abundant in both strains. (D) Increased half-life of Rum1 and Cdc18 proteins in Δsud1 strains. HA-tagged forms of Cdc18 (Upper) and Rum1 (Lower) were expressed under the control of the weak nmt1 promoter in either wild-type cells (lanes 1–4) or Δsud1 cells (lanes 5–8). Degradation of Cdc18 and Rum1 was monitored after acutely repressing de novo synthesis with thiamine and cycloheximide.
Rapid Proteolysis of Cdc18 and Rum1 Requires sud1+.
To determine the molecular basis of this re-replication phenotype, we examined the levels of several important cell-cycle regulatory proteins in Δsud1 cells by immunoblotting. We found that both the Rum1 CDK inhibitor and Cdc18 replication initiator proteins accumulated to abnormally high levels in the Δsud1 mutant strain (Fig. 2B). Interestingly, the increased abundance of Cdc18 and Rum1 was not a consequence of increased gene transcription or mRNA stability, as the abundance of cdc18+ and rum1+ transcripts was the same in Δsud1 and wild-type strains (Fig. 2C).
Formally, the accumulation of Cdc18 and Rum1 in Δsud1 cells could reflect either increased translation of the corresponding mRNAs or decreased degradation of the two polypeptides. To distinguish these two possibilities, HA epitope-tagged forms of cdc18+ and rum1+ were expressed from the weak nmt1 promoter, and the turnover of Cdc18 and Rum1 was monitored after adding thiamine and cycloheximide simultaneously to repress transcription and de novo translation (Fig. 2D). While Cdc18 and Rum1 were degraded quickly in wild-type cells, both proteins persisted in Δsud1 cells for more than one hour after shutoff. These data demonstrate that sud1+ is required for the rapid proteolysis of both Cdc18 and Rum1 proteins in vivo.
Because increased Rum1 expression itself can increase Cdc18 abundance (12), it was of interest to determine if Rum1 was required for the stable accumulation of Cdc18 in the Δsud1 mutant. A Δsud1 Δrum1 double mutant strain was constructed, and Cdc18 proteolysis examined as in the previous experiment. Again, Cdc18 proteolysis was just as defective in the Δsud1 Δrum1 mutant (Fig. 3A). We conclude that the requirement for sud1+ in Cdc18 degradation is not an indirect effect of increased Rum1 stability.
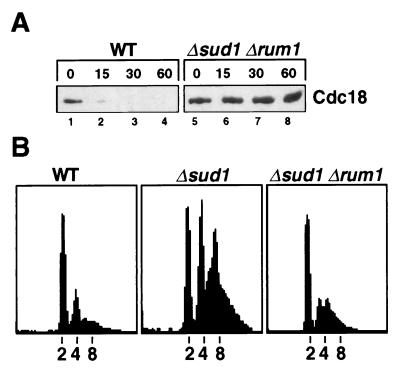
Figure 3.
Rum1 stabilization is required for re-replication of the genome in Δsud1 cells but does not cause the defect in rapid proteolysis of Cdc18. (A) Degradation of Cdc18 in wild-type and Δsud1 Δrum1 strains was measured as described above (see Materials and Methods). (B) Wild-type, Δsud1, and Δsud1 Δrum1 strains were grown overnight in minimal medium containing low nitrogen to induce re-replication. DNA content was analyzed by flow cytometry.
Rum1 Stabilization Plays a Major Role in the Δsud1 Re-Replication Phenotype.
The Δsud1 Δrum1 double mutant also allowed us to test the role of Rum1 stability in the re-replication phenotype caused by loss of sud1+. Wild-type, Δsud1, and Δsud1 Δrum1 strains were grown overnight in minimal medium containing low nitrogen (0.25% NH4Cl). This treatment produces high levels of re-replication in the Δsud1 strain, as shown by the appearance of 4C and tetraploid 8C cells (Fig. 3B). By contrast, the Δsud1 Δrum1 mutant strain largely retained the normal 2C DNA content. These data suggest that Rum1 stabilization plays a major role in the re-replication cycles of the Δsud1 mutant strain, although we have not excluded the possibility that increased Cdc18 abundance also contributes to the re-replication phenotype.
Characterization of the Sud1 Protein.
To address the biochemical role of Sud1 in Cdc18 and Rum1 proteolysis, we examined the properties of the Sud1 protein itself. For these studies, an HA-tagged form of Sud1 was expressed in fission yeast. This protein was shown to be functional, as it complemented the sterility and diploidization phenotypes of the Δsud1 mutant (data not shown) and was readily detected with anti-HA monoclonal antibody 12CA5.
