Abstract
Hematopoietic stem/progenitor cells (HSPCs) possess the potentials of self-renewal, proliferation, and differentiation toward different lineages of blood cells. These cells not only play a primordial role in hematopoietic development but also have important clinical application. Characterization of the gene expression profile in CD34+ HSPCs may lead to a better understanding of the regulation of normal and pathological hematopoiesis. In the present work, genes expressed in human umbilical cord blood CD34+ cells were catalogued by partially sequencing a large amount of cDNA clones [or expressed sequence tags (ESTs)] and analyzing these sequences with the tools of bioinformatics. Among 9,866 ESTs thus obtained, 4,697 (47.6%) showed identity to known genes in the GenBank database, 2,603 (26.4%) matched to the ESTs previously deposited in a public domain database, 1,415 (14.3%) were previously undescribed ESTs, and the remaining 1,151 (11.7%) were mitochondrial DNA, ribosomal RNA, or repetitive (Alu or L1) sequences. Integration of ESTs of known genes generated a profile including 855 genes that could be divided into different categories according to their functions. Some (8.2%) of the genes in this profile were considered related to early hematopoiesis. The possible function of ESTs corresponding to so far unknown genes were approached by means of homology and functional motif searches. Moreover, attempts were made to generate libraries enriched for full-length cDNAs, to better explore the genes in HSPCs. Nearly 60% of the cDNA clones of mRNA under 2 kb in our libraries had 5′ ends upstream of the first ATG codon of the ORF. With this satisfactory result, we have developed an efficient working system that allowed fast sequencing of 32 full-length cDNAs, 16 of them being mapped to the chromosomes with radiation hybrid panels. This work may lay a basis for the further research on the molecular network of hematopoietic regulation.
Hematopoietic stem/progenitor cells (HSPCs) are a group of immature blood cells capable of differentiating into all types of cells found in the peripheral blood. HSPCs can proliferate extensively so that a limited number of these cells give rise to millions of mature cells that are released to the circulation every day. HSPCs also are able to self-renew, allowing a small amount of these “seed” cells to repopulate a bone marrow transplant recipient (1–3). Although it is not yet possible to directly identify HSPCs, these cells are known to be contained in the mononucleated cells in hematopoietic tissues and are highly enriched in populations with characteristic immunophenotype, namely cells expressing cluster of differentiation 34 (CD34+) (4). By using a panel of differentiation markers, CD34+ cells can be divided further into different groups, the CD34+CD38− or CD34+thy1+lin− population being considered pluripotent stem cells whereas those of CD34+CD38+ are considered progenitors committed to a given orientation of differentiation. Functionally, HSPCs can be assessed by their ability to reconstitute the marrow function in vivo or to form colonies corresponding to different stages of differentiation and/or distinct lineages in in vitro culture system. Several sources exist for CD34+ cells, such as bone marrow, umbilical cord blood, and peripheral blood after mobilization with cytokines and/or chemotherapy. CD34+ cells account for ≈1–2% of human bone marrow cells and ∼1% of umbilical cord blood mononuclear cells (5). There are no major functional differences between HSPCs from different origins (6, 7). Clinically, in addition to classical bone marrow transplantation, peripheral blood HSPCs have been used increasingly in recent years. Meanwhile, umbilical cord blood HSPC transplantation has been applied successfully in pediatric patients and, more recently, also in adult cases (8–10). In view of the easy access and collection from a large population, attempts are being made in several countries to establish banks of HSPCs from umbilical cord blood (11). Although HSPC transplantation now is carried out mainly for hematological disorders, its potential use in gene therapy for genetic diseases has been proposed. Therefore, HSPC, in addition to playing an essential role in the hematopoietic development that has been known for decades, may take an important place in a broader area of clinical medicine.
Like any specified tissue and cell population in the human body, the biological features of HSPCs are determined largely at the level of gene expression. Sequencing of cDNA libraries to generate large numbers of partial cDNA sequences [or expressed sequence tags (ESTs)], combined with bioinformatics, has been shown to be a useful way to evaluate the gene expression profile in a given tissue, to compare transcription levels of many genes between different tissues or cells, and to identify novel genes (12–18). Characterization of the gene expression patterns in CD34 cells helps to understand the molecular basis of the function and the developmental regulation of these cells as well as the pathogenesis of hematological diseases deriving from the same cells. The present report is on the gene expression profile of CD34+ cells based on the analysis of 9,866 ESTs as well as the preliminary result of an attempt to clone, in a relatively efficient way, full-length cDNA clones for newly identified genes.
MATERIALS AND METHODS
Sources of Cells and RNA Preparation.
Umbilical cord blood samples (643 samples) collected within 2–4 hours after birth were obtained from five hospitals in Shanghai. Mononuclear cells were isolated by density gradient centrifugation using a lymphocytes separation medium. CD34+ cell populations were isolated by passing twice through the magnetic activated cell sorting separation column with CD34+ mAb-conjugated magnetic beads (Miltenyi Biotec, Auburn, CA). Total RNA was prepared from CD34+ cells by using TRIzol reagent (GIBCO/BRL). Total RNA (≈150 μg) was extracted from 2 × 108 CD34+ cells.
cDNA Library Construction.
cDNA libraries were constructed by using PCR-based protocol. In brief, cDNA synthesis was performed with a CapFinder PCR cDNA library construction kit (CLONTECH) by using 1–2 μg total RNA, followed by 20–24 rounds of PCR amplification and inserted into Lambda ZAP II vector (Stratagene). After construction, cDNA libraries were excised en masse and rescued as phagemid.
