Abstract
The Smith–Lemli–Opitz syndrome (SLOS) is an inborn disorder of sterol metabolism with characteristic congenital malformations and dysmorphias. All patients suffer from mental retardation. Here we identify the SLOS gene as a Δ7-sterol reductase (DHCR7, EC 1.3.1.21) required for the de novo biosynthesis of cholesterol. The human and murine genes were characterized and assigned to syntenic regions on chromosomes 11q13 and 7F5 by fluorescense in situ hybridization. Among the mutations found in patients with the SLOS, are missense (P51S, T93M, L99P, L157P, A247V, V326L, R352W, C380S, R404C, and G410S), nonsense (W151X), and splice site (IVS8–1G>C) mutations as well as an out of frame deletion (720–735 del). The missense mutations L99P, V326L, R352W, R404C, and G410S reduced heterologous protein expression by >90%. Our results strongly suggest that defects in the DHCR7 gene cause the SLOS.
The autosomal recessive Smith–Lemli–Opitz syndrome (SLOS, MIM 270400) is a remarkable example of an inborn error of metabolism that impairs morphogenesis. Phenotypic expression is variable and comprises congenital anomalies such as microcephaly, cleft palate, syndactyly of toes 2 and 3, polydactyly, and visceral malformations as well as postnatal failure to thrive and mental retardation (1). Genitalia in severely affected boys are either ambiguous or fail to become masculinized (1). A deficiency of the ultimate step of cholesterol biosynthesis is suspected to cause the SLOS because a decrease of plasma cholesterol and the accumulation of its precursor 7-dehydrocholesterol are always observed (2). Sterol metabolism depends on the tight regulation of endogenous biosynthesis and receptor mediated transport (3). Failure to remove the Δ7-bond in de novo-synthesized sterols could be caused by a deficiency of the catalytic subunit, a regulatory protein, or of a cofactor of the Δ7-sterol reductase (DHCR7). We recently cloned the cDNA encoding the catalytic subunit of the enzyme (4). Using this cDNA, we now isolated and characterized the corresponding human and mouse genes. Among the mutations found in patients with biochemically confirmed diagnosis of SLOS were ten missense, one nonsense, and one splice acceptor site mutation as well as a 16-bp deletion. Several of the missense mutations reduced heterologous protein expression by >90%. This result strongly suggests that DHCR7 defects cause the SLOS.
EXPERIMENTAL PROCEDURES
Isolation and Mapping of the Human DHCR7 Gene.
A P1 clone from a human genomic library (clone I.D. ICRFP700C2137Q06) was isolated by hybridizing a 32P-labeled human DHCR7 cDNA probe to prespotted filters obtained from the Resource Centre/Primary Database (Berlin, Germany). Intron–exon boundaries were identified by PCR amplification of intron sequences by using cDNA sequence-derived oligonucleotides and DNA sequencing. Sites for restriction enzymes were mapped by Southern hybridization of exon-specific probes to P1 clone digests and digestion of PCR-amplicons, respectively.
Cloning of the Murine cDNA and Gene.
Mouse Dhcr7 cDNAs were identified in the expressed sequence tag database with the tblastn algorithm. The clone with the longest 5′-sequence (GenBank accession no. AA003001) was sequenced entirely and found to contain a 262-bp deletion. Another clone (GenBank accession no. AA475208) as well as two reverse transcription-PCR products from mouse liver contained the deleted region that was replaced with a BamHI–XbaI fragment from clone AA475208. Vectors for heterologous expression in yeast (mD7-ORF and mD7-myc) were prepared with oligonucleotides CCGCTCGAGAAAATGGCTTCGAAATCCCAGC (introducing a XhoI site upstream of the start codon) and CCGCTCGACAGATCTTGCTTCGAAATCCCAGCAC (introducing a 5′ BglII site in frame with the 9E10 c-myc epitope cassette, ref. 4). For Northern blot analysis a 32P-labeled Dhcr7 cDNA was hybridized to mouse multiple tissue Northern blots (CLONTECH). A cosmid clone (clone I.D. MPMGc121P1353Q3) was isolated from a genomic library (strain 129Ola) by hybridizing the 32P-labeled Dhcr7 cDNA probe to prespotted filters obtained from the Resource Centre/Primary Database (Berlin, Germany).
Genetic Analysis.
All patients were diagnosed with SLOS by their physicians who provided DNA or blood samples for molecular analysis. The clinical diagnosis was biochemically confirmed by identification of 7-dehydrocholesterol in the serum by gas chromatography and mass spectrometry. Genomic DNA was prepared from cultured fibroblasts (SLO5 and SLO9) or peripheral lymphocytes. Exons 3–9 were amplified by using PCR (annealing temperature 65°C) with the oligonucleotides shown in Table 2. For single strand conformation polymorphism (SSCP) analysis, 7 μl of a PCR reaction was subjected to electrophoresis on a Mutation Detection Enhancement (FMC) gel that was stained with silver (5). PCR products were purified on QIAquick columns (Qiagen) and cycle sequenced on an ABI377 automated sequencer (Perkins–Elmer). PCR analysis of cDNA from fibroblasts with cDNA sequence-derived oligonucleotides was performed by isolating mRNA with RNAzol B (Molecular Research Center, Cincinnati, OH), and transcribing cDNA with a 1st strand cDNA synthesis kit (Pharmacia).