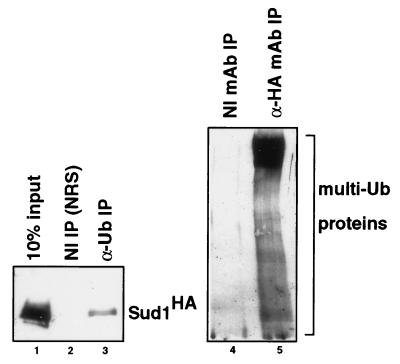
We performed immunoprecipitation experiments to determine if Sud1 was associated with ubiquitinated proteins in fission yeast. Extracts were incubated with antibodies to either ubiquitin or HA-Sud1, and the resulting immunoprecipitates immunoblotted with the reciprocal antibody. Interestingly, Sud1 was readily precipitated by ubiquitin-specific antibodies (Fig. 4 Left). Moreover, a ladder of large, ubiquitin-containing species was observed in HA-Sud1 immunoprecipitates, which represent cellular protein-multiubiquitin adducts (Fig. 4 Right). These data indicate that Sud1 associates with the ubiquitin pathway in fission yeast, most likely through interactions with cellular proteins that are undergoing multiubiquitination and degradation.
Figure 4.
Sud1 is physically associated with the ubiquitin pathway in fission yeast. (Left) Extracts containing HA-tagged Sud1 protein were immunoprecipitated with either control nonimmune rabbit serum (lane 2) or anti-ubiquitin antibodies (lane 3), separated by SDS/PAGE, and immunoblotted with anti-HA mAb 12CA5. One-tenth of the original extract was loaded on the same gel for comparison. (Right) Extracts containing HA-tagged Sud1 were immunoprecipitated with either control nonimmune mAb (lane 4) or HA-specific mAb 12CA5 (lane 5), and then immunoblotted with anti-ubiquitin antibodies. The position of high molecular weight protein-multiubiquitin adducts is indicated.
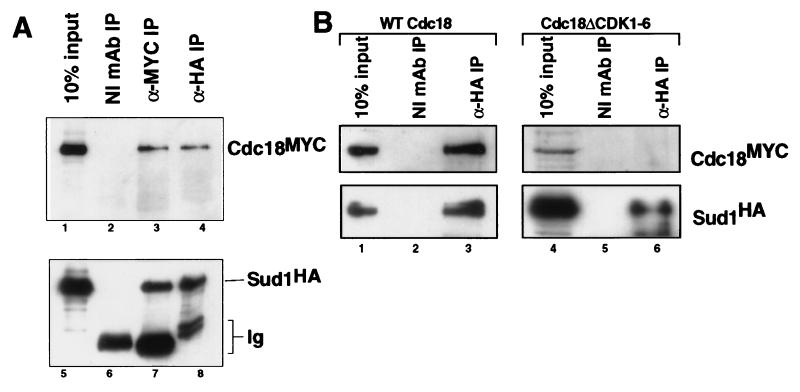
To test for associations with specific target proteins, we examined the ability of Sud1 to interact physically with Cdc18. Through coprecipitation studies, we found that Sud1 and Cdc18 bind one another in vivo (Fig. 5A). To determine the functional significance of this interaction, we examined the role of Cdc18 phosphorylation in the process. As we reported previously, mutation of the six CDK consensus sites in Cdc18 abolishes its in vivo phosphorylation pattern and renders it immune to rapid degradation (6). Interestingly, the Cdc18ΔCDK1–6 mutant protein is incapable of binding to Sud1, unlike wild-type Cdc18 (Fig. 5B). These data indicate that Sud1 binding requires phosphorylation of Cdc18 by fission yeast CDKs and is required for rapid proteolysis of Cdc18.
Figure 5.
Sud1 and Cdc18 bind one another in a phosphorylation-dependent manner. (A) Extracts derived from cells expressing HA epitope-tagged Sud1 and MYC epitope-tagged Cdc18 were immunoprecipitated with either control nonimmune mAb (lanes 2 and 6), anti-HA mAb 12CA5 (lanes 3 and 7), or anti-MYC mAb 9E10 (lanes 4 and 8). Immunoprecipitates and an aliquot of the input extract (lanes 1 and 5) were separated by SDS/PAGE and immunoblotted with either 9E10-horseradish peroxidase conjugate (9E10-HRP; Upper) or 12CA5 mAb (Lower) Ig, immunoglobulin. (B) HA-Sud1 was expressed together with either wild-type MYC-Cdc18 or MYC-Cdc18ΔCDK1–6. Extracts were immunoprecipitated with control nonimmune mAb (lanes 2 and 5) or 12CA5 mAb (lanes 3 and 6), then immunoblotted with 9E10-horseradish peroxidase conjugate (Upper) and 12CA5 mAb (Lower). Aliquots of the input extracts (lanes 1 and 4) were included on the same gel for comparison.
