Abstract
IL-5, IL-3, and GM-CSF are hematopoietic cytokines that are key mediators of the allergic inflammatory response. The receptors for these three cytokines consist of a cytokine-specific α (Rα) chain and a shared common β (βc) chain. Herein, we demonstrate that agonistic ligation of these receptor subunits rapidly induces proteasomal degradation of the βc, but not the Rα, cytoplasmic domain, resulting in termination of signal transduction and yielding a truncated βc isoform ligated to the Rα subunit. Proteasomal degradation of the βc cytoplasmic domain was also a prerequisite for endocytosis and lysosomal degradation of the ligated receptor subunits. Moreover, proteasome-dependent termination of signaling induced by one βc-engaging cytokine resulted in cellular desensitization to signal transduction by subsequent stimulation with another βc-engaging cytokine. These data provide the first evidence for ligand-dependent proteasomal degradation of the βc cytoplasmic domain, and they establish a novel mechanism for heterotypic desensitization of shared cytokine receptor signaling.
Introduction
IL-5, IL-3, and GM-CSF are related cytokines that regulate the growth, differentiation, and survival of hematopoietic cells (1). Whereas GM-CSF and IL-3 have broader overlapping hematopoietic activities, IL-5 is more eosinophil-specific, and is central to the pathophysiology of eosinophilic inflammation in atopy and asthma. The receptors for human IL-5, IL-3, and GM-CSF are members of the hematopoietin receptor superfamily (2). The high-affinity receptor complexes for each of these cytokines are composed of a ligand-specific α chain and a dimer of the common β subunit (βc) that is shared by all three cytokines (3–6). Alone, each specific α chain forms a low-affinity receptor when bound by its respective ligand, but its association with βc converts the binding to high affinity. The association of the α and βc subunits mediated by ligand binding initiates signal transduction, which includes the JAK/STAT, Ras/MAPK, and PI 3-K signaling pathways (reviewed in refs. 7, 8). Although βc is the main signaling component for these receptor complexes, the α chains are also required for signal transduction (7, 8).
Early progenitor granulocytic CD34+ cells coexpress βc and the Rα chains of IL-5, IL-3, and GM-CSF (9–11). However, it is not clear how these progenitor cells regulate their responsiveness to these three cytokines during stochastic differentiation into mature eosinophils, basophils, or even macrophages. Likewise, fully differentiated lineages may also remain responsive to IL-5, IL-3, and GM-CSF. Since βc is the common receptor subunit for these three cytokines, investigators have hypothesized that distinction between cytokine-specific signals is provided by the α chains. Indeed, a yeast two-hybrid screen with the IL-5Rα cytoplasmic domain revealed that syntenin, a protein containing tandem PDZ domains, directly associated with IL-5Rα (12). Moreover, IL-5 mediated a direct interaction between syntenin and the transcription factor Sox4.
Activation of the JAK/STAT, MAPK, and PI 3-K signaling pathways by IL-5, IL-3, or GM-CSF is transient. Therefore, efficient mechanisms must exist to terminate signaling. One mechanism is the activation of cytosolic phosphatases that control the level of ligand-induced phosphorylated substrates. Two phosphatases described for these three cytokines are Src homology protein tyrosine phosphatase-1 (SHP-1) and SHP-2 (13, 14). Whereas SHP-2 appears to be a positive regulator of IL-5 signaling (14), overexpression of SHP-1 results in suppression of cell growth in response to IL-3 (15).
Another mechanism that counteracts IL-5, IL-3, or GM-CSF agonistic signals is induction of the family of small SH2-containing proteins that includes cytokine-inducible SH2-containing protein (16), JAK-binding protein (17), suppressors of cytokine signaling (18), and STAT-induced STAT inhibitors (19). Members of this family negatively regulate cytokine signal transduction by specifically downregulating JAK/STAT signaling.
Another mechanism for terminating cytokine-induced signal transduction is receptor endocytosis and degradation (reviewed in ref. 20). For most cell surface receptors, ligand binding results in the internalization of the receptor/ligand complex into clathrin-coated vesicles. These vesicles fuse with early endosomes, where receptors are either sorted for recycling back to the plasma membrane or are targeted into late endosomes and ultimately degraded in lysosomes. Proteasomes have also been implicated in degradation of the c-kit, PDGF-β, Met tyrosine kinase, growth hormone, IL-2Rβ, IL-9Rα, and erythropoietin transmembrane receptors (21–26). Prolongation of JAK/STAT signal transduction induced by IL-2, IL-3, and erythropoietin in the presence of proteasome inhibitors has further suggested that proteasome degradation could be an important mechanism for terminating cytokine receptor signals (27–30).
Herein, we provide the first evidence for ligand-dependent proteasomal degradation of the βc cytoplasmic domain, resulting in a truncated receptor isoform designated βc intracytoplasmic proteolysis (βIP). Analysis of the functional consequences of the βIP isoform demonstrates that proteasomal degradation is a mechanism for both rapid termination of βc signaling and for heterotypic desensitization to all of the βc-engaging cytokines.
Methods
Cell culture.
The human erythroleukemic cell line TF1 (31) was cloned by limiting dilution in media containing 5 ng/ml IL-5. The TF1-F11 clone was selected for its strong proliferative responsiveness to IL-5, IL-3, and GM-CSF. It was maintained in RPMI 1640 supplemented with 10% FBS, 10 mM HEPES buffered saline solution (Sigma Chemical Co., St. Louis, Missouri, USA), 50 mM β-mercaptoethanol, 10 μg/ml gentamicin, and 5 ng/ml human IL-5. All experiments used the TF1-F11 clone, herein denoted TF1. For the kinetic analyses, TF1 cells were depleted of IL-5 for 24–48 hours in RPMI 1640 containing 10% FBS. After cytokine starvation, cells were harvested, counted, and placed in media supplemented with 5–10 ng/ml of IL-5, IL-3, or GM-CSF for the indicated times. Recombinant human IL-5 was baculovirus-expressed and affinity-purified as previously described (32). IL-3 and GM-CSF were purchased from R&D Systems Inc. (Minneapolis, Minnesota, USA). The proteasome inhibitor N-acetyl-L-leucinyl-L-leucinyl-norleucinal (LLnL) and the lysosomal inhibitor bafilomycin A1 were purchased from Sigma Chemical Co. and were used at final concentrations of 50 μmol/l and 200 nmol/l, respectively. MG132, clasto-lactacystin β-lactone, and cytochalasin D were purchased from Calbiochem-Novabiochem Corp. (San Diego, California, USA), and were used at final concentrations of 50 μmol/l, 20 μmol/l, and 10 μmol/l, respectively. All inhibitors were dissolved in 100% DMSO. All experiments were repeated at least three times with consistently reproducible results.
Immunoprecipitation and immunoblot assays.