Table 2.
Intron specific primers for exon amplification
| Exon | Sense primer | 5′-end | Size | Antisense primer | 3′-end |
|---|---|---|---|---|---|
| 3 | GGT GGA TGC AAC AGG GAA AGG TGG | IVS2-70 | 240 | CCA AAG GCT GGA AAG CTC TGA GAC C | IVS3+66 |
| 4 | CCC AGT GTG ACT GCC TGC ATC CG | IVS3-31 | 301 | ACG CTC CCC ACC TGC TGT GTC CC | IVS4+47 |
| 5 | CTG CTA TTC GTC CCC CTT TGC AGG | IVS4-67 | 250 | GTC TTA GGG ACA AAG CAG CGC TGG G | IVS5+92 |
| 6 | AGG CAT GCT TCA GCC CAG CCA AGC | IVS5-52 | 329 | CTT TCT ACA TCA GGC TGG ACC CGC | IVS6+63 |
| 7 | TGG GCT CTC GCT AAG TAA GGT GGC | IVS6-80 | 321 | CAT CGG CGT TTC ACC CTC TCC AGC | IVS7+36 |
| 8 | TGT GAT TTC CCC GAG GTC CAT GGG | IVS7-71 | 258 | GCT TAG CAT GTG TCT GCC AAA TGC | IVS8+55 |
| 9 | CAA AGC ACC GCT TGA CCC CTT CCC | IVS8-37 | 364 | AGT AGG GCA GCA GGT GGC CAC CG | c1291as |
| 9 | CAC AGC AAG CTG CTG GTG TCG GGC | c1172s | 308 | TGG CAG AAC ACG CTC TTG ACA GC | c1480as |
Product size (bp) is given.
Fluorescence in Situ Hybridization.
Fluorescence in situ hybridization was performed as described (6) by using DNA from the genomic human P1 and the murine cosmid clone, respectively. Chromosomal in situ suppression hybridization and detection by immunofluorescence was carried out on metaphase chromosome preparations from phytohemagglutinin-stimulated normal human lymphocytes and normal mouse embryo fibroblasts with biotinylated (human) or digoxigenin-labeled (mouse) probes. Fifty of 50 human and 66 of 80 mouse metaphase cells exhibited specific labeling of one or both homologs of chromosomes 11 and 7, respectively.
In Vitro Mutagenesis and Heterologous Expression of Mutated cDNAs.
A modified wild-type (wt) DHCR7 cDNA was constructed from mycD7-ORF (4) in pBluescriptKSII (Stratagene) by introducing a SpeI restriction site with oligonucleotide GGAATTCACTAGTCCTTAGAAGATTCCAGGC. Mutations were introduced in AccI–SacI (296T>C) and SacI–SpeI (976G>T, 1054C>T, 1210C>T, 1228G>A, 1158C>T, 1272T>C, and 976T>G) cassettes by two-step mutagenesis as described (7). Second site mutagenesis was ruled out by DNA sequencing of both strands. wt and mutated cDNAs were subcloned into pCIneo (Promega) with XhoI–NotI restriction sites. Cultivation of tsA-201 cells and calcium phosphate transfection (6 μg of specific vector DNA; 9 μg of pUC18 carrier DNA/100-mm dish) were performed as described (8, 9). The amount of specific vector DNA used for transfection was saturating, and no decrease in wt cDNA expression was observed when 10-fold lower DNA concentrations were used. For cotransfection, 3 μg of each DHCR7 construct (wt and mutants), 3 μg of human emopamil-binding protein/sterol Δ8-Δ7 isomerase (10) in pCIneo, and 9 μg of pUC18 carrier DNA/100-mm dish were used. Protein concentrations were determined as described (4) with BSA as a standard.
RESULTS
Structure and Localization of the Human Gene on Chromosome 11.
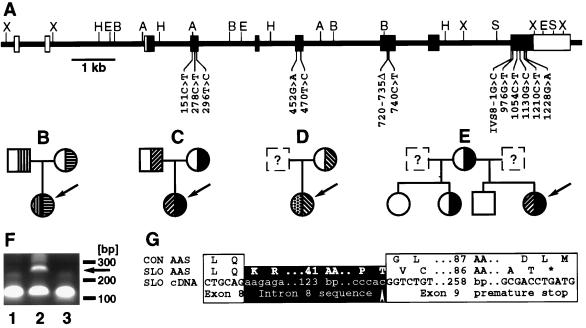
The DHCR7 cDNA (4) was used to identify a P1 clone in a filter-spotted human genomic library. Intron–exon boundaries and partial intron sequences were determined by PCR amplification of introns and sequencing. The isolated clone contained the entire ORF and the 5′- and 3′-nontranslated regions of the cDNA. Restriction enzyme digests of the P1 clone and subsequent Southern hybridization with exon-specific probes identified the restriction sites shown in Fig. 1A. The gene contains nine exons and eight introns and spans ∼14 kilobases (Fig. 1A, Table 1). Introns 4, 7, and 8 interrupt putative transmembrane segments (Fig. 2). The two first exons are noncoding, and the start codon is localized in exon 3. Exon 9 is the largest exon and contains the stop codon and the two polyadenylation sites recently described (4). The P1 clone was hybridized in situ to human metaphase chromosomes and detected by fluorescence on chromosomal region 11q13 (data not shown).