DISCUSSION
Degradation of cyclins and other regulatory proteins plays a major role in promoting cell-cycle progression in eukaryotic cells. As shown both in this paper and a recent report by Kominami and Toda (16), proteolysis can also play an important role in preventing reinitiation of DNA replication without mitosis, which otherwise would lead to abnormal increases in genome ploidy.
Recently we (6) and Benito et al. (26) demonstrated that phosphorylation of Cdc18 and Rum1 proteins by Cdc2 kinase (the major CDK in fission yeast) targets both substrates for rapid degradation. In this paper we verified that Cdc18 is degraded by the ubiquitin-proteasome pathway and have identified one of the cellular factors responsible for this process, which is encoded by the fission yeast sud1+ gene. Through genetic analysis we demonstrated that sud1+ stops unwanted diploidization and ensures stable maintenance of normal chromosomal DNA content in fission yeast. sud1+ is related to both S. cerevisiae CDC4 and S. pombe pop1+ and contains the F-box and WD-40 repeat motifs characteristic of this gene family (25). The Sud1 protein is required for rapid proteolysis of at least two important cell cycle regulators in fission yeast, namely the Cdc18 and Rum1 proteins (10, 27). Interestingly, we find that Sud1 interacts with ubiquitinated proteins in fission yeast and binds the Cdc18 polypeptide in a phosphorylation-dependent manner. These data suggest an attractive model for how CDK phosphorylation targets Cdc18 for destruction in vivo, namely by promoting the physical interaction of Cdc18 with Sud1 and other components of the ubiquitin-proteasome pathway. These additional components have not been identified yet in fission yeast but are likely to include homologs of the CDC34/UBC and CDC53/cullin protein families (25, 28, 29).
The sud1+ and pop1+ genes share many similarities in their biochemical and genetic properties. Cells lacking either gene undergo diploidization at increased frequency and accumulate increased levels of both Cdc18 and Rum1 polypeptides, demonstrating that both sud1+ and pop1+ are necessary to maintain these two critical cell-cycle regulators at physiological levels (16). Both Sud1 and Pop1 proteins bind Cdc18, and at least for Sud1 (but probably for Pop1 as well) this binding requires phosphorylation of the Cdc18 substrate at multiple CDK consensus sites. An important question is why fission yeast would have evolved two similar genes with apparently overlapping functions. One possibility is that while both Pop1 and Sud1 bind Cdc18 and target it for destruction, there may be other cellular phosphoproteins that are specifically targeted for degradation by Sud1 but not Pop1 (or vice versa). Alternatively, Pop1 and Sud1 may not have distinct substrate specificities, but may simply act additively to ensure rapid and quantitative degradation of target molecules.
Interestingly, degradation of CDC6 (the homolog of Cdc18 in budding yeast) also requires CDC4 (30, 31), although the role of CDK phosphorylation in this process has not yet been established. Given the evolutionary conservation of Sud1/Pop1/CDC4 homologs among not only budding and fission yeasts but also multicellular organisms (32–34), it seems likely that there will be additional important targets of this phosphorylation-coupled proteolysis pathway, particularly during developmental and neoplastic modulation of cell cycle progression in higher eukaryotes (32–34).
Acknowledgments
We thank Susan Forsburg and Kathy Gould for providing numerous S. pombe strains and plasmids, Sergio Moreno for Rum1 antiserum, Colin Gordon for mts2–1 and mts3–1 strains, and Tokashi Toda for exchanging sequence information before publication. We thank Stephen Desiderio and our colleagues in the Kelly laboratory for helpful discussions and comments on the manuscript. This work was supported by grants from the National Institutes of Health to T.J.K. and an M.D./Ph.D. Medical Scientist Training Program Award at the Johns Hopkins University School of Medicine to P.V.J.
ABBREVIATIONS
- CDK
cyclin-dependent kinase
- HA
hemagglutinin
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF064515).