Template Preparation and Sequencing.
Most of the sequencing templates, namely the double-stranded plasmids, were made by 96-well REAL prep 96 plasmid kit (Qiagen, Chatsworth, CA). Sequencing reactions were performed on a 9600 thermal reactor (Perkin–Elmer) by using a Dye Primer Cycle Sequencing Kit (Perkin–Elmer) with the −21M13 or M13Rev primers. Because the CapFinder cDNA library was cloned nondirectionally with regard to a cloning site, EST sequences were generated from either the 5′ or 3′ end of cDNA clones. In the cases of full-length cDNA sequencing, home-made oligonucleotides were synthesized on a 394 DNA Synthesizer (Perkin–Elmer) to drive the sequencing reaction. Reaction products were electrophoresed on a 377 DNA sequencer (Perkin–Elmer), and raw sequence data were automatically recorded.
Data Management and Bioinformatics Analysis.
factura software (Perkin–Elmer) and blast analysis were used to remove vector sequence from the ESTs and to identify “trash” sequences, defined as sequences from bacterial DNA, sequences from primer polymers, sequences containing >3% of ambiguous bases (N), or sequences <100 bp long. All sequence data were preserved on record tape. An in-house database for EST sequences generated from CD34+ cell libraries was established. These sequences were searched against GenBank and dbEST databases (Release 103.0) for homology comparison by using blast and fasta in the Genetics Computer Group program package.
Classification of ESTs Corresponding to Known Genes.
EST sequences were considered part of known genes if they shared at least 95% homology over at least 100 bp of DNA sequence on blast search. These ESTs were divided into eight categories according to the functions of corresponding genes as proposed in the literature (16) (see also Table 1).
Table 1.
Classification and distribution of 855 known genes and 158 EST-USSs homologous to known genes according to their functions
| Gene categories | Known genes (%) | EST-USSs homologous to known genes (%)* |
|---|---|---|
| (1) Hematopoiesis associated | 70 (8.2) | 2 (1.3) |
| (2) Cell division | 72 (8.4) | 12 (7.6) |
| (3) Cell signaling | 91 (10.6) | 9 (5.7) |
| (4) Cell structure/mobility | 40 (4.7) | 15 (9.5) |
| (5) Cell/organism defense | 39 (4.6) | 10 (6.3) |
| (6) Gene/protein expression | 224 (26.2) | 39 (24.7) |
| (7) Metabolism | 154 (18.0) | 48 (30.4) |
| (8) Unclassified | 165 (19.3) | 23 (14.5) |
| Total | 855 | 158 |
The provisionally functional classification was based on the homology of their structures to the known genes.
Further Analysis of ESTs Corresponding to Novel Genes.
For those ESTs representing previously unknown genes, GenBank and SWISSPROT searches were performed by using blast and fasta. Homology was considered meaningful if ESTs shared at least 50% homology over 100–200 bp of DNA or if their deduced protein sequences displayed meaningful structural similarity to the known genes. For those ESTs without significant homology, motif search was carried out by using Blocks database (version 9.0; http://www. blocks. fhcrc. org) (19).
Full-length cDNA Cloning.
Among ESTs corresponding to previously uncharacterized genes, 32 were chosen for further characterization. The following two methods were combined to facilitate the full-length cDNA cloning. The first was “silico cloning” using dbEST information (20). Starting from the known sequences obtained in the present work, overlapping EST sequences were assembled into contigs to obtain the ORF. Reverse transcription–PCR and sequencing were carried out if sequence ambiguity existed in these contigs. The second method was rapid amplification of cDNA ends (RACE) (21) performed by using a protocol of CLONTECH. Marathon ready human bone marrow and brain cDNAs were used as template for RACE. CLONTECH anchor primers and homemade gene-specific primers were used for the RACE PCR.
Radiation Hybrid (RH) Mapping of Full-Length cDNA Clones.
RH technique was used for chromosome mapping of the genes cloned (22). The test kit, G3 and G4 panels (Research Genetics, Huntsville, AL), contains 83 and 93 hybrid clones of hamster and human cells, respectively, each of which retains human large chromosomal fragment broken randomly with radiation. The primers for PCR tests were synthesized as mentioned above. The result analysis kindly was provided by the Stanford Human Genome Center (http://shgc-www.stanford.edu).
RESULTS
Library Construction and General Data of ESTs from CD34+.