Figure 1.
Mutations in the DHCR7 gene of patients with the SLOS. (A) Structure of the gene. Noncoding (open boxes) and coding exons (filled boxes) are shown. Restriction enzyme cleavage sites (A, AccI; B, BamHI; H, HindIII; S, SpeI; X, XhoI) and mutations are indicated. (B-E) Families with SLOS. The index patient is indicated by the arrow. Heterozygotes and homozygotes for mutations in the DHCR7 gene are shown. (B) Patient SLO8; mutations R404C (▥) and A247V (▤). (C) Patient SLO1; mutations V326L (▨) and W151X (▪). (D) Patient SLO6; mutations P51S (▩) and IVS8–1G>C (▧). (E) patient SLO2; mutations V326L (▨) and W151X (▪). (F) Abnormal reverse transcription-PCR-product (arrow) from fibroblasts from patient SLO5 (lane 2) not seen in cDNA from a control person (lane 1) and patient SLO9 (lane 3). Oligonucleotides c898s (GACCACTTCGGGTGGTACCTGGGC) and c999as (GACAGCTGCACGGGGTGGTACACC) are expected to give a 101-bp product. (G) Consequences of the splice site mutation found in patient SLO5. The amino acid sequence of a spliced transcript from a control person (CON AAS) is aligned with the amino acid sequence (SLO AAS) encoded by the transcript (SLO cDNA) from patient SLO5’s fibroblasts. The mutation (arrow) and the premature stop codon (∗) are indicated.
Table 1.
Intron–exon structure of the human and murine DHCR7 genes
| Intron | Species | cDNA position | Exon sequence | 5′-intron sequence | Intron length [bp] | 3′-intron sequence | Exon sequence | Amino acid interrupted |
|---|---|---|---|---|---|---|---|---|
| 1 | H | −132 | GCGCAG | gtgagtggcc | 556 | ttccttgcag | GACTTT | — |
| 1a | M | −7 | ACTCAG | gtgagaattc | 6,600 | cttccctcag | GGCCTG | |
| 2 | H | −7 | CTGGAG | gtaagtgtat | 2,400 | tttcttgcag | GGCCCA | — |
| 1b | M | −7 | AAGAGA | gtgagtattc | 6,500 | cttccctcag | GGCCTG | |
| 3 | H | 98 | TGCCTG | gtaaggatct | 840 | gtcctcgcag | GGAGGT | W33 |
| 2 | M | 86 | TGCCTG | gtaaggggct | 1,100 | cttctctcag | GGAAGT | W29 |
| 4 | H | 321 | TTCCAG | gtcagcagcc | 1,550 | tgtgtttcag | GTGCTT | — |
| 3 | M | 309 | TTCCAG | gtcagtccac | 2,650 | tccatttcag | GTGCTG | |
| 5 | H | 412 | CTGCAG | gtagcctcct | 800 | tcatctgcag | GGGTTG | G138 |
| 4 | M | 400 | CAGCAG | gtaacccttg | 633 | ctctctgcag | GGGTTG | G134 |
| 6 | H | 626 | AGACTG | gtatgttctt | 2,000 | tacttttaag | CAAATT | C209 |
| 5 | M | 614 | AGACTG | gtaagttcct | 1,750 | tgcttttaag | CAAATT | C205 |
| 7 | H | 831 | CTGCAG | gtgcctgtca | 975 | tgtcttgcag | GCCATC | — |
| 6 | M | 819 | TTGCAG | gtaacttggt | 2,100 | tgtcttacag | GCCATC | |
| 8 | H | 963 | CTGCAG | gtgaggaggt | 1,800 | cgccccccag | GGTCTG | — |
| 7 | M | 951 | CTGCAG | gtgaggagga | 1,450 | ttccccccag | GGCCTG |
Partial intron sequences for the human (H) and murine (M) gene are shown. Reverse transcription-PCR from mouse liver RNA revealed that exons 1a and 1b are used alternatively.
Figure 2.
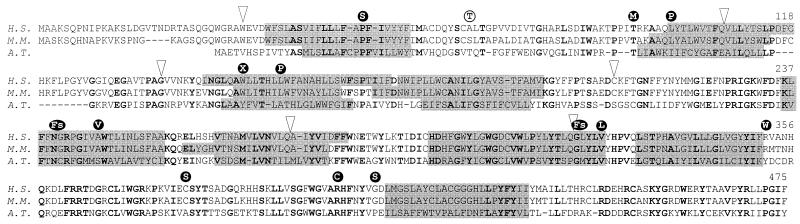
Alignment of DHCR7 amino acid sequences from man (Homo sapiens, H.S., GenBank accession no. AF034544), mouse (Mus musculus, M.M., GenBank accession no. AF057368), and cress (Arabidopsis thaliana, A.T., GenBank accession no. U49398). Identical amino acid residues are shown in bold. Putative transmembrane segments (grey boxes), introns (arrows) and mutated residues found in patients (•) and a healthy control person (○) are indicated.
