Abstract
Prokaryotic translational release factors, RF1 and RF2, catalyze polypeptide release at UAG/UAA and UGA/UAA stop codons, respectively. In this study, we isolated a bacterial RF2 mutant (RF2*) containing an E167K substitution that restored the growth of a temperature-sensitive RF1 strain of Escherichia coli and the viability of a chromosomal RF1/RF2 double knockout. In both in vivo and in vitro polypeptide termination assays, RF2* catalyzed UAG/UAA termination, as does RF1, as well as UGA termination, showing that RF2* acquired omnipotent release activity. This result suggests that the E167K mutation abolished the putative third-base discriminator function of RF2. These findings are interpreted as indicating that prokaryotic and eukaryotic release factors share the same anticodon moiety and that only one omnipotent release factor is sufficient for bacterial growth, similar to the eukaryotic single omnipotent factor.
Keywords: translation termination/stop codon recognition/discriminator/tRNA mimicry
The termination of protein synthesis takes place on the ribosomes as a response to a stop, rather than a sense, codon in the “decoding” site (A site). Translation termination generally requires two codon-specific polypeptide release factors (RFs), RF1 (for UAG/UAA) and RF2 (for UGA/UAA), in prokaryotes (1, 2) and one factor, eRF1 (omnipotent for the three stop codons), in eukaryotes (2–4) (Fig. 1A). However, the mechanism of stop codon recognition by release factors is unknown and represents a long-standing coding problem of considerable interest. It entails protein-RNA recognition rather than the well understood mRNA-tRNA interaction in codon-anticodon pairing (2, 5, 6).
Figure 1.
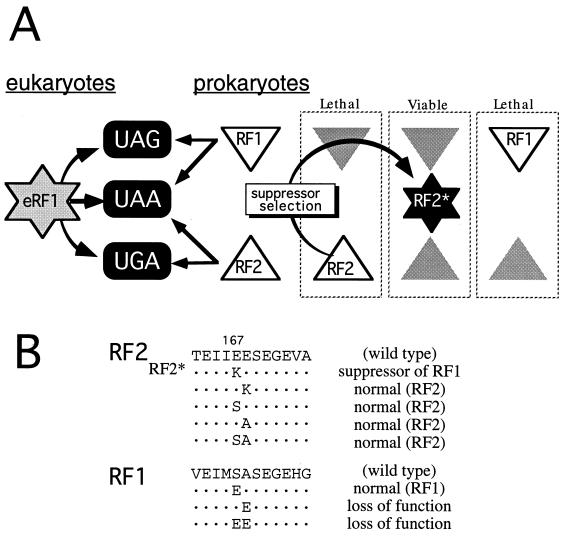
Bacterial RF2 mutant that substitutes for RF1 function. (A) Preference in stop codon recognition by RFs and rationale of RF2* selection. The plasmid-bearing RF2 gene was mutagenized in vitro, and the RF2* allele was isolated among temperature-resistant survivors of the Ts RF1 (prfA1) strain on transformation as described in Materials and Methods. Shaded triangles mean the loss of function by Ts or knockout mutations. It is assumed that RF1 and RF2 encode “second- and third-base discriminator” functions, respectively, and this paper provides genetic evidence for the existence of the third-base discriminator function in RF2. (B) Amino acid sequence around the mutation site (E167K) of RF2* and effects of relevant mutations on RF function. Substitutions generated by site-directed mutagenesis by using designed primers coding for the substitutions are indicated below the wild-type sequence of RF1 and RF2. These altered protein genes were cloned into plasmid pSUIQ and transformed and expressed in the Ts RF1 (prfA1) and RF2 (prfB286) strains, and the transformant growth was examined at 42°C on LB agar plates containing 1 mM IPTG (complementation assay). “normal” means that the mutation did not reduce or alter the activity to complement the relevant Ts allele, and “loss of function” means that the mutation abolished the activity.