The purity of CD34+ cells isolated for library construction reached 96–99%, as confirmed by immunophenotyping on flow cytometry. These cells were functionally intact as assessed with colony-forming unit–granulocyte macrophage, burst-forming unit–erythroid, and colony-forming unit–megakaryocyte (data not shown). According to in vitro plating efficiency, the progenitor cells in CD34+ cells were enriched ≈50-fold from the initial mononuclear cells. To synthesize sufficient amounts of cDNA inserts while not losing the representativity of cDNA with the CapFinder system, the condition of PCR amplification was adjusted so that no discrete bands in the DNA smear were seen. The Lambda ZAP-II was chosen as a cloning system because it gave rise to better titers of plaque forming units (0.2–0.5 × 106) and better sequencing templates in our hands. From this CD34+ cell cDNA library, 12,523 clones randomly picked were sequenced partially from one end by using M13 forward primer. Many sequences (9,866) were considered good ones whereas 2,657 were considered “trash.” The rate of successful sequences was thus 78.8%, which, to our knowledge, is among the best in the literature. Analysis of the 9,866 ESTs of satisfactory quality revealed four groups of sequences. Group I (4,697 ESTs, 47.6%) showed identity to the sequences in GenBank and should be considered labels of known genes, group II (2,603 ESTs, 26.4%) matched well to the EST sequences in the public domain database (dbEST), group III (1,415 ESTs, 14.3%) exhibited no significant homology to known genes or known ESTs in the public database and were thus defined as novel ESTs, and group IV (1,151 ESTs, 11.7%) were of either mitochondrial DNA, ribosomal RNA, or repetitive (Alu or L1) sequences. It should be pointed out that sequence redundancy, as defined by sequences with identical 5′ or 3′ ends, existed in our ESTs, probably as a result of the PCR amplification of cDNA inserts and/or clonal amplification during in vivo excision of the phage libraries. Hence, the ESTs of groups I, II, and III could be assembled into 2,201, 1,561, and 881 species, respectively, which are designated as unique sequence species (USSs). However, the distribution of the redundant sequences was relatively even because, in group I, only 54 (6.3%) USSs had a copy number of 3 or >3 (the highest copy number being 10.2), suggesting that the representativity of the library was not significantly affected by PCR amplification.
Gene Expression Profile in CD34+ cells.
After integration of overlapping sequences or sequences corresponding to different portions of the same gene, the group I (4,697 ESTs or 2,201 USSs) actually represented 855 known genes, of which the distribution into eight different functional categories is summarized in Table 1. A complete list of these genes will be available on the website at shgc.stc.sh.cn (the Shanghai Human Genome Center). The categories with the highest EST numbers are those related to gene/protein expression, metabolism, and unclassified, similar to EST databases previously generated from other tissues (16). The relative expression levels of these genes could be estimated according to the number of EST copies per gene. Thus, 37 genes (4.4%) were considered highly expressed because they possessed >20 ESTs, 148 genes with 6–20 ESTs were considered genes of intermediate level expression, and 670 genes with 1–5 ESTs were considered genes with low level expression. Among the 37 highly expressed genes, 27 (73.0%) were reported as ubiquitously expressed in most human tissues (16).
Table 2 shows 70 known genes (8.2% of the total identified in this work) that have the functions restricted to, or associated with, hematopoiesis and their relative expression level as reflected by the number of ESTs. These 70 genes could be divided further into several subgroups, such as differentiation antigens, cytokines, receptors and signal transduction molecules, transcription factors, and genes involved in lymphoid, erythroid, granulocytic, and megakaryocytic differentiation. Among the most abundant genes were those coding for γ and β globin, CDw52, and major histocompatibility complex proteins homologous to chicken B complex. It is worth noting that clusters of differentiation expressed were either early hematopoietic differentiation antigens such as CD34 or antigens expressed in multiple lineages, such as CD37, CD43, CD44, CD45, CDw52, CD53, CD59, CD62L, CD69, and CD82 whereas no expression of lineage specific antigens (for example, CD4, CD8, and CD3 for T cells or CD19, CD20, and CD22 for B cells) was found. Another group of differentiation antigens expressed includes the molecules of HLA class II antigens, known to be expressed in hematopoietic precursors of fetal origin.
Table 2.
ESTs for genes associated with hematopoiesis
| Gene classification | EST copy number |
|---|---|
| Differentiation antigen | |
| CD31 | 2 |
| CD34=glycoprotein expressed in lymphohematopoietic progenitor cells | 1 |
| CD37 | 1 |
| CD43 (sialophorin) | 3 |
| CD44 | 1 |
| CD45 (leukocyte common antigen T200) | 1 |
| CDw52 | 16 |
| CD53 | 1 |
| CD59 | 1 |