Assignment of the Murine Gene to Chromosome 7.
To confirm the chromosomal assignment and as a prerequisite for future in vivo studies, we cloned the murine ortholog by screening the expressed sequence tag database for sequences with homology to the human DHCR7. The protein is 89% identical and 98% similar with the human enzyme and lacks four amino acid residues in the N terminus (Fig. 2). In contrast to the human cDNA, no second polyadenylation site was found in line with the expression of a single 2.8-kilobase transcript in liver and other murine tissues (data not shown). Heterologous expression of the mouse cDNA in yeast confirmed that it was able to catalyze 7-dehydrocholesterol to cholesterol conversion (mD7-ORF 0.56 nmol/mg/min, mD7-myc 0.30 nmol/mg/min, no activity in microsomes from mock-transformed yeast cells). The tmpredict algorithm (11) identified nine putative transmembrane segments (TMS) in the amino acid sequences of the mouse, human, and plant orthologs (Fig. 2). With the mouse cDNA, a genomic cosmid clone was isolated and characterized. Within the coding sequence, intron–exon boundaries were the same as in the human gene (Table 1). In metaphase chromosomes from mouse fibroblast, the cosmid clone hybridized to the most distal region of chromosome 7 corresponding to 7F5 (data not shown).
Mutations in the Gene from SLOS-Affected Patients.
Our search for mutations in the DHCR7 gene was focused on the coding regions in exon 3–9. Exons 3–8 and their intron–exon boundaries were analyzed entirely. To amplify the coding region of exon 9, we used oligonucleotides corresponding to the sequence of intron 8 and the cDNA sequence, respectively (Table 2). Amplicons were analyzed for SSCP and sequenced if polymorphic. Thirteen nonconsanguinous patients with the clinical signs of the SLOS and biochemical confirmation of the diagnosis (7-dehydrocholesterol serum concentrations 174-1124 μM, Table 3) were recruited in seven different German centers. All eleven patients for which clinical information was available suffered from syndactyly and/or hexadactyly, facial dysmorphias, and mental retardation.
Table 3.
Mutations found in the coding exons or intron-exon boundaries from patients with SLOS
| Pat. | 7-DHC (μM) | Sex | Exon | Nucleotide change | Effect on coding sequence |
|---|---|---|---|---|---|
| Patients with two mutations | |||||
| SLO1 | 406 | F | 6 | 452G>A§ | • W151X |
| 9 | 976G>T‡ | ○ V326L | |||
| SLO2 | 1124 | F | 6 | 452G>A§ | • W151X |
| 9 | 976G>T | ○ V326L | |||
| SLO3a* | 304 | M | 6 | 452G>A | • W151X |
| 9 | 1130G>C | C380S | |||
| SLO3b* | 207 | M | 6 | 452G>A | • W151X |
| 9 | 1130G>C | C380S | |||
| SLO4 | 260 | F | 4 | 278C>T | T93M |
| 7 | 720-735del | • Fs | |||
| SLO5† | 174 | M | 4 | 296T>C | ○ L99P |
| — | IVS8-1G>C | • Fs | |||
| SLO6 | 262 | M | 4 | 151C>T | P51S |
| — | IVS8-1G>C§ | • Fs | |||
| SLO7 | 282 | M | 6 | 452G>A‡ | • W151X |
| 6 | 470T>C§ | L157P | |||
| SLO8 | 231 | F | 7 | 740C>T§ | A247V |
| 9 | 1210C>T‡ | ○ R404C | |||
| SLO9† | 231 | F | 4 | 296T>C | ○ L99P |
| 9 | 1228G>A | ○ G410S | |||
| SLO10 | 422 | M | 9 | 976G>T‡ | ○ V326L |
| 9 | 1054C>T§ | ○ R352W | |||
| Patients with single mutations | |||||
| SLO11 | 291 | M | 9 | 976G>T | ○ V326L |
| SLO12 | 928 | F | 9 | 976G>T | ○ V326L |
| SLO13 | 330 | M | 9 | 1054C>T | ○ R352W |
7-dehydrocholesterol concentrations (7-DHC) were determined in patients’ serum by gas chromatography and mass spectrometry. Mutations that are expected to completely abolish the enzymatic activity (•; FS, frameshift) or that reduce protein expression upon heterologous expression (○) are indicated.
Siblings.
Ref. 19.
Paternal origin.
Maternal origin.
In amplicons from all 13 patients, SSCP analysis identified additional conformers. Subsequent DNA sequencing identified 13 different mutations (Fig. 1A, Table 3). Among the mutations are one nonsense mutation (W151X), ten missense mutations (P51S, T93M, L99P, L157P, A247V, V326L, R352W, C380S, R404C, and G410S) and one 16-bp deletion (720–735 del). This deletion of a GC-rich nucleotide stretch in exon 7 shifts the reading frame in the codon for N240. The G>C mutation in the last base of intron 8 in patients SLO5 and SLO6 (IVS8–1G>C) disrupts the splice acceptor site. Reverse transcription-PCR analysis demonstrated the presence of an additional PCR product in fibroblast derived cDNA from patient SLO5 (Fig. 1F). Subcloning and DNA sequencing of this PCR product revealed the use of an alternative splice acceptor site upstream from the mutated intron 8 splice site. The insertion of 134 bp of intron DNA sequence shifted the reading frame and led to a premature stop (Fig. 1G).