The fact that two RFs from prokaryotes exhibit codon specificity suggests that they must interact directly with the codon. On accumulation of RF sequences from different organisms, the conservation of protein motifs has emerged in prokaryotic and eukaryotic RFs, as well as in the C-terminal portion of elongation factor EF-G, a translocase protein that forwards peptidyl tRNA from the A site to the P site on the ribosome (7). The three-dimensional structure of Thermus thermophilus EF-G comprises five subdomains; the C-terminal part, domains III–V, appears to mimic the shapes of the acceptor stem, the anticodon helix, and the T stem of tRNA, respectively (8–10). Furthermore, it appears that an RF region shares homology with domain IV of EF-G, thus constituting a putative “tRNA-mimicry” domain necessary for RF binding to the ribosomal A site (7). This mimicry model would explain why RFs recognize stop codons by assuming an anticodon-mimicry element in the protein and further suggest that all prokaryotic and eukaryotic RFs evolved from the progenitor of EF-G.
RF1 and RF2 are known to be structurally similar, and both read the UAA codon. It might be possible, therefore, to alter mutationally either factor so that its stop codon specificity is altered. In the present study, we mutationally altered RF2 and show that a single amino acid substitution permits it to terminate translation at the UAG stop codon as well as the UGA and UAA stop codons, providing genetic support for the existence of the anticodon mimicry element in protein release factors.
MATERIALS AND METHODS
Plasmids and Manipulations.
Plasmid pSUIQ-RF2 is an isopropyl 1-thio-β-d-galactoside (IPTG)-controllable RF2 expression plasmid equivalent to pSUIQ-RF3 (11) except that the Salmonella RF2 gene was substituted for the RF3 insert in pSUIQ-RF3. pSUIQT-RF2* carries the mutant (E167K) RF2 and a tetracycline-resistant marker. A C-terminal histidine tag was marked to RF2 and RF2* by using histidine-tagged PCR primers as described (12, 13). Site-directed mutagenesis of RF1 and RF2 was performed by using designed primers coding for the substitutions (see Fig. 1B) according to the standard procedure (14).
Mutagenesis of prfB and Selection of Suppressors.
The pSUIQ-RF2 DNA was mutagenized by incubation with 0.4 M hydroxylamine at pH 6.0 for 20 h at 37°C or by the error-prone PCR method (14). The plasmid then was precipitated with ethanol and rinsed several times with Luria–Bertani (LB) broth. The Escherichia coli K12 strain RM695 [W3110 prfA1 (Ts) recA∷Tn10; ref. 13] was transformed with the mutagenized DNA, and temperature-resistant colonies were selected at 42°C on LB agar plates containing 1 mM IPTG. Plasmid DNAs were recovered from these revertants and retransformed into the same parental strain, and those that gave a reproducible phenotype (i.e., growth at 42°C) were characterized further.
Construction of prfA prfB Knockout Strains.
The chromosomal prfA∷KmR and prfB∷CmR disruptants were made by transformation of recD or recBC sbc E. coli cells lysogenic for λprfA or λprfB transducing phage with linear DNAs containing each knockout construct (see Fig. 2A). After DNA hybridization analyses to confirm each chromosomal disruption, knockout alleles were transferred into the E. coli test strains containing pSUIQ-RF2 or pSUIQT-RF2* by P1 phage transduction by selecting for KmR and CmR with 0.1 mM IPTG.
Figure 2.