TF1 cells were cytokine-starved for 24–48 hours and stimulated with 5–10 ng/ml IL-5, IL-3, or GM-CSF for the indicated times. For all assays done with proteasome, endocytosis, and lysosome inhibitors, cells were cytokine-starved for 24–48 hours, pretreated with each of the inhibitors for 1 hour at 37°C, and then stimulated with IL-5 (10 ng/ml). For the restimulation assays, cells were cytokine-starved for 24 hours and treated with 20 ng/ml IL-5 for the indicated times. At the end of each incubation period, cells pretreated with IL-5 were harvested, washed once in PBS, and transferred to IL-3–containing media (20 ng/ml) for 5 minutes. For all immunoprecipitation and immunoblot (IP/IB) assays, protein lysates were prepared after lysis in buffer containing 50 mM Tris (pH 8.0), 0.5% NP-40, 280 mM NaCl, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, 1 mM DTT, 1 mM PMSF, 1 mM sodium orthovanadate, 1 μg/ml leupeptin, and 1 μg/ml pepstatin. Approximately 2–3 mg total protein was incubated with 2 μg of anti-βc monoclonal antibody (S-16; Santa Cruz Biotechnology Inc., Santa Cruz, California, USA), 2–3 μg of anti–soluble-IL-5Rα (anti–sIL-5Rα) polyclonal antibody (R&D Systems Inc.), or 2–3 μg of the monoclonal antibody 4G10 (Upstate Biotechnology Inc., Lake Placid, New York, USA) for 2 hours on ice. Immunocomplexes were collected on protein G–agarose beads, separated by 8 or 12.5% SDS-PAGE, and electroblotted to Immobilon-P membrane (Millipore Corp., Bedford, Massachusetts, USA). Proteins were detected by incubating the blots with the antibodies described above or with anti-βc amino terminal N-20 polyclonal antibody and carboxy terminal C-20 polyclonal antibody from Santa Cruz Biotechnology Inc., and carboxy terminal polyclonal antibody from Upstate Biotechnology Inc.; anti-ubiquitin monoclonal antibodies from Santa Cruz Biotechnology Inc. and Zymed Laboratories (South San Francisco, California, USA); or with anti-JAK2 antiserum and anti-STAT5 polyclonal antibodies (Upstate Biotechnology Inc.), followed by incubation with enhanced chemiluminescence reagents (Amersham Pharmacia Biotech, Piscataway, New Jersey, USA).
Deglycosylation of IL-5Rα isoforms.
Deglycosylation of anti–IL-5Rα immunocomplexes was performed with peptide N-glycosidase F (PNGase F) following the manufacturer’s instructions (New England Biolabs Inc., Beverly, Massachusetts, USA). Briefly, anti–IL-5Rα immunocomplexes were collected on protein G–agarose beads and incubated for 10 minutes in 1× denaturation buffer (0.5% SDS and 1% β-mercaptoethanol). Ten percent NP-40 and 10× G7 buffer (0.5 M sodium phosphate, pH 7.5) were added to each reaction at 1/10 the volume. PNGase F (1–2 μl) was added, and the reaction mixture was incubated at 37°C for 1–2 hours. The deglycosylated proteins were analyzed by SDS-PAGE, followed by Western blotting with anti–IL-5Rα antibodies.
RT-PCR.
One microgram of total RNA from TF1 cells stimulated for 1 hour with 5 ng/ml IL-5 was reverse-transcribed with 250 ng of oligo(dT) primer (Stratagene, La Jolla, California, USA) according to the manufacturer’s instructions. A 3′ primer corresponding to the βc poly-A+ tail (bp 2929–2951) and two different 5′ primers, bp 1281–1301 (33) and bp 1376–1397, were used to amplify a βc fragment containing its transmembrane and cytoplasmic domain (approximately 1.7 kb), as well as other alternatively spliced isoforms containing deletions in their cytoplasmic domains.
Northern blotting.
Samples containing 30 μg of total RNA were subjected to Northern blot analysis following standard protocols. The membranes were probed with 32P-labeled DNA probes generated from βc PCR fragments (5′ primer, bp 1281–1301; and 3′ primer, bp 1600–1621) (33), IL-5Rα (5′ primer, bp 217–242; and 3′ primer, bp 737–758 of the IL-5Rα sequence), sIL-5Rα (same primers as for IL-5Rα), and GAPDH. Each of the PCR fragments was randomly primed with the Rediprime II kit (Amersham Pharmacia Biotech, Piscataway, New Jersey, USA) in the presence of 32P-dCTP.
Results
IL-5, IL-3, and GM-CSF downregulate full-length βc protein expression.
TF1-F11, a clone responsive to IL-5, IL-3, and GM-CSF, was generated from the human erythroleukemic cell line TF1 (30) to investigate regulatory signaling mechanisms used by these three cytokines. This clone was used to test the hypothesis that, as a shared receptor subunit, βc might be a physiologic target for regulating a cell’s responsiveness to sequential stimulation by IL-5, IL-3, or GM-CSF, and thereby may also be an important mechanism for regulation of lineage differentiation by progenitor hematopoietic cells expressing receptors for these three cytokines.
Initial studies examined whether βc levels changed after cytokine stimulation. Kinetic analyses of βc expression were performed using cells that were cytokine-starved for 24 hours, then transferred to media containing either IL-5, GM-CSF, or IL-3 (Figure 1a). Whole-cell lysates were prepared from cells harvested at sequential timepoints after initiation of stimulation with each cytokine, and βc protein expression was assayed by IP/IB analysis. Stimulation with each cytokine resulted in full-length βc protein levels that decreased over the first 4 hours, then returned almost to basal levels by 24 hours (Figure 1a, top arrows).
Figure 1.
IL-5 reduces βc protein expression, while transiently increasing faster-migrating βc bands. (a) TF1 cells that were cytokine-starved for 48 hours were stimulated with 5 ng/ml of either IL-5, GM-CSF, or IL-3 for the indicated times (107 cells/lane). Whole-cell lysates were prepared and immunoprecipitated with anti-βc monoclonal antibody S-16. Immune complexes were resolved on 8% SDS-PAGE gels and immunoblotted with anti-βc polyclonal antibodies. The top arrow in each panel represents full-length βc receptors, and the bottom arrow indicates truncated βc receptors (Δβc). (b) TF1 cells that were cytokine-starved for 48 hours were stimulated with 5 ng/ml of either IL-5, IL-3, or GM-CSF for the indicated times (107 cells/lane). Total RNA was prepared for each timepoint, and 30 μg of RNA/lane was analyzed with a βc probe corresponding to residues 417–530, which includes a piece of the membrane-proximal extracellular domain, the transmembrane domain, and part of the intracytoplasmic domain. (c) Total RNA from the IL-5–stimulated (for 1 hour) TF1 cells was reverse-transcribed using oligo(dT) primers, and amplified by PCR using a 3′ βc primer corresponding to its poly-A+ tail, a 5′ primer to a region flanking the βc transmembrane domain (lane 3), and another 5′ primer located in its transmembrane domain (lane 4). As a negative control, no cDNA was included in lane 2. Lane 1 is a DNA size ladder.