In ten patients, we detected two mutations as expected for an autosomal recessive disorder (Table 3). The three remaining patients were heterozygous for the mutations V326L (SLO11, SLO12) and R352W (SLO13), respectively. SSCP analysis of the coding exons 3–9 in all three patients as well as DNA sequencing of exons 3–9 in patient SLO13 failed to reveal the presumed second mutation.
The SLOS is an autosomal-recessive disorder and the mutations are expected to be in trans. To clarify whether the patients are compounds, we sequenced parental DNA and established the germline origin and the inheritance of mutations in patients SLO1, SLO7, SLO8, and SLO10 (Fig. 1 B and C, Table 3). In patients SLO6 and SLO2 (Fig. 1 D and E, Table 3), only maternal DNA was available. These two patients had inherited the nonsense mutation (W151X) or the splice acceptor site mutation (IVS8–1G>C) but not the second mutation from their mothers. Moreover, healthy siblings of patient SLO2 were analyzed (Fig. 1E). Whereas in two siblings no mutations were identified, one sister also had inherited the mutation W151X from the mother, suggesting that this girl is a healthy carrier of the SLOS. Two patients (SLO3a and SLO3b) were brothers and carried the same mutations (Table 3). Thus in four patients (SLO1, SLO7, SLO8, and SLO10), the mutations were unequivocally, and in two others (SLO2 and SLO6), the mutations were most likely in trans.
Polymorphisms and Rare Variants.
In the patients, SSCP analysis had revealed no variants in exons 3, 5, and 8. In healthy controls, these three exons were therefore not further analyzed. A variety of conformers were identified in exons 4, 6, 7, and 9 (see above). Some of them also were frequently observed among >80 alleles from healthy controls. Sequencing revealed mutations in exons 4 (189A>G 0.64, 0.36; 231C>T 0.87, 0.13; n = 80), 6 (438C>T 0.90, 0.10; n = 82), and 9 (1158C>T 0.76, 0.24; 1272T>C 0.92, 0.08; n = 84), respectively (allelic frequencies refer to conformers). No additional conformers were seen in exon 7 (n = 126). These mutations among control persons most likely represent polymorphisms because they are silent and frequent. Two mutations were found once (199G>A 0.99, 0.01; 207C>T 0.99, 0.01; n = 80) and most likely are rare variants. One of these mutations (199G>A) results in an amino acid substitution (A67T) but was not found among the patients.
Heterologous Expression of Mutated cDNAs.
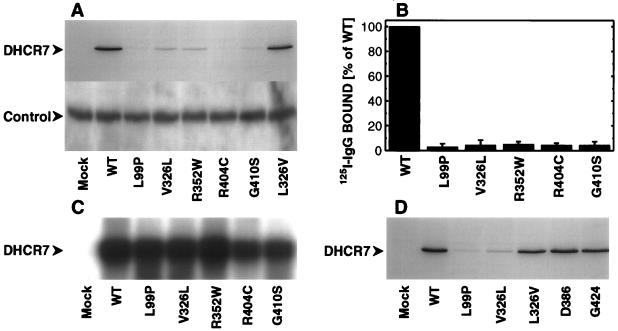
To confirm the pathogenicity of the missense mutations, we introduced the mutations L99P (296T>C), V326L (976G>T), R352W (1054C>T), R404C (1210C>T), and G410S (1228G>A) in the c-myc epitope tagged DHCR7 cDNA (4). wt and mutant cDNAs were transfected into the HEK293 (human embryonic kidney) derived cell line tsA-201 (8) to rule out improper processing upon heterologous expression in nonhuman cells. Immunoblotting revealed a decrease of c-myc immunoreactivity of the mutated proteins (Fig. 3A Upper). For none of the mutant constructs protein expression was completely abolished (not shown). Similar protein loads were verified by Coomassie staining of gels run in parallel (not shown) and by coimmunostaining endogenous sigma1-receptors (12) (Fig. 3A Lower). Reduction of c-myc immunoreactive protein was quantified and found to be 95–97% (n = 3) for all five missense mutations (Fig. 3B). Northern blot analysis revealed similar transcript levels for wt and mutant cDNAs (Fig. 3C). To control for errors during transfection, we cotransfected a second plasmid carrying the c-myc-tagged human epomamil-binding protein/sterolΔ8-Δ7 isomerase (10), which expressed at identical levels throughout mock-, wt-, and mutant-transformed cells. In contrast to wt, no DHCR7 associated 9E10 c-myc immunoreactivity was detectable in microsomes from cells that were cotransfected with the mutant cDNAs (data not shown). To rule out an artifact of in vitro mutagenesis or cloning, we introduced the silent mutations 1158C>T and 1272T>C and remutated the mutation 976G>T (V326L) back to wt sequence (976T>G). All three mutants expressed at the same level as the wt cDNA (Fig. 3 A and D).