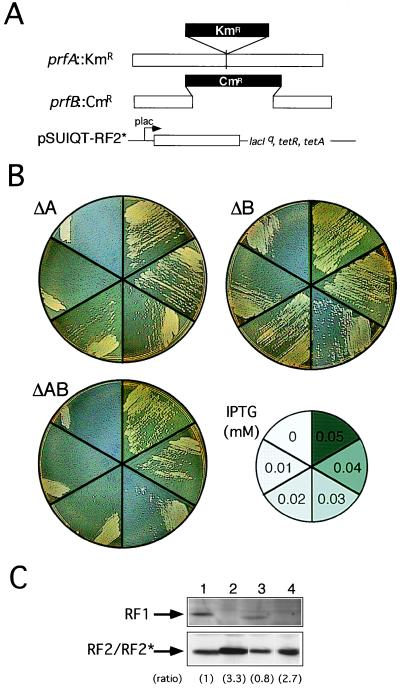
Replacement of RF1 and RF2 function with RF2* by chromosomal gene disruption. (A) Disruption of RF1 (prfA) and RF2 (prfB) genes by KmR and CmR cassettes, respectively, on the E. coli chromosome. RF2* was cloned in plasmid pSUIQT so as to be expressed by the addition of IPTG. (B) Viability of pSUIQT-RF2* transformants of RF1 knockout (ΔA, prfA∷KmR), RF2 knockout (ΔB, prfB∷CmR), and RF1/RF2 double knockout (ΔAB, prfA∷KmR prfB∷CmR) strains. Cells were grown at 37°C on LB agar plates containing different IPTG concentrations as indicated. Cell viability can be judged by single colony formability, not by appearance of a cell zone (caused by cell density effect carrying over some IPTG) at the top of the cell suspension streak. (C) Western immunoblot analysis of the level of RF2* required for suppression of prfA and/or prfB knockout strains. Equal amounts (10 μg of bulk protein) of cell lysates were analyzed by SDS/PAGE and subjected to immunoblot staining with anti-RF1 (Top) and anti-RF2 (Bottom) antibodies. Lanes: 1, E. coli W3110 (nontransformant control); 2, pSUIQT-RF2* transformant of prfA∷KmR disruptant (0.04 mM IPTG); 3, pSUIQT-RF2* transformant of prfB∷CmR disruptant (0.01 mM IPTG); and 4, pSUIQT-RF2* transformant of prfA∷KmR prfB∷CmR double disruptant (0.04 mM IPTG). Lanes 2–4 represent RF2* synthesized with the minimal concentrations of IPTG sufficient to restore viability of prfA and/or prfB knockout strains. Relative intensities of immunoblots compared with that of lane 1 are shown in parentheses.
Analysis of Protein Products of the 3A′ Gene.
E. coli test strains were transformed with the 3A′ reporter plasmid pAB96 (15, 16). Transformants were grown in LB media containing selective antibiotics and IPTG (1 mM), and exponentially growing cells were examined for the synthesis of 3A′ and 2A′ proteins as described (13).
Protein Overproduction and Purification.
Histidine-tagged RF genes were cloned downstream of a T7 RNA polymerase promoter in plasmid pET30a (Novagen) according to the manufacturer’s instructions as described (13). The resulting plasmids were transferred to BL21 (DE3). BL21 (DE3) contains a lysogenic λ phage derivative, DE3, carrying the gene for T7 RNA polymerase under the control of an inducible lacUV5 promoter. Overexpression of recombinant proteins was achieved by T7 RNA polymerase in BL21 (DE3) transformants in the presence of 0.5 mM IPTG for 2.5 h, and histidine-tagged RF proteins were purified to homogeneity from cell lysates by affinity chromatography by using Ni-NTA Agarose (Qiagen). RF2 and RF2* proteins used for in vitro fMet release sustained a Glu-to-Lys change at position 157 because it generally enhances or stabilizes histidine-tagged RF activity in vitro (unpublished work).
RESULTS
Isolation of RF2 Mutant That Suppresses RF1 Allele.
A genetic selection was used to isolate a mutant RF2 protein that substitutes for RF1 function (see Materials and Methods; Fig. 1A). This selection had two components: a temperature-sensitive (Ts) RF1 mutant of E. coli (prfA1: ref. 17) that is lethal at 42°C and a Salmonella RF2 gene (prfB: ref. 18) fused to an IPTG-inducible lac promoter in plasmid pSUIQ-RF2. Because the activity of E. coli RF2 is weak and its overexpression is toxic to cells, we used the Salmonella prfB gene, which does not show such phenotypes (13). The plasmid DNA was mutagenized in vitro with hydroxylamine or with the error-prone PCR method (14) and transformed into the prfA1 (Ts) strain.