| CD62L (Leu-8, leukocyte adhesion molecule-1) | 6 |
| CD69 | 3 |
| CD82 | 4 |
| DC class II histocompatibility antigen α chain | 1 |
| Fragment for class II histocompatibility antigen β chain (pII-β-3) | 2 |
| HLA class II SB-3 β chain | 6 |
| HLA-DR-α | 11 |
| HLA-DR antigens associated invariant chain (p33) | 20 |
| HLA-F gene for human leukocyte antigen F | 1 |
| Membrane-associated protein (HEM-1) | 1 |
| Ia-associated invariant γ chain | 10 |
| LPAP protein | 1 |
| MHC protein homologous to chicken B complex | 31 |
| MHC class II HLA-SB-β chain mRNA (dr4, w6) | 3 |
| MHC class II HLA-DPw4b-β chain | 1 |
| RING6 mRNA for HLA class II α chain-like | 2 |
| Bone-marrow proteoglycan | 2 |
| Hematopoietic proteoglycan core protein | 4 |
| HS1 gene for hematopoietic lineage cell specific | 1 |
| Cytokine | |
| Allograft inflammatory factor-1 | 5 |
| Angiopoietin-1 | 1 |
| IK factor | 2 |
| Interleukin-2 | 1 |
| Macrophage inflammatory protein (G0S19-1) | 5 |
| Monocyte chemotactic protein-3 | 1 |
| Monocyte-derived neutrophil-activating protein | 2 |
| Natural killer cell enhancing factor A | 6 |
| Natural killer cell enhancing factor B | 1 |
| Putative cytokine 21 (HC21) | 4 |
| Receptor and signaling | |
| BCR | 1 |
| Fc-epsilon-receptor γ chain | 4 |
| FcERI γ chain | 1 |
| IgG Fc receptor hFcRn | 3 |
| IgE receptor beta chain | 2 |
| Hematopoietic progenitor kinase | 1 |
| Interleukin 2 receptor γ chain | 14 |
| Lymph node homing receptor | 5 |
| CD87 (urokinase-type plasminogen receptor) | 3 |
| Transcription regulation | |
| AF1q | 3 |
| GATA-2 | 1 |
| Nuclear factor erythroid 2 isoform f (basic leucine zipper protein) | 1 |
| STAT5 | 2 |
| Lymphoid differentiation | |
| Cyclophilin (T-cell) | 13 |
| Immunoglobulin heavy chain variable region V3-9P | 1 |
| κ light chain | 1 |
| ω light chain protein 14.1 | 4 |
| Recombination activating protein | 1 |
| T cell receptor β chain (germline) | 1 |
| T cell receptor γ V region 5 | 1 |
| T cell receptor germline γ chain | 1 |
| TCR gene for β chain leader protein (V β 8.1) | 1 |
| Erythroid differentation | |
| β-globin | 10 |
| G-γ-globin | 30 |
| A-γ-globin | 13 |
| Granulocytic differentiation | |
| Defensin | 2 |
| Leukotriene A-4 hydrolase | 2 |
| Myeloperoxidase | 5 |
| Neutrophil gelatinase associated lipocalin | 14 |
| Megakaryocytic differentiation | |
| Platelet activating factor acetylhydrolase IB γ subunit | 6 |
| Others | |
| B4-2 protein | 5 |
| Plasminogen activator inhibitor 2 | 1 |
Bioinformatics Analysis of USSs of Unknown Genes.
The 2,603 ESTs of group II could be assembled further into 1,561 USSs whereas the 1,415 ESTs of group III could be assembled into 881 USSs. blast and fasta analysis, as well as motif search, showed that 127 USSs (8.1%) of group II and 31 USSs (3.5%) of group III exhibited homology to the sequences of known genes or protein functional domains. These 158 USSs, also classified into eight groups (Table 1), provide a good basis for molecular cloning of novel genes with functional hint.
Full-length cDNA Cloning of Novel Genes Expressed in CD34+ Cells.
Getting the complete 5′ end is the most difficult step in full-length cDNA cloning. To evaluate the efficacy of cDNA cloning by using the 5′ cap adopted in the construction of CD34+ cell cDNA library, we analyzed 1,261 ESTs sequenced from the 5′ extremity and corresponding to 112 known genes from our libraries. The 5′ part was considered complete when the first nucleotide of EST was within 10 bp or upstream of the published initiation site of transcription. The results showed that the chance for ESTs to reach the 5′ end was determined mainly by the sizes of mRNA because, for mRNAs <1 kb and between 1 and 2 kb, the percentages of the ESTs having initiation site of transcription were 40.9% and 55.1%, respectively, whereas for genes with mRNAs longer than 2 kb, only 2.8% of ESTs fell into this group (Table 3). When the initiation site of translation (the first ATG of ORF) was searched, the percentages of ESTs containing the first ATG for genes with mRNA of <1 kb, 1–2 kb, and >2 kb were 57.8%, 67.6%, and 24.6%, respectively (Table 3). On the other hand, the percentages of the genes covered by ESTs having the initiation site of transcription or the first ATG were even higher (Table 4).
Table 3.
Status of the 5′ extremity of 1261 ESTs of known genes
| mRNA size | ESTs, no. | First ATG included (%) | 5′ end reached (%)* |
|---|---|---|---|
| <1 kb | 735 | 425 (57.8) | 301 (40.9) |
| 1–2 kb | 457 | 320 (67.8) | 252 (55.1) |
| >2 kb | 69 | 17 (24.6) | 5 (2.8) |
| Total | 1261 | 762 (60.4) | 558 (44.2) |
Defined as the first nucleotide of EST being within 10 bp or upstream of published initiation site of transcription.
Table 4.
Coverage of the 5′ end of known genes by cDNA clones
| mRNA size | Genes, no. | First ATG included (%) | 5′ end reached (%)* |
|---|---|---|---|
| <1 kb | 64 | 54 (84.3) | 37 (57.8) |
| 1–2 kb | 37 | 27 (72.9) | 22 (59.4) |
| >2 kb | 11 | 6 (54.5) | 2 (18.1) |
| Total | 112 | 87 (77.6) | 61 (54.4) |
Defined as the first nucleotide of EST being within 10 bp or upstream of published initiation site of transcription.