Figure 3.
Heterologous expression of mutated cDNAs. (A) Western blot analysis of 40 μg of microsomal protein from tsA-201 cells overexpressing the wt DHCR7 cDNA and cDNAs with the mutations L99P, V326L, R352W, R404C, G410S, and L326V (mutation V326L reverted to wt sequence). (Upper) DHCR7 immunostaining with antibody 9E10 (50 ng/ml) against the c-myc epitope; (Lower) control immunostaining with anti PBP45 (12) (300 ng/ml) against the endogenous sigma1-receptor. Antibody 9E10 and anti PBP45 were visualized with purified 125I-labeled anti mouse IgG (NEN, Vienna, Austria; 35 μCi/ml; 24 h exposure) and alkaline phospatase-conjugated anti rabbit IgG (Sigma; 10 μg/ml; CDP-Star, Boehringer Mannheim), respectively. (B) Quantitative analysis of DHCR7 protein expression. Twenty or 40 μg of microsomal protein were separated on 12% (wt/vol) SDS-polyacrylamide gels and transferred to polyvinyldifluoride membranes. 9E10 c-myc immunoreactivity was quantified by membrane slicing and γ-counting. Specific immunostaining was defined as the difference between microsomes from DHCR7-transfected cells (8, 970–24,060 cpm, n = 3) and mock-transformed cells (540–3,000 cpm, n = 3). Data shown are the mean ± SD from three separate transfections. (C) Northern blot analysis of 5 μg of total RNA hybridized with 32P-labeled DHCR7 (8 h exposure). (D) Western blot analysis of 40 μg of microsomal protein from cells overexpressing the wt DHCR7 cDNA and cDNAs with the mutations L99P, V326L, and L326V (mutation V326L reverted to wt sequence) and the silent mutations 1158C>T (D386) and 1272T>C (G424). After immunostaining with mAb 9E10 (50 ng/ml), 125I-labeled anti mouse IgG was added (24-h exposure).
DISCUSSION
In vertebrates, sterols are required as major membrane constituents and precursors for the synthesis of bile acids and steroid hormones. Unexpectedly, cholesterol is also pivotal for morphogenesis. Mutations in men and mice that impair cholesterol transport, cholesterol biosynthesis, or the cholesterol requiring hedgehog-signaling pathway cause malformations (13). Patients with SLOS fail to synthesize cholesterol to a variable extent and accumulate the ultimate cholesterol precursor 7-dehydrocholesterol (2). Here, we isolate the SLOS gene and show that 10 of 13 patients with SLOS are heterozygous for two mutations that alter the amino acid sequence of the enzyme Δ7-sterol reductase (DHCR7).
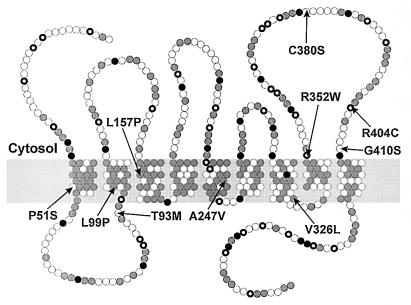
Analysis of the amino acid sequences of enzyme orthologs from man, mouse, and cress identified nine putative TMS (Fig. 2). The lamin B receptor of the nuclear membrane is not only homologous with sterol reductases (4) but also shares similar hydropathy plots (not shown). It differs from the enzymes in its long hydrophilic N terminus, which binds to DNA and lamin B in the nuclear lamina and is oriented toward the nucleosol (14). Because the inner nuclear membrane is continuous with the membrane of the endoplasmic reticulum, the nucleosolic orientation of the N terminus of the lamin B receptor implies that the N terminus of sterol reductases is oriented toward the cytosol. In the model derived from these assumptions, most of the short linkers between TMS are oriented toward the lumen of the endoplasmic reticulum whereas the domains between TMS2–3, TMS4–5, and TMS8–9 face the cytosol (Fig. 4). NADPH is an indispensable cofactor of 7-dehydrocholesterol reduction suggesting that the NADPH-binding domain is localized in the N terminus or one of the hydrophilic segments connecting TMS2–3, TMS4–5, or TMS8–9.
Figure 4.
Proposed topology model of the DHCR7 protein grey circle, hydrophobic residues; , positively charged residues; •, negatively charged residues; ○, others). Mutations identified in patients with the SLOS are indicated (arrows).
Our search for mutations in the DHCR7 gene of patients with SLOS was focused on the coding exons and their splice sites. To rule out that the mutations in exons 4, 6, 7, and 9 of the patients represent frequent variants, we also analyzed these same exons from healthy controls. In control persons, mutations occurred with a frequency of 1–36%. All but one of these mutations (199G>A; A67T) were silent. This mutation is considered to be not pathogenic because the mouse and plant isoenzymes carry serine and threonine residues in this same position (Fig. 2). Thirteen different mutations within the coding exons or the adjacent intron sequences were identified in 13 nonconsanguinous patients with the SLOS. Ten patients carried two mutations. Failure to identify the expected second mutation in three of the patients could be caused by (i) a silent mutation in the coding sequence modifying gene transcription, RNA splicing, or RNA stability, (ii) mutations in the gene regions omitted from analysis (exons 1–2 and their splice sites, the 3′ noncoding sequence of exon 9 and intron sequences), or (iii) partial gene deletions (complete deletions are ruled out by the heterozygous nature of the mutations).