An additional and striking observation from this analysis was the presence of a smaller, 75-to 85-kDa doublet (designated as Δβc in Figure 1a, bottom arrow) that was present 1 hour after initiation of cytokine stimulation and disappeared by 4 hours after initiation of cytokine stimulation. Further kinetic analyses consistently demonstrated the appearance of this 75- to 85-kDa Δβc doublet within 15 minutes of stimulation with each of the three cytokines; the doublet was maximal at 1 hour and disappeared by 2–3 hours after initiation of stimulation (Figures 2 and 4). We predicted that the rapid induction and disappearance of the Δβc doublet was indicative of a tightly controlled mechanism that might serve an important regulatory function for signaling by these cytokines. Based on the Δβc bands’ molecular mass and their reactivity with a panel of domain-specific βc antibodies (anti-βc peptide polyclonal antibody N-20, encompassing amino acids 21–40, from Santa Cruz Biotechnology Inc.; polyclonal antibody against the complete βc cytoplasmic domain from Upstate Biotechnology Inc.; and anti-βc peptide polyclonal antibody C-20, encompassing amino acids 861–881, Santa Cruz Biotechnology Inc.), the Δβc bands were characterized as βc truncations containing a complete extracellular domain, a transmembrane domain, and a small part of the intracellular membrane-proximal domain (about 25–35 amino acids of its intracytoplasmic domain). PNGase F treatment resolved the Δβc doublet into three bands (63, 66, and 67 kDa), of which the 63-kDa and 66-kDa bands were predominant (data not shown).
Figure 2.
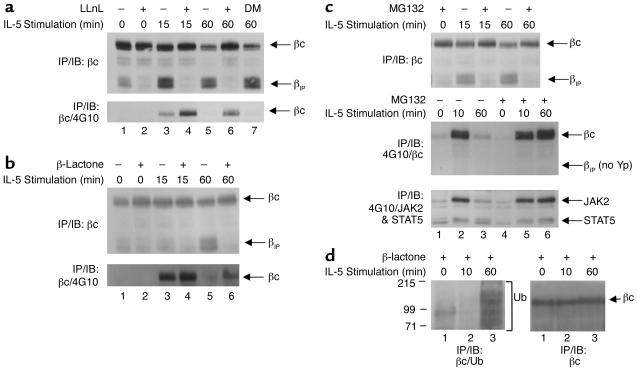
IL-5 induces proteasomal degradation of the βc cytoplasmic domain. (a) TF1 cells that were cytokine-starved for 24 hours were left untreated (lanes 1, 3, and 5) or were pretreated with 50 μM LLnL (lanes 2, 4, and 6) or 0.01% (vol) DMSO (lane 7) for 1 hour prior to IL-5 stimulation (10 ng/ml) (top and bottom panels). Whole-cell lysates were immunoprecipitated with anti-βc monoclonal antibody S-16, and immunoblotted with anti-βc polyclonal antibodies (top panel). The top arrow indicates full-length βc receptors; the bottom arrow corresponds to βIP. The membrane was stripped and reprobed with monoclonal antibody 4G10 (bottom panel). (b) Same as in a except the cells were pretreated with 20 μM β-lactone for 1 hour, followed by IL-5 stimulation. The top panel shows analysis by IP/IB with the same anti-βc antibodies described in a. The membrane was then stripped and reprobed with 4G10 (bottom panel). (c) Same as in a and b, except that the cells were pretreated with 50 μM MG132 for 1 hour, followed by IL-5 stimulation (all panels). The top panel was analyzed by IP/IB with the same anti-βc antibodies used in a and b. The middle panel was immunoprecipitated with 4G10, and immunoblotted with the anti-βc polyclonal antibodies. The blot was then stripped and reprobed with both anti-JAK2 and anti-STAT5 antibodies (bottom panel). (d) TF1 cells (3 × 107 cells/IP) that had been cytokine-starved for 24 hours were pretreated with 30 μM β-lactone for 1 hour, followed by IL-5 stimulation (15 ng/ml). Whole-cell lysates were immunoprecipitated with anti-βc antibodies and immunoblotted with anti-ubiquitin monoclonal antibodies (left panel). The bracket designates βc ubiquitination, as well as other ubiquitinated proteins that coimmunoprecipitated with anti-βc antibodies. The blot in the left panel was stripped and reprobed with anti-βc antibodies (right panel). DM, DMSO, Yp, tyrosine phosphorylation; Ub, ubiquitin.
Figure 4.
Proteasome inhibitors stabilize IL-5Rα and βc protein/protein interaction. TF1 cells that were cytokine-starved for 48 hours were either left untreated (lanes 1, 3, and 5) or were pretreated with 50 μM LLnL (lanes 2, 4, and 6) or 0.01% (vol) DMSO (lane 7) for 1 hour prior to IL-5 stimulation (10 ng/ml) for the indicated times. Whole-cell lysates were prepared, and IP/IB analysis was performed with anti–IL-5Rα polyclonal antibody (top panel). The blot in the top panel was stripped and reprobed with anti-βc polyclonal antibodies (bottom panel). Note that both full-length βc receptor and βIP coimmunoprecipitate with IL-5Rα antibodies (bottom panel).
To determine whether the cytokine-induced decrease in βc protein levels was due to a decrease in βc mRNA levels, Northern blot analyses were performed with total RNA from IL-5–, IL-3–, or GM-CSF–stimulated cells. βc transcripts were detected with a probe that spanned the membrane-proximal extracellular domain, the transmembrane domain, and a small piece of the membrane-proximal cytoplasmic domain (amino acids 417–530). As seen in Figure 1b, βc mRNA levels were not significantly altered by IL-5, IL-3, or GM-CSF stimulation, indicating that the decrease in βc total protein was not due to a corresponding reduction in its mRNA. Since regulation of βc mRNA and protein expression was the same for IL-5, IL-3, and GM-CSF, our studies hereafter used IL-5 to further investigate βc regulation.
Δβc is not an alternatively spliced product of βc.
The possibility existed that the induced Δβc was an alternatively spliced βc isoform. RT-PCR analysis was performed (Figure 1c) with a 3′ primer corresponding to the βc poly-A+ tail (bp 2929–2951) and two different 5′ primers, one that flanked the transmembrane domain (bp 1281–1301; lane 3), and one that was located within the transmembrane domain (bp 1376–1397; lane 4). Both sets of primers detected only the expected βc mRNA transcript. No alternatively spliced βc isoforms were detected. Hence, internal alternative splicing was not the basis for Δβc expression.
Reduction in full-length βc protein expression is mediated by proteasomal degradation.
Since βc and Δβc protein levels were not regulated by cytokine-induced transcriptional mechanisms, we next investigated whether Δβc was generated by proteolysis of the βc cytoplasmic domain. The established role of proteasomes in degrading cytoplasmic proteins, as well as several mammalian plasma membrane receptors (21–25), led us to speculate that Δβc might be generated by proteasomal degradation of the βc cytoplasmic domain. Indeed, pretreatment of cells with 50 μM of the proteasome inhibitor LLnL for 1 hour, followed by IL-5 stimulation, inhibited induction of Δβc (Figure 2a, top panel, bottom arrow) and resulted in stable levels of full-length βc protein (Figure 2a, top panel, top arrow). Since Δβc was a result of IL-5–induced proteasomal degradation of the βc cytoplasmic domain, we renamed the Δβc bands βIP.