Of >1 million transformant colonies screened, three colonies grew at 42°C dependent on 1 mM IPTG, which induces plasmid-bearing RF2 expression. Plasmid DNA altered prfA1 cells to grow at 42°C, and sequencing revealed one mutation in each within the prfB gene, which caused a Glu(GAA)-to-Lys(AAA) change at amino acid position 167 (E167K: Fig. 1B). This substitution conferred on RF2 the ability to compensate the Ts lethal defect of RF1, not only in prfA1 of E. coli but also in prfA101 (19) of S. typhimurium (data not shown). This mutation, designated RF2*, also restored the growth of the RF2 Ts strain prfB286 (20) at 42°C (data not shown), suggesting that the mutation did not switch stop codon selectivity from RF2 to RF1 but conferred on RF2 omnipotent stop codon recognition.
Viability of RF1-RF2 Double Knockout Cells in the Presence of Omnipotent RF2*.
To firmly establish that RF2* replaces both RF1 and RF2 functions in vivo, we made RF1-gene-knockout (prfA∷KmR) and RF2-gene-knockout (prfB∷CmR) strains (see Fig. 2A) and tested whether RF2* supported growth of the double knockout. Note that the chromosomal prfA∷KmR and prfB∷CmR knockout is viable only when both wild-type RF1 and RF2 proteins are provided from plasmids or phages (data not shown). First, prfA∷KmR and prfB∷CmR alleles were transduced successfully by P1 phage into E. coli cells containing a lac promoter-controlled RF2* expression plasmid, pSUIQT-RF2*, with 0.1 mM IPTG. Then, the single or double knockout transformants were tested for growth at different IPTG concentrations. As shown in Fig. 2B, RF2 knockout cells grew at IPTG concentrations higher than 0.01 mM (Fig. 2B, Right), whereas RF1 knockout (Fig. 2B, Left) and RF1/RF2 double knockout (Fig. 2B, Left) cells grew at concentrations higher than 0.04 mM.
The cellular level of RF2* expressed in each transformant was investigated by Western blot analyses by using anti-RF2 antibody. As shown in Fig. 2C, the level of RF2* produced in RF2 knockout transformants with 0.01 mM IPTG (Fig. 2C, lane 3) was nearly equivalent to that of RF2 synthesized in wild-type haploid cells (Fig. 2C, lane 1). On the other hand, the level of RF2/RF2* or RF2* alone increased threefold in transformants of the RF1 knockout strain (Fig. 2C, lane 2) or the RF1/RF2 double knockout strain (Fig. 2C, lane 4) with 0.04 mM IPTG, suggesting that recognition of UAG by RF2* was weaker compared with UGA. Western blot analyses using anti-RF1 antibody confirmed that the prfA∷KmR allele abolished RF1 synthesis (Fig. 2C, lanes 2 and 4).
Omnipotent Release Activity.
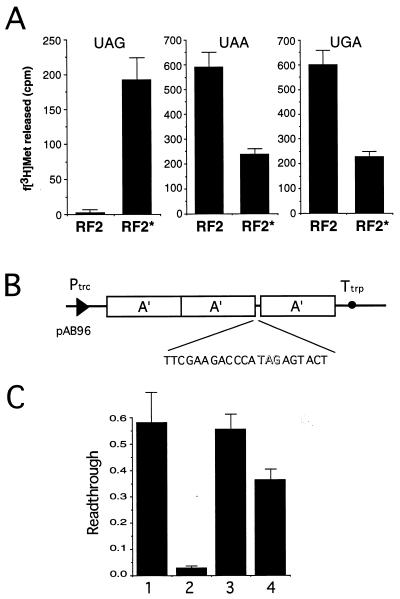
To explore the activity of RF2* in vitro, wild type as well as the E167K prfB gene product was marked with a C-terminal histidine tag (which does not affect the activity of wild-type RF2 in vivo and in vitro; ref. 13), and the tagged proteins were purified to homogeneity by affinity chromatography by using Ni-NTA agarose. The ability to terminate protein synthesis was monitored by the rate of N-formylmethionine (fMet) release at stop codons (21). As expected, RF2* catalyzed UAG termination in vitro in contrast to wild-type RF2, as well as normal UGA/UAA termination although the specific activity was significantly lower than that of wild-type RF2 (Fig. 3A) or RF1 (data not shown).