Like in many other genes, the mutations are heterogenous. Recurrent among the 13 patients were five mutations (V326L, 5; W151X, 4; IVS8–1G>C, 2; L99P, 2; R352W, 2). Three of the mutations identified most likely represent functional null alleles because they introduce a premature stop codon (W151X) or shift the reading frame (720–735 del, IVS8–1G>C). Mutant proteins are therefore expected to lack essential catalytic domains indispensable for enzymatic activity. The majority of mutations are missense mutations that are either localized within (P51S, L99P, L157P, A247V, and V326L) or close (T93M, R352W, R404C, and G410S) to the putative TMS (Figs. 2 and 4). Only one mutation was found in the cytoplasmic loop that connects TMS8–9 (C380S). The conservative mutation A247V also was found in a TMS of the proteolipid protein (A242V) and becomes pathogenic by impairing protein folding (15). The P51S mutation removes an evolutionarily conserved proline residue from TMS1 whereas two other mutations introduce proline residues in putative transmembrane α-helices (L99P and L157P, Fig. 2). In the vasopressin V2 receptor, a leucine to proline mutation in the first TMS (L44P) caused receptor retention and degradation in the endoplasmic reticulum (16). Two mutations exchange evolutionarily conserved amino acid residues within conserved stretches of the amino acid sequence (V326L and R404C).
Heterologous expression of the mutations L99P, V326L, R352W, R404C, and G410S in a human cell line resulted in similar transcript but reduced protein expression as compared with the wt cDNA (Fig. 3 A and C). The silent mutations 1158C>T and 1272T>C had no effect on protein expression (Fig. 3D). This result indicates that the missense mutations impair protein stability because of misfolding or defective protein transport. We therefore suggest that the missense mutations L99P, V326L, R352W, R404C, and G410S are pathogenic because they decrease expression of the DHCR7 protein by >90% (Fig. 3B). This implies that three major classes of DHCR7 mutant alleles exist: (class A) null mutations which abolish catalytic activity completely (W151X, 720–735del, IVS-8G>C), (class B) missense mutations that reduce protein expression, and (class C) yet to be discovered missense mutations that impair substrate to product conversion. It remains to be clarified whether mutations such as L99P, V326L, R352W, R404C, and G410S are of mixed type, i.e., whether their pathogenicity is in part due to reduced catalytic activity (class D).
Liver microsomes from patients with fatal SLOS showed a residual 7-dehydrocholesterol metabolism of 10% (17). A likely explanation is that one of the two DHCR7 alleles is not completely inactivated in line with the results of the molecular analysis: in seven patients, a putative null allele is combined with a missense mutation. The remaining six patients carry one or two missense mutations. Because none of the patients carried two null alleles, it is tempting to speculate that an individual with a disruption of both DHCR7 genes and thus a complete deficiency of the endogenous cholesterol biosynthetic pathway is not viable.
Our data conclusively establish that mutations in the DHCR7 gene underly the SLOS for the following reasons. (i) The biochemical anomaly found in all patients with SLOS is the accumulation of 7-dehydrocholesterol (2). This intermediate is converted into cholesterol upon heterologous expression of the DHCR7 cDNA (4). (ii) None of the 13 mutations identified in the patients with SLOS was found among >80 alleles from healthy control persons. Selection of the 13 nonconsanguinous patients was unbiased except for the diagnosis of SLOS with an accumulation of 7-dehydrocholesterol. Healthy siblings of patient SLO2 carried none or one but not both mutations (Fig. 1E) whereas in two brothers with the SLOS both carried the same mutations (patient SLO3a, b, Table 3). Moreover, in each patient analyzed at least one mutation was found. (iii) Three of the mutations are predicted to be null mutations and were found in seven patients. However, none of the 13 patients was a homozygote for two null mutations, which we predict to be lethal. (iv) Upon heterologous expression of five of the ten missense mutations, protein expression was substantially reduced as compared with wt.
Previously, the SLOS gene was assigned to chromosome 7q32.1 because of a balanced de novo translocation in a patient with clinical and biochemical features of SLOS (18). Here, we localized the DHCR7 genes of men and mice in syntenic regions on chromosomes 11q13 and 7F5, respectively, in line with the previous assignment of WI-16597 and WI-13500 (4). FGF-3, FGF-4, and CCND1 genes are localized in these same regions (Mouse Genome Informatics, URL:http://www.informatics.jax.org; March 1998). Because most other genes found in human chromosome 11q13 are localized on mouse chromosome 19, the DHCR7 gene is suggested to be colocalized with FGF-3, FGF-4, and CCND1. The identification of mutations in the DHCR7-catalytic subunit in all 13 patients analyzed so far suggests that the DHCR7 gene is the major SLOS locus. Therefore, the 7q32 region is not expected to contain a widespread SLOS gene if any.