We next explored the cellular consequences of inhibiting IL-5–induced proteasomal degradation of the βc cytoplasmic domain. We hypothesized that prolonged βc tyrosine phosphorylation would be detected if the βc cytoplasmic tail was not degraded, as was the case in the presence of LLnL. When the membrane in Figure 2a (top panel) was stripped and reblotted with anti-phosphotyrosine monoclonal antibody (4G10), there was an accumulation of βc tyrosine phosphorylation in the presence of LLnL after 15 minutes of IL-5 stimulation (bottom panel, lane 4). Moreover, βc tyrosine phosphorylation was still detectable, even after 1 hour of IL-5 stimulation in the presence of LLnL (lane 6), whereas the untreated and DMSO-treated lanes returned to basal βc tyrosine phosphorylation levels (lanes 5 and 7).
To further confirm that proteasomes mediated degradation of the βc cytoplasmic domain, two additional proteasome-specific inhibitors were used. Treatment of cells with clasto-lactacystin β-lactone (β-lactone) or MG132 also inhibited the generation of βIP (Figure 2, b and c, top panels). In addition, each of the proteasome inhibitors prolonged tyrosine phosphorylation of full-length βc receptors (Figure 2b, bottom panel; Figure 2c, middle panel). In contrast, βIP was not tyrosine phosphorylated (Figure 2c, middle panel), since its cytoplasmic domain that contained the substrate tyrosine residues was degraded by the proteasomes. To examine the effect of proteasome inhibitors on downstream signaling events, the membrane shown in Figure 2c, middle panel, was stripped and reprobed with anti-JAK2 and anti-STAT5 antibodies (Figure 2c, bottom panel). In the absence of MG132, tyrosine phosphorylation of JAK2 and STAT5 returned to about basal levels after 1 hour of IL-5 stimulation (Figure 2c, bottom panel, lane 3). In contrast, JAK2 and STAT5 tyrosine phosphorylation was still at peak levels in the presence of the inhibitor at 1 hour (Figure 2c, lane 6, lower panel), indicating that proteasome inhibitors also prolong IL-5–stimulated JAK/STAT signaling. Taken together, these data indicate that after IL-5 stimulation, the cytoplasmic domains of activated βc receptors are rapidly degraded by proteasomes, resulting in the generation of βIP. The data further indicate that proteasomal degradation of the βc cytoplasmic tail is an important mechanism for termination of JAK2 and STAT5 signaling.
Since most proteins that are degraded by proteasomes are modified first by ubiquitination (reviewed in ref. 34), we investigated whether βc or βc-associated molecules were ubiquitinated in response to IL-5. Immunoblotting of βc immunoprecipitates with anti-ubiquitin antibodies (Figure 2d, left panel) demonstrated a ladder of ubiquitinated proteins 1 hour after IL-5 stimulation in the presence of β-lactone (lane 3). Stripping and reprobing the membrane with anti-βc antibodies (Figure 2d, right panel) demonstrated that the ladder of ubiquitinated proteins precipitated by anti-βc antibodies was comprised of several βc-associated molecules, as well as βc.
IL-5 downregulates IL-5Rα expression and simultaneously upregulates sIL-5Rα expression.
Human IL-5R is a transmembrane heterotrimer consisting of a ligand-specific, 60-kDa α subunit (IL-5Rα) and a dimer of the 130-kDa βc (3, 6). Cloning of IL-5Rα cDNA revealed alternatively spliced transcripts that correspond to the transmembrane (IL-5Rα) and soluble (sIL-5Rα) isoforms of the receptor (35, 36). In solution, both isoforms bind IL-5 with similar affinities; therefore, the proposed role for sIL-5Rα is that of an antagonist (37–39).
Since βc associates with IL-5Rα after IL-5 ligation, and since βc expression was downregulated after IL-5 stimulation, we investigated whether IL-5Rα and sIL-5Rα expression was regulated in a similar manner. Kinetic analyses of IL-5Rα and sIL-5Rα expression were performed using cells that were cytokine-starved for 48 hours, then transferred to media containing IL-5. At sequential timepoints, whole-cell extracts were analyzed by IP/IB with anti–IL-5Rα polyclonal antibodies specific for the extracellular domain, and thus capable of recognizing both IL-5Rα (60 kDa) and intracellular sIL-5Rα (48 kDa). IL-5Rα expression decreased in a time-dependent manner (Figure 3a, top arrow). In contrast, intracellular sIL-5Rα expression increased 4–6 hours after initiation of IL-5 stimulation, then returned to basal levels by 24 hours after stimulation was begun (Figure 3a, bottom arrow). Treatment of the anti–IL-5Rα immunoprecipitates with PNGase F demonstrated that the doublet bands for IL-5Rα and sIL-5Rα were due to differential glycosylation (Figure 3b, lanes 1 and 2). The IL-5Rα doublet resolved to a single 46-kDa unglycosylated band, and the sIL-5Rα doublet resolved to a single 38-kDa unglycosylated band. PNGase F treatment of baculovirus-expressed recombinant sIL-5Rα confirmed the 38-kDa band as sIL-5Rα (Figure 3b; compare lane 2 and lane 4, bottom arrow). The difference in migration between sIL-5Rα that was immunoprecipitated from cells, and the recombinant sIL-5Rα (Figure 3b, lanes 1 and 3), appears to be due to differences in glycosylation by mammalian cells and the insect cells used for baculovirus expression of recombinant proteins.
Figure 3.
IL-5 regulation of transmembrane IL-5Rα and sIL-5Rα expression. (a) TF1 cells (107/lane) were cytokine-starved for 48 hours and stimulated with 5 ng/ml human IL-5 for the indicated times. Whole-cell lysates were prepared and subjected to IP/IB with anti–IL-5Rα antibodies. The top arrow represents IL-5Rα (60 kDa), and the bottom arrow corresponds to sIL-5Rα (50 kDa). The multiple bands represent differential glycosylation (see below). (b) TF1 cells continuously cultured in IL-5 were immunoprecipitated with anti–IL-5Rα antibodies (lanes 1 and 2), and either left untreated (lane 1) or treated with PNGase F (lane 2) for 1–2 hours to remove N-linked glycosyl groups. Lanes 3 and 4 are controls for sIL-5Rα migration, using 20 ng of baculovirus-expressed, recombinant sIL-5Rα protein (brsIL-5Rα), which was either untreated (lane 3) or treated with PNGase F (lane 4) for 1–2 hours, separated by 12.5% SDS-PAGE, and immunoblotted with anti–IL-5Rα antibodies. Note how the multiple bands in lane 1 resolve to two distinct bands in lane 2 (PNGase F–treated). (c) TF1 cells (107/lane) were cytokine-starved for 48 hours and stimulated with 5 ng/ml IL-5 for the indicated times. Total RNA was prepared for each timepoint, and 30 μg of RNA/lane were analyzed with a probe specific to the extracellular domain of IL-5Rα (detects both isoforms). GAPDH was used as an internal control.
As a negative control, kinetic analyses were also performed in the absence of IL-5 for the same timepoints. No changes in the protein expression of IL-5Rα or sIL-5Rα were detected (data not shown), confirming that the decrease in IL-5Rα and the increase in sIL-5Rα protein expression following addition of IL-5 to cultures were both induced by IL-5.
IL-5 decreases IL-5Rα mRNA expression.