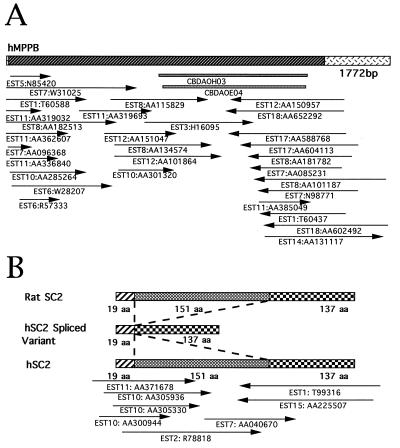
Based on the bioinformatics analysis, 32 cDNA clones with relatively high homology to known genes of functional importance have been chosen initially for full-length cloning. When the sequences from both ends of cDNA clones were examined, 26 (81%) already contained a complete ORF, confirming that our library indeed had high percentages of clones which could really be synthesized from both 3′ and 5′ ends of mRNA. Sequencing of these 26 cDNA clones was relatively rapid when conventional techniques such as subcloning, deletion using exonucleases, and primer extension were applied. RACE PCR with adapter-added cDNA pools allowed us to get the 5′ end of ORF in one of the six remaining cDNA clones whereas incorporation of the sequences from dbEST, followed by reverse transcription–PCR confirmation, helped to complete the ORF in the other five. A typical example of silico cloning of full-length cDNA (human MPPB gene) is shown in Fig. 1A. Table 5 gives the list of the 32 genes cloned and sequenced from this work, and their sequences are available in the GenBank database (see the accession numbers on Table 5). The chromosomal localizations of 16 genes have been obtained by RH technique. It is interesting to point out that the human b(2)gcn homolog gene (AF047433) was mapped by RH on chromosome 20q12 whereas an EST was labeled previously also by using RH and actually corresponded to the 3′ untranslatable region of this gene.
Figure 1.
(A) Silico cloning of the full-length cDNA of human MPPB gene. CBDAOH03 and CBDAOE04 are two clones from the CD34+ cell cDNA library. Thirty overlapping dbESTs, as indicated by arrows for their positions with respect to the cDNA structure (top bar), were assembled into a contig containing an ORF (the striated section of the bar). (B) mRNA isoforms for the human SC2 gene. The hSC2 spliced variant was identified from a clone of the CD34+ cell cDNA library whereas hSC2 sequence was obtained by dbEST assembly.
Table 5.
List of the full-length cDNA cloned and sequenced in the present work
| Accession no. | Gene name | cDNA length | Amino acids in ORF | Homology to known gene sequences | Chromosome localization |
|---|---|---|---|---|---|
| AF047433 | Human b(2)gcn homolog gene | 1112 bp | 245 aa | Fruit fly b(2)gcn gene | 20q12 |
| AF054177 | Human chd-1-like gene | 1147 bp | 220 aa | Mouse chd-1 gene | F |
| AF054174 | Human histone macroH2A1.2 | 1472 bp* | 371 aa | Rat histone macroH2A1.2 | 5q31.3–32 |
| AF054176 | Human AII/AVP-like gene | 2218 bp | 514 aa | Rat angiotensin/vasopressin receptor (AII/AVP) | 1q43–44 |
| AF038955 | Human G protein γ 5 subunit | 520 bp | 68 aa | Cattle G protein γ 5 subunit | 11q25 |
| AF038950 | Human ABC-7 gene | 2384 bp | 752 aa | Mouse ABC transporter-7 | Xq21.3 |
| AF047437 | Human FSA-1 homolog gene | 1027 bp | 293 aa | Fox sperm acrosomal protein (FSA-1) | ND |
| AF038953 | Human E25 gene | 1082 bp | 263 aa | Mouse E25 gene | ND |
| AF038958 | Human SC2 gene | 1146 bp | 308 aa | Rat SC2 = synaptic glycoprotein | ND |
| AF038959 | Human SC2 gene, short form | 629 bp | 157 aa | Rat SC2 = synaptic glycoprotein | ND |
| AF047442 | Human vesicle trafficking protein sec22b | 1462 bp | 215 aa | Mouse vesicle trafficking protein sec22b | 1q21.2–21.3 |
| AF038966 | Human SCAMP gene | 1926 bp | 338 aa | Rat SCAMP 37 | 5q13.3–14.1 |
| AF047438 | Human GOS28/P28 gene | 1012 bp | 255 aa | Rat cis-golgi GOS28 | |
| AF038957 | Human initiation factor 4e (IF4e) homolog gene | 989 bp | 236 aa | Caenorhabditis elegans peptide similar to initiation factor 4e | ND |
| AF038952 | Human cochaperonin “cofactor A” | 574 bp | 108 aa | Mouse cochaperonin “cofactor A” | ND |
| AF047440 | Human ribosomal protein L33 like gene | 512 bp | 65 aa | Escherichia coli ribosmal protein L33 and yeast L39 | 2p22 |
| AF047441 | hRPA40, isoform of hRPA39 | 1103 bp | 342 aa | Human RNA polymerase I subunit hRPA39 | ND |
| AF054186 | Human P18 gene | 840 bp | 174 aa | Rat elongation factor P18 | 6p23–25.1 |
| AF054180 | HD-ZNF1 | 1619 bp | 353 aa | Human Kruppel related zinc finger protein (HTF10) | 19p13.11–13.12 |
| AF038961 | Human SL15 gene | 1410 bp | 247 aa | Rat SL15 gene | 17p12–13.1 |
| AF047432 | Human ARF6 | 1356 bp* | 175 aa | Mouse ARF6 | ND |
| AF038954 | Human vacuolar H(+)-ATPase subunit | 1078 bp | 118 aa | Cattle vacuolar H(+)-ATPase subunit | F |
| AF038962 | HD-VDAC3 | 1414 bp | 283 aa | Mouse VDAC3 gene | 8q12–13 |
| AF047436 | Human F1Fo-ATPase synthase f subunit gene | 452 bp | 94 aa | Cattle F1Fo-ATP synthase complex f subunit | ND |
| AF047435 | Human CI-KFYI homolog gene | 419 bp | 76 aa | Cattle CI-KFYI | 4q28.2–31.1 |
| AF047434 | HD-CI-15 | 540 bp* | 106 aa | Cattle CI-15 (IP) | ND |
| AF054181 | Human CI-MNLL homolog gene | 437 bp* | 58 aa | Cattle CI-MNLL | ND |
| AF054182 | Human MPPB | 1772 bp* | 489 aa | Rat MPPB | ND |
| AF054175 | Human MP68 homolog gene | 627 bp | 58 aa | Rat 6.8kd mitochondrial proteolipid | 14q32.33 |
| AF038956 | Human GMF β homolog gene | 561 bp | 142 aa | Human glia maturation factor β | ND |
| AF038960 | Human SKD1 gene | 1978 bp | 444 aa | Mouse SKD1 | 18q21.32–21.33 |
| AF047439 | HSPC001 | 1138 bp | 300 aa | Mouse tropomyosin† | ND |
| AF054179 | Human H beta 58 homolog | 2669 bp‡ | 327 aa | Mouse H beta 58 gene | 10q21.1 |
ND, not done; F, failure.