Figure 3.
Polypeptide release activity of RF2* at a UAG codon. (A) in vitro fMet release assay with histidine-tagged RF2 and RF2*. f[3H]Met release from the [f[3H]Met-tRNAf⋅AUG⋅ribosome] complex on addition of RFs and terminator triplets was determined (21). Reactions contained 20 μM UAG (Left), UAA (Center), and UGA (Right), as well as equal molar amounts (50 pmol) of RF proteins. The relative fMet-release activity of RF2* to RF1 at UAG is approximately one-fourth or one-fifth under these experimental conditions (data not shown). (B) The 3A′ reporter gene construct (pAB96) for UAG readthrough assay (15, 16). (C) Influence on UAG readthrough of expression of various RF constructs. Readthrough (RT) values, i.e., molar amounts of 3A′ domain protein (translation readthrough) relative to 2A′ domain protein (translation termination), were measured as described (13, 15, 16). Bars: 1, pSUIQ (vector control); 2, pSUIQT-RF1; 3, pSUIQT-RF2; 4, pSUIQT-RF2*. Experiments were performed independently at least five times in A and C, and the values are expressed with SDs.
Evidence for in vivo termination at UAG also was demonstrated by using an assay based on a gene that codes for three identical engineered antibody binding B domains of protein A from Staphylococcus aureus (15, 16). The sequence with the UAG codon was inserted into a linker between the segments coding for the second and third identical IgG binding domains (Fig. 3B). The predicted influence of RF2* on termination at UAG in growing bacteria was shown by the appearance of the two-domain (2A′) protein product in addition to the three-domain (3A′) protein, a readthrough product. As shown in Fig. 3C, overexpression of RF2* stimulated termination at UAG compared with RF2 but much less than RF1, which is consistent with the above observation that the acquired capacity of RF2* to release fMet at UAG as well as to suppress prfA1 (Ts) growth was weak compared with RF1 (see Figs. 2 B and C and 3A).
DISCUSSION
The present study showed that a single amino acid substitution (E167K) in bacterial release factor RF2 permits it to terminate translation at all three stop codons. This finding suggests that Glu-167 or its vicinity in bacterial RF2 is involved, directly or indirectly, in stop codon recognition by means of third-base discrimination (that is, guanine rejection and adenine acceptance) and that the E167K mutation abolished this discriminator function (see Fig. 1A). Consistent with this model, RF2* tends to release fMet, slightly but significantly, at some sense triplets in vitro including UGG (unpublished work).
RF1 and RF2 proteins have overall sequence similarities, and all known bacterial RFs share seven well conserved regions, designated A through G (7). Glu (or rarely Asp) at position 167 is well conserved among RF2 proteins from different bacteria but much less so among RF1 proteins, where Ser often is encoded at the equivalent position (Fig. 4). Glu-167 of E. coli and Salmonella RF2 is followed by another Glu, but neutralization of the charge of this dipeptide is not the simple cause of the loss of the third-base discriminator function because the E168K substitution protein failed to restore the growth of prfA1 strain at 42°C (see Fig. 1B). Swapping two doublet sequences, Ser-Ala and Glu-Glu, between RF1 and RF2 did not confer the counterpart specificity (see Fig. 1B). These results indicate that the residues around position 167 may not represent the direct discriminator element, even though the E167K substitution specifically interferes with the third-base discriminator of RF2. Similarly, RF1 must have a second-base discriminator somewhere in the protein (Fig. 1A). Presumably, the same, but reciprocal, selection strategy that we used in this study would uncover a residue(s) that affects the second-base discriminator of RF1.
Figure 4.