To determine whether the decrease in IL-5Rα and the increase in sIL-5Rα protein expression corresponded to changes in their mRNA levels, Northern blot analyses were performed. Total RNA from IL-5–stimulated cells was analyzed with a probe that recognized both transcripts. Stimulation of cells with IL-5 resulted in an approximately 80% reduction of IL-5Rα mRNA by 4 hours, which persisted through 24 hours of continuous stimulation (Figure 3c, top panel). This is in agreement with the progressive decline in IL-5Rα protein levels over 24 hours (Figure 3a). One interpretation of this finding is that after IL-5 stimulation was begun, there was a reduction in IL-5Rα mRNA stability. Alternatively, since IL-5Rα is the splice product of the IL-5Rα gene, IL-5 may have induced a decrease in splicing of this transcript. By comparison, sIL-5Rα mRNA levels were similar at baseline and after 4 hours of IL-5 stimulation, then declined by approximately 50% after 24 hours of continuous stimulation (Figure 3c, middle panel). The increase in sIL-5Rα protein over the first 4 hours, coupled with stable sIL-5Rα mRNA levels, is consistent with increased sIL-5Rα translation.
Proteasome inhibitors stabilize IL-5Rα and βc receptor interaction.
Since proteasome inhibitors stabilized βc protein expression in response to IL-5, we explored LLnL’s effect on IL-5Rα expression. Pretreatment of cells with LLnL, followed by IL-5 stimulation for 1–3 hours, did not result in the dramatic accumulation of IL-5Rα expression at 1 hour as was seen with βc. Moreover, LLnL had no discernible effect on the faster-migrating sIL-5Rα bands (Figure 4, top panel, bottom arrow). Thus, the IL-5Rα isoforms were not directly regulated by proteasomes. However, there was a slight persistence of IL-5Rα over 3 hours, compared with cells not treated with LLnL (Figure 4, top panel, compare lanes 5 and 6). Since this slight accumulation of IL-5Rα protein occurred over a period of 3 hours of IL-5 stimulation in the presence of LLnL, we suspected that its persistence was an indirect effect of its association with βc. Therefore, we predicted that βc would still be associated with IL-5Rα at the 3-hour timepoint. To test this hypothesis, the membrane in Figure 4, top panel, was stripped and reblotted with anti-βc antibodies (bottom panel). In the absence of LLnL, both βc and βIP coimmunoprecipitated with IL-5Rα 1 hour after initiation of IL-5 stimulation (lane 3). This interaction was undetectable by 3 hours after initiation of stimulation (lane 5). In contrast, in the presence of LLnL, there was a sustained protein/protein interaction of only full-length βc with IL-5Rα (since generation of βIP was inhibited) that was still detectable by the 3-hour timepoint (lane 6). Therefore, in addition to inhibiting degradation of βc cytoplasmic domains in response to IL-5, proteasome inhibitors also prolonged the association of full-length βc receptors with IL-5Rα. Together, these data elucidate the sequence of events leading to the generation of βIP. First, there is the IL-5–induced ligation of IL-5Rα with full-length βc, as indicated by coimmunoprecipitation of both receptor subunits in the absence or presence of LLnL (Figure 4, bottom panel, lanes 3 and 4). The ligation of IL-5Rα and βc is then followed by proteasomal degradation of the βc cytoplasmic domain, thereby generating βIP, as seen in the absence of LLnL (lane 3), but not in its presence (lane 4).
Components of IL-5R are endocytosed and degraded in lysosomes following βc proteasomal cleavage.
The concomitant disappearance of βIP and decrease in IL-5Rα led us to further predict that proteasomal degradation of the βc cytoplasmic domain was a prerequisite for initiation of endocytosis and lysosomal degradation of the IL-5–ligated IL-5Rα and βc. This hypothesis was tested using inhibitors that block internalization of plasma membrane proteins. Pretreatment of cells for 1 hour with 10 μM cytochalasin D, a potent inhibitor of actin filament function, did not affect proteasomal degradation of the βc cytoplasmic domain, since βIP was still induced by IL-5 (Figure 5a, lanes 3 and 4). However, treatment of the cells with cytochalasin D did result in accumulation of βIP protein (and βc to a lesser extent) after 2 hours of IL-5 stimulation, whereas levels of both proteins were reduced in the absence of the inhibitor (Figure 5a, lanes 5 and 6). Reprobing the membrane with anti–IL-5Rα antibodies revealed that cytochalasin D also caused accumulation of IL-5Rα protein that coimmunoprecipitated with βc and βIP (Figure 5a, bottom panel). Thus, endocytosis of the IL-5R complex occurs after proteasomal degradation of the βc cytoplasmic domain. Identical results were seen with two other inhibitors of endocytosis, cytochalasin B and wortmannin (data not shown).
Figure 5.
The ligated IL-5R complex is endocytosed and degraded in lysosomes. (a) TF1 cells that had been cytokine-starved for 24 hours were left untreated (lanes 1, 3, and 5) or were pretreated with 10 μM cytochalasin D (lanes 2, 4, and 6) for 1 hour prior to IL-5 stimulation (10 ng/ml) for the indicated times. Whole-cell lysates were prepared, and proteins were immunoprecipitated with anti-βc monoclonal antibody and immunoblotted with anti-βc polyclonal antibodies (top panel). The membrane was stripped and reprobed with anti–IL-5Rα antibodies (bottom panel). Note the accumulation of βc, βIP, and IL-5Rα in the presence of the inhibitor in lane 6. (b) Same as in a, top panel, except that cells were pretreated with 200 nM bafilomycin A1 in lanes 2, 4, and 6. Note the accumulation of βIP in the presence of bafilomycin A1 in lane 6 (bottom arrow).
To determine the fate of the βIP/IL-5Rα complex after endocytosis, we analyzed the disappearance of βc and βIP in the presence of bafilomycin A1 (Figure 5b). Bafilomycin A1 is a specific inhibitor of the vacuolar proton pump ATPase, and is known to prevent acidification of endocytic structures, thereby inhibiting lysosomal degradation (40, 41). Cells were pretreated for 1 hour with 200 nM bafilomycin A1, followed by IL-5 stimulation for the indicated times. Figure 5b (lanes 5 and 6) demonstrates that in the absence of bafilomycin A1, βIP returned to basal levels (lane 1) after 2 hours of IL-5 stimulation (lane 5). However, in the presence of bafilomycin A1, βIP persisted (lane 6). To analyze the fate of IL-5Rα that coimmunoprecipitated with βc and βIP, the blot in Figure 5b was stripped and reblotted with anti–IL-5Rα antiserum. Like βc and βIP, coimmunoprecipitated IL-5Rα protein persisted at the same timepoints in the presence of bafilomycin A1 (data not shown), indicating that degradation of the ligated receptor complex was blocked. In aggregate, these data demonstrate that following IL-5 binding to its full-length receptors, βc is cleaved by the proteasomes and endocytosed, and then the truncated receptor complex is degraded in the lysosomes.
Reduced βc protein expression results in heterotypic desensitization of cytokine signaling.