Full-length obtained from electroextension and confirmed with reverse transcription–PCR.
Homology exists for some regions in the sequence.
Full-length obtained with RACE.
Identification of cDNA Isoforms.
The information from dbEST was not only important for extending the 5′ cDNA sequences but also was useful in finding some variation in mRNA processing. For example, a cDNA was found to be homologous to the synaptic glycoprotein (SC2) in rats (Table 5, AF038959). Nevertheless, a stretch of 453 bp encoding 151 amino acids was lacking in this sequence. Because this deletion of 453 bp was still in frame, it was possible that the cDNA isoform resulted from an alternative splicing of mRNA. When dbESTs were searched, a group of overlapping ESTs were found corresponding to the same gene. This contig had the sequence of the 453 bp missing in our clone (Fig. 1B, Table 5 AF038958), suggesting, thus, a special splicing pattern in the CD34+ cells.
DISCUSSION
Sequencing ESTs in different organs, tissues, or cells of the human body is complimentary to the genomic DNA sequencing in the human genome project (23). During the past years, tens of thousands of EST sequences were mapped on chromosomes by using RH to create sequence tagged sites, which serve both as a scaffold of a physical map and as gene labels in a transcription map (24). Currently, systematic screening of transcription units builds up the link between the structural and the functional genomics (25). Although a huge amount of EST from different tissues has been accumulated in databases, sequences from CD34+ cells are still rare, possibly because these cells are difficult to be obtained and purified. An attempt was reported at cloning novel genes that are expressed specifically in murine hematopoietic stem cells through subtractive hybridization (26). Here, we have been undertaking the EST sequencing of cDNA libraries of umbilical cord blood CD34+ cells as the first step of a long term effort to explore the genes expressed in HSPCs. A preliminary profile of gene expression in this cell population emerged based on the analysis of 9,866 ESTs. Interestingly, the proportions of EST-USSs of group I (known genes), group II (dbEST), and group III (novel sequences) were quite similar to a recent report for ESTs of cardiovascular system (27). The fact that groups I and II contained about three-fourths of all HSPC ESTs indicates that most genes with high expression level already could be identified. This indication is in line with the recent reports that over half of the human genes already were labeled by dbESTs (24, 28) and that the proportion could be even higher in the databases of some genomics industries (29). However, the poor representativity of some well known and important genes, such as erythropoietin, in dbEST suggests that completion of the list of human genes, especially those with low-level expression or temporally and/or spatially restricted expression, needs continuous efforts. Therefore, the group III ESTs, although accounting for only 14.3% of all ESTs obtained, is worth paying particular attention in the future discovery of new genes.
The expression pattern of 855 known genes in CD34+ cells, as estimated by the distribution of Group I ESTs into different functional categories, was similar to that reported for other tissues (Tables 1) (16). Homology comparison and motif searches also led to provisional, functional classification of 158 group II and group III ESTs. In addition, we observed characteristic gene expression restricted to or associated with hematopoiesis (Table 2). Among the differentiation antigen markers expressed, only early antigens and crosslineage antigens were expressed whereas lineage-specific antigen expression was absent. This is good evidence that the purified CD34+ cell population used to construct cDNA library was not contaminated by the mature lymphocytes that dominate in the mononuclear cells in umbilical cord blood. The expression of cytokines and receptors suggests regulatory pathways within this heterogeneous population whereas that of GATA-2 is in line with the transcriptional regulation in developing hematopoietic tissue. Of note, HPK1, a hematopoietic protein kinase that activates the stress-activated protein kinase pathway was found expressed at low level (30). The expression of two genes involved in leukemogenesis at a very early stem/progenitor cell level, namely BCR and AF1q, implies a possible link between the stage of differentiation and chromosomal translocations. The presence of ESTs of recombination activating gene (RAG2), Ig, and T-cell receptor β or γ chain genes corresponding to lymphoid differentiation may be a consequence of the inclusion of the lymphoid precursors in the CD34+ population. However, we also found expression of some genes with specific functions in relatively differentiated blood cells, such as the globin and myeloperoxidase. Because these proteins are not expressed in CD34+ cells, it is possible that regulatory mechanisms exist at both transcriptional and translational levels. Meanwhile, the simultaneous expression of both γ (HbF) and β (HbA) globin reflects adequately the developmental stage of erythroid differentiation at time of birth (31, 32). From this point, we speculate that the ESTs of groups II and III also should represent genes of which the expression and function are related to the developmental and differentiation stages of CD34+ cells from umbilical cord blood. The next step will be to perform the EST analysis in CD34+ CD38− cells to get insight into the gene expression profile in pluripotent hematopoietic stem cells.