We anticipate that more mutations in the coding but also the noncoding regions of the DHCR7 gene will be identified. Our work paves the way to investigate the genotype-phenotype correlation and to determine the carrier frequency in various populations. Moreover, identification of the SLOS gene and isolation of its murine counterpart will help to further investigate the molecular pathogenesis of the disorder in patients and animal models. Not least, prenatal diagnosis by molecular analysis and carrier identification in families now become possible.
Acknowledgments
We are grateful to Drs. P. Bittigau (Berlin), G. F. Hoffmann, and D. Haas (Marburg), U. Seedorf (Münster), U. Wendel (Düsseldorf), and G. Gillessen-Kaesbach (Essen), and P. Meinecke (Hamburg) for providing patient DNA and biochemical data. We thank Dr. H. J. Menzel for help with DNA-sequencing and B. Fiechtner for outstanding technical assistance. Supported by grants from Fonds zur Förderung der wissenschaftlichen Forschung (P11636 to H.G.; P11695 to G.U.), the Dr. Legerlotz Foundation (to F.F.M.) and the Österreichische Nationalbank (P6515 to H.G.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: DHCR7, Δ7-sterol reductase; SLOS, Smith–Lemli–Opitz syndrome; wt, wild type; TMS, transmembrane segment; SSCP, single strand conformation polymorphism.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF057368).
References
- 1.Smith D W, Lemli L, Opitz J M. J Pediatr. 1964;64:210–217. doi: 10.1016/s0022-3476(64)80264-x. [DOI] [PubMed] [Google Scholar]
- 2.Tint G S, Irons M, Elias E R, Batta A K, Frieden R, Chen T S, Salen G. N Engl J Med. 1994;330:107–113. doi: 10.1056/NEJM199401133300205. [DOI] [PubMed] [Google Scholar]
- 3.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 4.Moebius F F, Fitzky B U, Lee J N, Paik Y-K, Glossmann H. Proc Natl Acad Sci USA. 1998;95:1899–1902. doi: 10.1073/pnas.95.4.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Budowle B, Chakraborty R, Giusti A M, Eisenberg A J, Allen R C. Am J Hum Genet. 1991;48:137–144. [PMC free article] [PubMed] [Google Scholar]
- 6.Weiskirchen R, Erdel M, Utermann G, Bister K. Genomics. 1997;44:83–93. doi: 10.1006/geno.1997.4855. [DOI] [PubMed] [Google Scholar]
- 7.Sinnegger M J, Wang Z, Grabner M, Hering S, Striessnig J, Glossmann H, Mitterdorfer J. J Biol Chem. 1997;272:27686–27693. doi: 10.1074/jbc.272.44.27686. [DOI] [PubMed] [Google Scholar]
- 8.Neuhuber B, Gerster U, Mitterdorfer J, Glossmann H, Flucher B E. J Biol Chem. 1998;273:9110–9118. doi: 10.1074/jbc.273.15.9110. [DOI] [PubMed] [Google Scholar]
- 9.Mitterdorfer J, Sinnegger M J, Grabner M, Striessnig J, Glossmann H. Biochemistry. 1995;34:9350–9355. doi: 10.1021/bi00029a010. [DOI] [PubMed] [Google Scholar]
- 10.Hanner M, Moebius F F, Weber F, Grabner M, Striessnig J, Glossmann H. J Biol Chem. 1995;270:7551–7557. doi: 10.1074/jbc.270.13.7551. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann K, Stoffel W. Biol Chem Hoppe-Seyler. 1993;347:166. doi: 10.1515/bchm3.1992.373.1.187. [DOI] [PubMed] [Google Scholar]
- 12.Hanner M, Moebius F F, Flandorfer A, Knaus H-G, Striessnig J, Kempner E, Glossmann H. Proc Natl Acad Sci USA. 1996;93:8072–8077. doi: 10.1073/pnas.93.15.8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herz J, Willnow T E, Farese R V. Nat Genet. 1997;15:123–124. doi: 10.1038/ng0297-123. [DOI] [PubMed] [Google Scholar]
- 14.Ye Q, Worman H J. J Biol Chem. 1994;269:11306–11311. [PubMed] [Google Scholar]
- 15.Jung M, Sommer I, Schachner M, Nave K-A. J Neurosci. 1996;16:7920–7929. doi: 10.1523/JNEUROSCI.16-24-07920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oksche A, Schuelein R, Rutz C, Liebenhoff U, Dickson J, Mueller H, Birnbaumer M, Rosenthal W. Mol Pharmacol. 1996;50:820–828. [PubMed] [Google Scholar]
- 17.Shefer S, Salen G, Batta A K, Honda A, Tint G S, Irons M, Elias E R, Chen T C, Holick M F. J Clin Invest. 1995;96:1779–1785. doi: 10.1172/JCI118223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alley T L, Scherer S W, Huizenga J J, Tsui L-C, Wallace M R. Am J Med Gen. 1997;68:279–281. [PubMed] [Google Scholar]
- 19.Ullrich K, Koch H G, Meschede D, Flotmann U, Seedorf U. Neuropediatrics. 1996;27:111–112. doi: 10.1055/s-2007-973760. [DOI] [PubMed] [Google Scholar]