Comparison of the amino acid sequences of prokaryotic RF1 and RF2. The similarity alignments of RFs were accomplished as described (7). Relevant sequences around positions 157 (general enhancer allele; in shade) and 167 (omnipotent allele; in black) are shown.
To our understanding, prokaryotes have two codon-specific RFs whereas eukaryotes have only one. Mycoplasma genitalium is a small genome-size bacteria but has only one factor, RF1 (22). This should not be an exception to the rule of distinct stop codon selection by two RFs in prokaryotes. We assume that an equivalent to RF2 should have disappeared during evolution because Mycoplasma reassigned the UGA codon to the glutamine codon during evolution, making RF2 harmful.
With regard to the “one or two RF scenario,” it is intriguing that RF2 acquired omnipotent release activity for all three stop codons by a single amino acid substitution. The consequence of this change is to eliminate the specificity, not change it (in the strictest sense), for stop codon recognition: hence, a “loss of specificity” rather than a “gain of function.” Nevertheless, this point clearly demonstrates that, despite the highly conserved “one or two RF scenario” in eukaryotes and prokaryotes, respectively, prokaryotic RFs encode potentially the same protein motif(s) to read the stop codon, as does eukaryotic RF, which we previously proposed as an “RF-tRNA mimicry” hypothesis (7). In other words, all prokaryotic and eukaryotic RFs might have evolved from a common progenitor, and RF2 E167K may be more like the progenitor. The common progenitor theory is also consistent with several pieces of biochemical evidence for common catalytic and binding properties on the ribosome. We speculate that the ancestral RF had a primitive anticodon moiety that recognized more than the common three stop triplets and only later evolved to facilitate precise recognition of these three stop triplets. In prokaryotes, this recognition occurred by generating the two proteins RF1 and RF2, with a distinct discriminator function for the second and third bases, whereas in eukaryotes, it occurred by generating another mechanism to reject all nonstop triplets by using one protein, eRF1.
Why do prokaryotes have two RFs, while eukaryotes have only one? To date, two archaebacteria, Methanococcus jannaschii (23) and Archaeoglobus fulgidus (24), whose genome sequences have been completed, have only single eukaryotic RFs, which is consistent with the view that eukaryotes and archaebacteria have a common ancestor and share essential genes for macromolecular syntheses (25). Therefore, prokaryotic RF1 and RF2 should have diverged from the progenitor before eukaryotes and archaebacteria diverged. In view of the present finding that RF2 can revert (easily) to an omnipotent factor, there should have been some evolutionary bias, rather than a simple “frozen accident,” to force prokaryotes to maintain two RFs. One possibility might be a distinct stop codon usage of biological significance. Compared with UAG, UGA occasionally is programmed as an alternate decoding signal for selenocysteine incorporation or frameshifting. In fact, RF2 genes of prokaryotes are associated with autogenous +1 frameshift control at the internal UGA signal (26). If this mechanism is essential to control RF2 distinctly from RF1, it is biased in favor of the “two RF scenario” in prokaryotes, although other biases also might have been involved.
The present findings provide genetic support that a single ancestral RF existed for prokaryotes and eukaryotes as well as for the direct reading of three stop codons by RFs. To our knowledge, RF2* is a protein that was altered in stop codon selectivity. Based on the “RF-tRNA mimicry” hypothesis, amino acids clustered around positions 200–213 may mimic the anticodon loop of tRNA (2, 7) and selectively recognize the stop codon in conjunction with the predicted discriminator element. A true test of this model, however, awaits a structural study as well as further functional studies of release factors.
Acknowledgments
We thank K. Matsumura for participation in the early part of prfB gene knockout; L. Isaksson for the 3A′ reporter system; W. P. Tate for anti-RF1 antibody; J. Hershey, D. Court, and C. Yanofsky for critical reading of the manuscript and valuable comments; and C. Y. Nakamura for editing the manuscript. This work was supported in part by grants from The Ministry of Education, Science, Sports and Culture, Japan; the Human Frontier Science Program (awarded in 1993 and 1997); and the Program for Promotion of Basic Research Activities for Innovation Biosciences (PROBRAIN).