Since total βc levels are reduced after IL-5 stimulation, we predicted that a physiologic consequence of this effect would be decreased cellular responsiveness to further stimulation by any βc-ligating cytokine. To test our hypothesis, cells were cytokine-starved for 24 hours, then either left unstimulated or stimulated with IL-5 for 5 minutes or 1 hour, and then restimulated with IL-3 for 5 minutes (after washing away the remaining IL-5 in the culture media). Cellular responsiveness to cytokine stimulation was determined by tyrosine phosphorylation of βc. Whole-cell lysates were prepared and immunoprecipitated with anti-βc antibodies, then immunoblotted with anti-phosphotyrosine monoclonal antibody (4G10). As seen in Figure 6a (lane 4, top panel), when cells were pretreated with IL-5 for 1 hour, followed by a 5-minute pulse with IL-3, βc tyrosine phosphorylation was barely detectable compared with lane 5, which represents cells that have only been stimulated with IL-3 for 5 minutes. Furthermore, following IL-5 activation of JAK2 and STAT5, restimulation with IL-3 did not induce tyrosine phosphorylation of JAK2 and STAT5 (data not shown). Reblotting the membrane in the top panel of Figure 6a with anti-βc antibodies revealed that after 1 hour of IL-5 stimulation, total βc protein levels were reduced by about 50% and βIP was generated (Figure 6a, bottom panel, lanes 3 and 4). In addition, identical results were seen with cells that were pretreated with GM-CSF and then restimulated with IL-5 (Figure 6b). These data are consistent with a model of heterotypic desensitization in which cells recently stimulated with one βc-engaging cytokine are less responsive to further signaling by any cytokine that requires βc for signal transduction.
Figure 6.
IL-5 stimulation results in cellular refractoriness to other βc-engaging cytokines. (a) TF1 cells that had been cytokine-starved for 24 hours were unstimulated (lane 1), or were stimulated with 20 ng/ml IL-5 for 5 minutes (lane 2) or 1 hour (lane 3). After 1 hour of incubation in IL-5, cells in lane 4 were harvested, washed once in 1× PBS (to remove IL-5), and restimulated with 20 ng/ml IL-3 for 5 minutes. As a positive control (lane 5), cells were stimulated only with 20 ng/ml IL-3 for 5 minutes. Whole-cell lysates were prepared and immunoprecipitated with anti-βc monoclonal antibody and immunoblotted with monoclonal antibody 4G10 (upper panel). The membrane was stripped and reprobed with anti-βc polyclonal antibodies (bottom panel). The migration of βc and βIP are indicated. (b) Same as in a, except that cells were pretreated with 20 ng/ml GM-CSF for 5 minutes or 1 hour, washed once in 1× PBS (to remove GM-CSF), and restimulated with 20 ng/ml IL-5 for 5 minutes. As a positive control (lane 5), cells were stimulated with 20 ng/ml IL-5 for 5 minutes only.
Discussion
These data demonstrate a novel mechanism for heterotypic desensitization of the βc transmembrane receptor. In this mechanism, agonistic ligation is terminated by proteasomal degradation of the βc cytoplasmic domain, followed by endocytosis and degradation of the βc external domain by lysosomes. This model is depicted in Figure 7, which illustrates five sequential steps described in this report involving IL-5R. First, IL-5 engagement of IL-5Rα leads to the recruitment of βc to form the high-affinity IL-5R. Second, agonistic signaling pathways are activated within seconds, with phosphorylation of tyrosine and serine residues in the βc cytoplasmic domain (7, 8, 42). Third, proteasomes rapidly degrade the cytoplasmic domain of the ligated βc, thereby terminating signal transduction by elimination of the phosphorylated βc residues. This βc remnant, termed βIP, remains associated with IL-5Rα on the cell surface. Fourth, IL-5Rα and βIP are endocytosed, and fifth, they are degraded in lysosomes. It was speculated that such a model was a potential mechanism for modulation of some mammalian receptors (43), and our data confirm the existence of such a mechanism. Moreover, inhibition of proteasomal degradation of the βc cytoplasmic domain was accompanied by persistence of βc tyrosine phosphorylation with ongoing downstream signaling, as evidenced by persistence of JAK2 and STAT5 tyrosine phosphorylation. Such prolonged signaling suggests that proteasomal degradation of the βc cytoplasmic domain is an important, and possibly predominant, mechanism for rapid termination of βc signal transduction.
Figure 7.
Model of IL-5R subunit regulation following IL-5 stimulation. IL-5 engagement of IL-5Rα leads to the recruitment of βc to form the high-affinity IL-5R (step 1). This protein/protein interaction (IL-5Rα/βc/IL-5) leads to the activation of intracellular signaling pathways that result in the activation of βc by both tyrosine and serine phosphorylation (step 2). Once activated, the cytoplasmic domains of βc are rapidly degraded by proteasomes to terminate agonistic signaling, resulting in the generation of βIP/IL-5Rα/IL-5 complexes (step 3). This molecular complex is then endocytosed (step 4) and degraded in lysosomes (steps 4 and 5). In addition, IL-5 induces a reduction in IL-5Rα mRNA and protein expression, while simultaneously increasing sIL-5Rα expression (upper left corner). S-p, serine phosphorylation.
Several studies have also investigated a role for proteasomes in dampening cytokine signaling (27–29). However, the precise mechanisms by which proteasomes might mediate such effects have remained speculative. Suggested alternative models of such proteasomal modulation of cytokine receptors have included: (a) proteasomal degradation of the cytokine receptor after receptor endocytosis, as a part of proteolytic clearance of an expended receptor (21–26), (b) proteasomal degradation of cytokine receptor adapter or downstream signaling molecules, and (c) proteasomal degradation of the signaling cytoplasmic domain of the cytokine receptor prior to endocytosis for lysosomal proteolysis of the residual expended receptor. Our studies provide direct evidence for the latter mechanism, but do not exclude the possibility that the second alternative also occurs.
Further insight into the mechanisms of proteasomal modulation of cytokine receptors is also provided by the targeted proteasomal degradation of the βc, but not the IL-5Rα, cytoplasmic domain. This is consistent with the selective proteasomal degradation of only the IL-2Rβ and the IL-9Rα subunits of IL-2R and IL-9R, respectively (25). None of the previously reported proteasome-regulated receptors have been a shared cytokine receptor subunit, as βc is. Thus, the regulatory consequence of proteasomal degradation of IL-2Rβ and IL-9Rα was cytokine specific. By contrast, our studies demonstrate that the consequence of proteasomal degradation of βc is both homotypic and heterotypic desensitization to signaling by all of the βc-engaging cytokines.
Based on βIP’s molecular mass and characterization with domain-specific antibodies, the termination of proteasomal degradation of βc’s cytoplasmic domain is predicted to occur between residues 490–505. These amino acids are membrane-proximal to important regulatory sequences required for activation of the Ras/MAPK, JAK/STAT, and PI 3-K signaling pathways, including the binding/activation sites for Shc (Y-577), SHP-2 (Y-612), and STAT5 (Y-612, Y-695, and Y-750). The absence of phosphorylated tyrosines on βIP is in agreement with this predicted structure and loss of signaling capacity.