EST production in large scale has laid a milestone for the discovery of novel genes whereas getting the full-length cDNA is still time-consuming work. Expression genomics is now at a turning point from ESTs to full-length cDNA cloning and sequencing (33). In this work, we have developed an efficient working system to clone full-length cDNA of novel genes by combining different approaches. Our major attempt was to use a cloning system that has high probability to get the 5′ end of cDNA. Among several systems reported in the literature (34–36), the CapFinder protocol was chosen because it also allows us to make a library with a limited amount of RNA. The actual result was quite satisfactory when ESTs for a group of 112 genes were analyzed. Among 1,261 cDNA clones corresponding to these genes, 60.4% contained sequences upstream of the initiation site of translation (first ATG of ORF). Conversely, 87 of 112 genes (77.6%) had their initiation site of translation covered by cDNA clones (Tables 3 and 4). These data are in strong contrast to those obtained from conventional cDNA libraries in which full-length clones are usually <20% (35). Indeed, obtaining the full-length cDNA sequences of novel genes in the present work was much favored in that an entire ORF was included in one cDNA clone for 26 of 32 novel genes (Table 5). One disadvantage of our system, however, could be the under-representation of cDNAs larger than 2 kb caused by PCR amplification, as evidenced by the clear difference in full-length cDNA coverages between genes whose mRNA sizes are between 0.5 and 2 kb and genes with mRNA larger than 2 kb (Tables 3 and 4). Technical innovation for selective cloning of large cDNA is thus required in the future.
Bioinformatics as a tool is of multiple interest with respect to full-length cDNA cloning. It helps to analyze massive data so that novel sequences can be quickly identified and possible structure and function can be predicted through homology comparison and motif search (19, 37, 38). As a result, ESTs with high homology to functionally important known genes then could be selected in priority for full-length cDNA cloning. Moreover, incorporation of the dbEST helped to get the 5′ end sequences in five genes and to identify mRNA isoforms of one gene we characterized. Finally, the use of adapter-ligated cDNA for RACE provides another mean to obtain the full-length cDNA, which was used in the study of one gene in the present work (Table 5). It is anticipated that the throughput of full-length cDNA cloning can be enhanced further with the improvement of existing methods and development of new tools, especially those related to the sequence assembled from dbEST. The rapid characterization of the novel genes in CD34+ populations will found the basis for the studies of gene expression/function that are essential for hematopoiesis.
Acknowledgments
The authors are grateful to Prof. Zhen-Yi Wang for his continuous support, Dr. M. D. Chen and Dr. L. Zhu for their constructive discussion, Ms. M Yu, Q. Cao, and X. Y. Su for their excellent technical assistance and all members of the Shanghai Institute of Hematology for their encouragement. This work was supported in part by SmithKline Beecham Pharmaceuticals, the Chinese High Tech Program 863, National Natural Science Foundation of China, Ministry of Public Health, and Shanghai Commission for Science and Technology.
ABBREVIATIONS
- EST
expressed sequence tag
- HSPC
hematopoietic stem/progenitor cells
- RACE
rapid amplification of cDNA end
- RH
radiation hybrid
- USS
unique sequence species
Footnotes
References
- 1.Morrison S J, Uchida N, Weissman I L. Annu Rev Cell Dev Biol. 1995;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- 2.Ogawa M. Blood. 1993;81:2844–2853. [PubMed] [Google Scholar]
- 3.Morrison S J, Wright D E, Cheshier S H, Weissman I L. Curr Opin Immunol. 1997;9:216–221. doi: 10.1016/s0952-7915(97)80138-0. [DOI] [PubMed] [Google Scholar]
- 4.Krause D S, Fackler M J, Civin C I, May W S. Blood. 1996;87:1–13. [PubMed] [Google Scholar]
- 5.D’Arena G, Musto P, Cascavilla N, Di Giorgio G, Zendoli F, Carotenuto M. Haematologica. 1996;81:404–408. [PubMed] [Google Scholar]
- 6.Almici C, Carlo-Stella C, Wagner J E, Rizzoli V. Haematologica. 1995;80:473–479. [PubMed] [Google Scholar]
- 7.Hao Q L, Shah A J, Thiemann F T, Smogorzewska E M, Crooks G M. Blood. 1995;86:3745–3753. [PubMed] [Google Scholar]
- 8.Kurtzberg J, Laughlin M, Grahan M L, Smith C, Olson J F, Casey J R, Halperin E D, Ciocci G, Carrier C, Stevens C E, et al. N Engl J Med. 1996;335:157–166. doi: 10.1056/NEJM199607183350303. [DOI] [PubMed] [Google Scholar]