References
- 1.King R W, Deshaies R J, Peters J-M, Kirschner M W. Science. 1996;274:1652–1658. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 2.Lanker S, Valdivieso M H, Wittenberg C. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 3.Willems A R, Lanker S, Patton E E, Craig K L, Nason T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 4.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 5.Feldman R M R, Correll C C, Kaplan K B, Deshaies R J. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 6.Jallepalli P V, Brown G W, Muzi-Falconi M, Tien D, Kelly T J. Genes Dev. 1997;11:2767–2779. doi: 10.1101/gad.11.21.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jallepalli P V, Kelly T J. Curr Opin Cell Biol. 1997;9:358–363. doi: 10.1016/s0955-0674(97)80008-7. [DOI] [PubMed] [Google Scholar]
- 8.Muzi-Falconi M, Brown G W, Kelly T J. Curr Biol. 1996;6:229–233. doi: 10.1016/s0960-9822(02)00464-5. [DOI] [PubMed] [Google Scholar]
- 9.Hayles J, Fisher D, Woollard A, Nurse P. Cell. 1994;78:813–822. doi: 10.1016/s0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 10.Moreno S, Nurse P. Nature (London) 1994;367:236–242. doi: 10.1038/367236a0. [DOI] [PubMed] [Google Scholar]
- 11.Martin-Castellanos C, Labib K, Moreno S. EMBO J. 1996;15:839–849. [PMC free article] [PubMed] [Google Scholar]
- 12.Jallepalli P V, Kelly T J. Genes Dev. 1996;10:541–552. doi: 10.1101/gad.10.5.541. [DOI] [PubMed] [Google Scholar]
- 13.Correa-Bordes J, Nurse P. Cell. 1995;83:1001–1009. doi: 10.1016/0092-8674(95)90215-5. [DOI] [PubMed] [Google Scholar]
- 14.Muzi-Falconi M, Brown G W, Kelly T J. Proc Natl Acad Sci USA. 1996;93:1566–1570. doi: 10.1073/pnas.93.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishitani H, Nurse P. Cell. 1995;83:397–495. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- 16.Kominami K, Toda T. Genes Dev. 1997;11:1548–1560. doi: 10.1101/gad.11.12.1548. [DOI] [PubMed] [Google Scholar]
- 17.Brown G W, Jallepalli P V, Huneycutt B J, Kelly T J. Proc Natl Acad Sci USA. 1997;94:6142–6147. doi: 10.1073/pnas.94.12.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreno S, Klar A, Nurse P. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 19.Ohi R, Feoktistova A, Gould K L. Gene. 1996;174:315–318. doi: 10.1016/0378-1119(96)00085-6. [DOI] [PubMed] [Google Scholar]
- 20.Burke J D, Gould K L. Mol Gen Genet. 1994;242:169–176. doi: 10.1007/BF00391010. [DOI] [PubMed] [Google Scholar]
- 21.Gordon C, McGurk G, Dillon P, Rosen C, Hastie N D. Nature (London) 1993;366:355–357. doi: 10.1038/366355a0. [DOI] [PubMed] [Google Scholar]
- 22.Gordon C, McGurk G, Wallace M, Hastie N D. J Biol Chem. 1996;271:5704–5711. doi: 10.1074/jbc.271.10.5704. [DOI] [PubMed] [Google Scholar]
- 23.Hirano T, Hiraoka Y, Yanagida M. J Cell Biol. 1988;106:1171–1183. doi: 10.1083/jcb.106.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samejima I, Yanagida M. J Cell Biol. 1994;127:1655–1670. doi: 10.1083/jcb.127.6.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goebl M G, Yochem J, Jentsch S, McGrath J P, Varshavsky A, Byers B. Science. 1988;241:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 26.Benito J, Martin-Castellanos C, Moreno S. EMBO J. 1998;17:482–497. doi: 10.1093/emboj/17.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly T J, Martin G S, Forsburg S L, Stephen R J, Russo A, Nurse P. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- 28.Kipreos E T, Lander L E, Wing J P, He W W, Hedgecock E M. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 29.Mathias N, Johnson S L, Winey M, Adams A E M, Goetsch L, Pringle J R, Byers B, Goebl M G. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drury L S, Perkins G, Diffley J F. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piatti S, Bohm T, Cocker J H, Diffley J F X, Nasmyth K. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 32.Kao C T, Lin M, O’Shea-Greenfield A, Weinstein J, Sakamoto K M. Oncogene. 1996;13:1221–1229. [PubMed] [Google Scholar]
- 33.Hubbard E J A, Wu G, Kitajewski J, Greenwald I. Genes Dev. 1997;11:3182–3193. doi: 10.1101/gad.11.23.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang J, Struhl G. Nature (London) 1998;391:493–498. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]