The ligand-induced activation signal that targets βc’s cytoplasmic domain for proteasomal degradation is not known. However, ubiquitination of βc suggests a ubiquitin-dependent proteasomal degradation pathway. It is possible that phosphorylation of residues in βc’s cytoplasmic domain might be a prerequisite for its ubiquitination and degradation. It is also tempting to speculate that βc’s association with IL-5Rα leads to a change in receptor conformation that exposes a previously masked ubiquitination site that allows for proteasomal recruitment. Indeed, coimmunoprecipitation analyses revealed that c-Cbl (44), a ubiquitin protein ligase, associates with βc after IL-5 stimulation (data not shown).
Another finding from our studies was the existence of IL-5–induced differential regulation of IL-5Rα and sIL-5Rα expression. Such differential regulation of receptor isoforms has been recently suggested for IL-5Rα (45) and for GM-CSFRα (46). The observed reduction in IL-5Rα mRNA levels is in agreement with a previous report using eosinophils (47), and may be due to decreased mRNA stability; this would account for a reduction in IL-5Rα protein expression. Alternatively, IL-5 stimulation could lead to termination or repression of IL-5Rα mRNA splicing, since this transcript is the splice product of the gene (36). In either case, the result would be additive to the reduction of IL-5Rα protein resulting from endocytic degradation of the ligated receptor.
The physiologic consequences of IL-5–induced changes in βc, IL-5Rα, and sIL-5Rα may be dependent on the differentiation stage of the responding cell. The decreased expression of IL-5Rα and βc, coupled with increased expression of sIL-5Rα, would likely provide autocrine downregulation of IL-5 signaling during cellular differentiation. Proteasomal degradation of activated βc receptors would also provide a mechanism that enables a cell that expresses receptors to more than one of the three βc-ligating cytokines (IL-5, IL-3, and GM-CSF) to limit paracrine cytokine responsiveness. Such paracrine regulation may be crucial for enabling progenitor cells to differentiate along divergent pathways, and may be a common mechanism used by other shared cytokine receptor subunits for heterotypic desensitization.
Acknowledgments
This research was supported in part by grants from NIH (AI-36936 and AI-07495), an American Academy of Allergy, Asthma and Immunology Research Fellowship Award (M. Martinez-Moczygemba), the State of Texas Advanced Technology Program (grant no. 004949-0180-1999), the Cullen Foundation, the Mendenhall Foundation, and the Trammell Foundation. We appreciate the advice and manuscript review by Richard Cook, Steve Elledge, Mark Entman, Jeff Rosen, and David Tweardy.
References
- 1.Metcalf D. Hematopoietic regulators: redundancy or subtlety? Blood. 1993;82:3515–3523. [PubMed] [Google Scholar]
- 2.Arai KI, et al. Cytokines: coordinators of immune and inflammatory responses. Annu Rev Biochem. 1990;59:783–836. doi: 10.1146/annurev.bi.59.070190.004031. [DOI] [PubMed] [Google Scholar]
- 3.Tavernier J, et al. A human high affinity interleukin-5 receptor (IL-5R) is composed of an IL-5-specific α chain and a β chain shared with the receptor for GM-CSF. Cell. 1991;66:1175–1184. doi: 10.1016/0092-8674(91)90040-6. [DOI] [PubMed] [Google Scholar]
- 4.Kitamura T, Sato N, Arai K, Miyajima A. Expression cloning of the human IL-3 receptor cDNA reveals a shared β subunit for the human IL-3 and GM-CSF receptors. Cell. 1991;66:1165–1174. doi: 10.1016/0092-8674(91)90039-2. [DOI] [PubMed] [Google Scholar]
- 5.Hayashida K, et al. Molecular cloning of a second subunit of the receptor for human granulocyte-macrophage colony-stimulating factor (GM-CSF): reconstitution of a high-affinity GM-CSF receptor. Proc Natl Acad Sci USA. 1990;87:9655–9659. doi: 10.1073/pnas.87.24.9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carr PD, et al. Structure of the complete extracellular domain of the common beta subunit of the human GM-CSF, IL-3, and IL-5 receptors reveals a novel dimer configuration. Cell. 2001;104:291–300. doi: 10.1016/s0092-8674(01)00213-6. [DOI] [PubMed] [Google Scholar]
- 7.Adachi T, Alam R. The mechanism of IL-5 signal transduction. Am J Physiol. 1998;275:C623–C633. doi: 10.1152/ajpcell.1998.275.3.C623. [DOI] [PubMed] [Google Scholar]
- 8.DeGroot RP, Coffer PJ, Koenderman L. Regulation of proliferation, differentiation and survival by the IL-3/IL-5/GM-CSF receptor family. Cell Signal. 1998;9:619–628. doi: 10.1016/s0898-6568(98)00023-0. [DOI] [PubMed] [Google Scholar]
- 9.Yamada T, et al. IL-3, IL-5, granulocyte-macrophage colony-stimulating factor receptor alpha-subunit, and common beta-subunit expression by peripheral leukocytes and blood dendritic cells. J Allergy Clin Immunol. 1998;101:677–682. doi: 10.1016/S0091-6749(98)70177-0. [DOI] [PubMed] [Google Scholar]
- 10.Denburg JA, et al. Regulation of IL-5 and IL-5 receptor expression in the bone marrow of allergic asthmatics. Int Arch Allergy Immunol. 1999;118:101–103. doi: 10.1159/000024040. [DOI] [PubMed] [Google Scholar]
- 11.Miltiti S, et al. Expression of interleukin-3 and granulocyte-macrophage colony-stimulating factor receptor common chain βc, βIT in normal haematopoiesis: lineage specificity and proliferation-independent induction. Br J Haemtol. 2000;111:441–451. doi: 10.1046/j.1365-2141.2000.02348.x. [DOI] [PubMed] [Google Scholar]
- 12.Geijsen N, et al. Cytokine-specific transcriptional regulation through an IL-5Rα interacting protein. Science. 2001;293:1136–1138. doi: 10.1126/science.1059157. [DOI] [PubMed] [Google Scholar]
- 13.Bone H, Dechert U, Jirik F, Schrader JW, Welham MJ. SHP1 and SHP2 protein-tyrosine phosphatases associate with βc after interleukin-3-induced tyrosine phosphorylation. J Biol Chem. 1997;272:14470–14476. doi: 10.1074/jbc.272.22.14470. [DOI] [PubMed] [Google Scholar]