ABBREVIATIONS
- RF
release factor
- eRF1
eukaryotic release factor 1
- IPTG
isopropyl 1-thio-β-d-galactoside
- LB
Luria–Bertani
References
- 1.Scolnick E, Tompkins R, Caskey T, Nirenberg M. Proc Natl Acad Sci USA. 1968;61:768–774. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakamura Y, Ito K, Isaksson L A. Cell. 1996;87:147–150. doi: 10.1016/s0092-8674(00)81331-8. [DOI] [PubMed] [Google Scholar]
- 3.Konecki D S, Aune K C, Tate W, Caskey C T. J Biol Chem. 1977;252:4514–4520. [PubMed] [Google Scholar]
- 4.Frolova L, Le Goff X, Rasmussen H H, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni A-L, Celis J E, Philippe M, et al. Nature (London) 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 5.Tate W P, Brown C M. Biochemistry. 1992;31:2443–2450. doi: 10.1021/bi00124a001. [DOI] [PubMed] [Google Scholar]
- 6.Stansfield I, Tuite M. Curr Genet. 1994;25:385–395. doi: 10.1007/BF00351776. [DOI] [PubMed] [Google Scholar]
- 7.Ito K, Ebihara K, Uno M, Nakamura Y. Proc Natl Acad Sci USA. 1996;93:5443–5448. doi: 10.1073/pnas.93.11.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ævarsson A, Brazhnikov E, Garber M, Zheltonosova J, Chirgadze Yu, Al-Karadaghi S, Svensson L A, Liljas A. EMBO J. 1994;13:3669–3677. doi: 10.1002/j.1460-2075.1994.tb06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czworkowski J, Wang J, Steitz T A, Moore P B. EMBO J. 1994;13:3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark B F C, Nyborg J. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 11.Matsumura K, Ito K, Kawazu Y, Mikuni O, Nakamura Y. J Mol Biol. 1996;258:588–599. doi: 10.1006/jmbi.1996.0271. [DOI] [PubMed] [Google Scholar]
- 12.Ito K, Nakamura Y. Biochimie. 1997;79:287–292. doi: 10.1016/s0300-9084(97)83516-x. [DOI] [PubMed] [Google Scholar]
- 13.Uno M, Ito K, Nakamura Y. Biochimie. 1996;78:935–943. doi: 10.1016/s0300-9084(97)86715-6. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Björnsson A, Isaksson L A. J Mol Biol. 1993;232:1017–1029. doi: 10.1006/jmbi.1993.1457. [DOI] [PubMed] [Google Scholar]
- 16.Mottagui-Tabar S, Björnsson A, Isaksson L A. EMBO J. 1994;13:249–257. doi: 10.1002/j.1460-2075.1994.tb06255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rydén S M, Isaksson L A. Mol Gen Genet. 1984;193:38–45. doi: 10.1007/BF00327411. [DOI] [PubMed] [Google Scholar]
- 18.Kawakami K, Nakamura Y. Proc Natl Acad Sci USA. 1990;87:8432–8436. doi: 10.1073/pnas.87.21.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott T, Wang X. J Bacteriol. 1991;173:4144–4154. doi: 10.1128/jb.173.13.4144-4154.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawakami K, Inada T, Nakamura Y. J Bacteriol. 1988;170:5378–5381. doi: 10.1128/jb.170.11.5378-5381.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikuni O, Ito K, Moffat J, Matsumura K, McCaughan K, Nobukuni T, Tate W, Nakamura Y. Proc Natl Acad Sci USA. 1994;91:5798–5802. doi: 10.1073/pnas.91.13.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 23.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, Fitzgerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 24.Klenk H-P, Clayton R A, Tomb J-F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, et al. Nature (London) 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 25.Martin W, Müller M. Nature (London) 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 26.Craigen W J, Cook R G, Tate W P, Caskey C T. Proc Natl Acad Sci USA. 1985;82:3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]