- 9.Broxmeyer H, Hangoc G, Cooper S, Ribeiro R, Graves V, Yoder M, Wagner J, Vadhan-Raj S, Benninger L, Rubinstein P, et al. Proc Natl Acad Sci USA. 1992;89:4109–4113. doi: 10.1073/pnas.89.9.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairo M S, Wagner J E. Blood. 1997;90:4665–4678. [PubMed] [Google Scholar]
- 11.Stone R. Science. 1992;257:615. [Google Scholar]
- 12.Adams M D, Kelley J M, Gocayne J D, Dubnick M, Polumeropoulos M H, Xiao H, Merril C R, Wu A, Olde B, Moreno R F, et al. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 13.Okubo K, Hori N, Matoba R, Niiyama T, Fukushima A, Kojima Y, Matsubara K. Nat Genet. 1992;2:173–179. doi: 10.1038/ng1192-173. [DOI] [PubMed] [Google Scholar]
- 14.Adams M D, Kerlavage A R, Fields C, Venter J C. Nat Genet. 1993;4:256–267. doi: 10.1038/ng0793-256. [DOI] [PubMed] [Google Scholar]
- 15.Liew C C, Hwang D M, Fung Y W, Laurenssen C, Cukeman E, Tsui S, Lee C Y. Proc Natl Acad Sci USA. 1994;91:10645–10649. doi: 10.1073/pnas.91.22.10645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams M D, Kerlavage A R, Fleischmann R D, Fuldner R A, Bult C J, Lee N H, Kirkness E F, Weinstock K G, Gocayne J D, White O, et al. Nature (London) 1995;377:3–174. [PubMed] [Google Scholar]
- 17.Hillier L, Lennon G, Becker M, Bonaldo M F, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, et al. Genome Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- 18.Weinstock K G, Kirkness E F, Lee N H, Earle-Hughes J A, Venter J C. Curr Opin Biotechnol. 1994;5:599–603. doi: 10.1016/0958-1669(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 19.Henikoff S, Greene E A, Pietrokovski S, Bork P, Attwood T K, Hood L. Science. 1997;278:609–614. doi: 10.1126/science.278.5338.609. [DOI] [PubMed] [Google Scholar]
- 20.Capone M C, Gorman D M, Ching E P, Zlotnik A. J Immunol. 1996;157:969–973. [PubMed] [Google Scholar]
- 21.Chenchik A, Moqadam F, Siebert P. CLONTECHniques. 1995;X(1):5–7. [Google Scholar]
- 22.Cox D R, Burmeister M, Price E R, Kim S, Myers R M. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- 23.Strachan T, Abitbol M, Davidson D, Beckmann J S. Nat Genet. 1997;16:126–132. doi: 10.1038/ng0697-126. [DOI] [PubMed] [Google Scholar]
- 24.Schuler G D, Boguski M S, Stewart E A, Stein L D, Gyapay G, Rice K, White R E, Rodriguez-Tome P, Aggarwal A, Bajorek E, et al. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- 25.Hieter P, Boguski M. Science. 1997;278:601–602. doi: 10.1126/science.278.5338.601. [DOI] [PubMed] [Google Scholar]
- 26.Brady G, Billia F, Knox J, Hoang T, Kirsch I R, Voura E B, Hawley R G, Cumming R, Buchwald M, Siminovitch K. Curr Biol. 1995;5:909–922. doi: 10.1016/S0960-9822(95)00181-3. [DOI] [PubMed] [Google Scholar]
- 27.Hwang D M, Dempsey A A, Wang R X, Rezvani M, Barrans J D, Dai K S, Wang H Y, Ma H, Cukerman E, Liu Y Q, et al. Circulation. 1997;96:4146–4203. doi: 10.1161/01.cir.96.12.4146. [DOI] [PubMed] [Google Scholar]
- 28.Wolfsberg T G, Landsman D. Nucleic Acids Res. 1997;25:1626–1632. doi: 10.1093/nar/25.8.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haseltine W A. Sci Am. 1997;276:92–97. doi: 10.1038/scientificamerican0397-92. [DOI] [PubMed] [Google Scholar]
- 30.Kiefer F, Tibbles L A, Anafi M, Janssen A, Zanke B W, Lassam N, Pawson T, Woodgett J R, Iscove N N. EMBO J. 1996;15:7013–7025. [PMC free article] [PubMed] [Google Scholar]
- 31.Zon L I. Blood. 1995;86:2876–2891. [PubMed] [Google Scholar]
- 32.Morrison S J, Shah N M, Anderson D J. Cell. 1997;88:287–298. doi: 10.1016/s0092-8674(00)81867-x. [DOI] [PubMed] [Google Scholar]
- 33.Marshall E. Science. 1996;274:1456. doi: 10.1126/science.274.5292.1456a. [DOI] [PubMed] [Google Scholar]
- 34.Okayama H, Berg P. Mol Cell Biol. 1982;2:161–170. doi: 10.1128/mcb.2.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato S, Sekine S, Oh S-W, Kim N-S, Umezawa Y, Abe N, Yokoyama-Kobayashi M, Aoki T. Gene. 1994;150:243–250. doi: 10.1016/0378-1119(94)90433-2. [DOI] [PubMed] [Google Scholar]
- 36.Carnnci P, Kvam C, Kiramura A, Ohsumi T, Okazaki Y, Itoh M, Kamiya M, Shibata K, Sasaki N, Izawa M, et al. Genomics. 1996;37:327–336. doi: 10.1006/geno.1996.0567. [DOI] [PubMed] [Google Scholar]
- 37.Altschul S F, Boguski M S, Gish W, Wootton J C. Nat Genet. 1994;6:119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- 38.Botstein D, Cherry J M. Proc Natl Acad Sci USA. 1997;94:5506–5507. doi: 10.1073/pnas.94.11.5506. [DOI] [PMC free article] [PubMed] [Google Scholar]