- 14.Pazdrak K, Adachi T, Alam R. Src homology 2 protein tyrosine phosphatase (SHPTP2)/Src homology 2 phosphatase 2 (SHP2) tyrosine phosphatase is a positive regulator of the interleukin 5 receptor signal transduction pathways leading to the prolongation of eosinophil survival. J Exp Med. 1997;186:561–568. doi: 10.1084/jem.186.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yi T, Mui AL, Krystal G, Ihle JN. Hematopoietic cell phosphatase associates with the interleukin-3 (IL-3) receptor beta chain and down-regulates IL-3-induced tyrosine phosphorylation and mitogenesis. Mol Cell Biol. 1993;13:7577–7585. doi: 10.1128/mcb.13.12.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshimura A, et al. A novel cytokine-inducible gene CIS encodes an SH-2-containing protein that binds to tyrosine-phosphorylated interleukin 3 and erythropoietin receptors. EMBO J. 1995;14:2816–2826. doi: 10.1002/j.1460-2075.1995.tb07281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Endo TA, et al. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 18.Starr R, et al. A family of cytokine-inducible inhibitors of signaling. Nature. 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 19.Naka T, et al. Structure and functions of a new STAT-induced STAT inhibitor. Nature. 1997;387:924–929. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 20.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 21.Miyazawa K, et al. Ligand-dependent polyubiquitination of c-kit gene product: a possible mechanism of receptor down modulation in M07e cells. Blood. 1994;83:137–145. [PubMed] [Google Scholar]
- 22.Mori S, Tanaka K, Omura S, Saito Y. Degradation process of ligand-stimulated platelet-derived growth factor β-receptor involves ubiquitin-proteasome proteolytic pathway. J Biol Chem. 1995;270:29447–29452. doi: 10.1074/jbc.270.49.29447. [DOI] [PubMed] [Google Scholar]
- 23.Jeffers M, Taylor GA, Weidner KM, Omura S, Vande Woude GF. Degradation of the Met tyrosine kinase receptor by the ubiquitin-proteasome pathway. Mol Cell Biol. 1997;17:799–808. doi: 10.1128/mcb.17.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Kerkhof P, Govers R, Alves dos Santos CM, Strous GJ. Endocytosis and degradation of the growth hormone receptor are proteasome-dependent. J Biol Chem. 2000;275:1575–1580. doi: 10.1074/jbc.275.3.1575. [DOI] [PubMed] [Google Scholar]
- 25.Yen C-H, et al. Involvement of the ubiquitin-proteasome pathway in the degradation of nontyrosine kinase-type cytokine receptors of IL-9, IL-2, and erythropoietin. J Immunol. 2000;165:6372–6380. doi: 10.4049/jimmunol.165.11.6372. [DOI] [PubMed] [Google Scholar]
- 26.Yu A, Malek TR. The proteasome regulates receptor-mediated endocytosis of interleukin-2. J Biol Chem. 2001;276:381–385. doi: 10.1074/jbc.M007991200. [DOI] [PubMed] [Google Scholar]
- 27.Yu C, Burakoff SJ. Involvement of proteasomes in regulating Jak-STAT pathways upon interleukin-2 stimulation. J Biol Chem. 1997;272:14017–14020. doi: 10.1074/jbc.272.22.14017. [DOI] [PubMed] [Google Scholar]
- 28.Callus BA, Mathey-Prevot B. Interleukin-3-induced activation of the JAK/STAT pathway is prolonged by proteasome inhibitors. Blood. 1998;91:3182–3192. [PubMed] [Google Scholar]
- 29.Verdier F, et al. Proteasomes regulate erythropoietin receptor and signal transducer and activator of transcription 5 (STAT5) activation. Possible involvement of the ubiquitinated Cis protein. J Biol Chem. 1998;273:28185–28190. doi: 10.1074/jbc.273.43.28185. [DOI] [PubMed] [Google Scholar]
- 30.Aman MJ, et al. CIS associates with the interleukin-2 receptor β chain and inhibits interleukin-2-dependent signaling. J Biol Chem. 1999;274:30266–30272. doi: 10.1074/jbc.274.42.30266. [DOI] [PubMed] [Google Scholar]
- 31.Kitamura T, et al. Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3, or erythropoietin. J Cell Physiol. 1989;140:323–334. doi: 10.1002/jcp.1041400219. [DOI] [PubMed] [Google Scholar]
- 32.Dickason RR, Huston DP. Creation of a biologically active interleukin-5 monomer. Nature. 1996;379:652–655. doi: 10.1038/379652a0. [DOI] [PubMed] [Google Scholar]
- 33.Gale RE, Freeburn RW, Khwaja A, Chopra R, Linch DC. A truncated isoform of the human β chain common to the receptors for GM-CSF, IL-3, and IL-5 with increased mRNA expression in some patients with acute leukemia. Blood. 1998;91:54–63. [PubMed] [Google Scholar]
- 34.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 35.Takaki S, et al. Molecular cloning and expression of the murine interleukin-5 receptor. EMBO J. 1990;9:4367–4374. doi: 10.1002/j.1460-2075.1990.tb07886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavernier J, et al. Molecular basis of the membrane-anchored and two soluble isoforms of the human interleukin 5 receptor α subunit. Proc Natl Acad Sci USA. 1992;89:7041–7045. doi: 10.1073/pnas.89.15.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Devos R, et al. Recombinant soluble human interleukin-5 (hIL-5) receptor molecules: cross-linking and stoichiometry of binding to IL-5. J Biol Chem. 1993;268:6581–6587. [PubMed] [Google Scholar]
- 38.Brown PM, et al. Epitope-labeled soluble human interleukin-5 (IL-5) receptors. J Biol Chem. 1995;270:29236–29243. doi: 10.1074/jbc.270.49.29236. [DOI] [PubMed] [Google Scholar]
- 39.Monahan J, et al. Attenuation of IL-5-mediated signal transduction, eosinophil survival, and inflammatory mediator release by a soluble human IL-5 receptor. J Immunol. 1997;159:4024–4034. [PubMed] [Google Scholar]
- 40.Thomsen P, Rudenko O, Berezin V, Norrild B. The HPV-16 E5 oncogene and bafilomycin A1 influence cell motility. . Biochim Biophys Acta. 1999; 1452:285–295. doi: 10.1016/s0167-4889(99)00132-9. [DOI] [PubMed] [Google Scholar]
- 41.Moore RH, et al. Agonist-induced sorting of human beta2-adrenergic receptors to lysosomes during downregulation. J Cell Science. 1999;112:329–338. doi: 10.1242/jcs.112.3.329. [DOI] [PubMed] [Google Scholar]
- 42.Stomski FC, et al. Identification of a 14-3-3 binding sequence in the common β chain of the granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin-3 (IL-3), and IL-5 receptors that is serine-phosphorylated by GM-CSF. Blood. 1999;94:1933–1942. [PubMed] [Google Scholar]
- 43.Hicke L. Getting down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 1999;9:107–112. doi: 10.1016/s0962-8924(98)01491-3. [DOI] [PubMed] [Google Scholar]
- 44.Anderson SM, Burton EA, Koch BL. Phosphorylation of Cbl following stimulation with interleukin-3 and its association with Grb2, Fyn, and phosphatidylinositol 3-kinase. J Biol Chem. 1997;272:739–745. doi: 10.1074/jbc.272.2.739. [DOI] [PubMed] [Google Scholar]
- 45.Tavernier J, et al. Interleukin 5 regulates the isoform expression of its own receptor alpha-subunit. Blood. 2000;95:1600–1607. [PubMed] [Google Scholar]
- 46.Heaney ML, Vera JC, Raines MA, Golde GW. Membrane-associated and soluble granulocyte/macrophage-colony-stimulating factor receptor α subunits are independently regulated in HL-60 cells. Proc Natl Acad Sci USA. 1995;92:2365–2369. doi: 10.1073/pnas.92.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang P, et al. Selective inhibition of IL-5 receptor alpha-chain gene transcription by IL-5, IL-3, and granulocyte-macrophage colony-stimulating factor in human blood eosinophils. J Immunol. 1998;160:4427–4432. [PubMed] [Google Scholar]