Abstract
The study of bacterial viruses (bacteriophages or phages) proved pivotal in the nascence of the disciplines of molecular biology and microbial genetics, providing important information on the central processes of the bacterial cell (DNA replication, transcription and translation) and on how DNA can be transferred from one cell to another. As a result of the pioneering genetics studies and modern genomics, it is now known that phages have contributed to the evolution of the microbial cell and to its pathogenic potential. Because of their ability to transmit genes, phages have been exploited to develop cloning vector systems. They also provide a plethora of enzymes for the modern molecular biologist. Until the introduction of antibiotics, phages were used to treat bacterial infections (with variable success). Western science is now having to re-evaluate the application of phage therapy – a therapeutic modality that never went out of vogue in Eastern Europe – because of the emergence of an alarming number of antibiotic-resistant bacteria. The present article introduces the reader to phage biology, and the benefits and pitfalls of phage therapy in humans and animals.
Key Words: Bacterial virus, Bacteriophage, Diagnostic tools, Human and animal studies, Novel therapies, Phage therapy, Phage typing, Phagotherapy
The discovery of viruses specific to bacteria (bacteriophages or phages) is credited to the Englishman, Frederick Twort (1), and the French Canadian, Felix d’Herelle (2), but it is the latter scientist who probably more accurately recognized what he was dealing with and who is responsible for naming these agents of bacterial death. He is also responsible for recognizing their potential clinical significance. He noted (3):
“Another thought came to me also. If this is true, the same thing will have probably occurred in the sick man. In his intestine, as in my test-tube, the dysentery bacilli will have dissolved away under the action of their parasite. He should now be cured”.
It is interesting to note that phage therapy ceased to be used in the West with the advent of the antibiotic era but has been rediscovered because of the rise in antimicrobial-resistant bacteria. While the study and exploitation of phages flourished in the West, particularly in the development of molecular tools, its use in the former Soviet Union as a therapeutic tool has remained steady for over 80 years.
Phage therapy has been the subject of numerous recent review articles (4-16). (Regrettably, the present article largely ignores the literature from the former Soviet Union because of language problems and lack of detail provided in the published studies.) To simplify the discussion, the present article focuses almost exclusively on the tailed lytic (also known as virulent) bacteriophages belonging to the viral order Caudovirales (17).
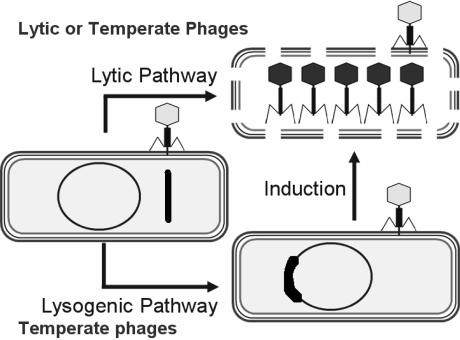
Members of the Caudovirales order are divided morphologically into three families (based on the length and complexity of their tails) and functionally into two groups (based on their effect on infection of host cells) (Figure 1). Lytic phage infection leads exclusively to cell death, lysis and the release of progeny phage particles. While infection by temperate phages may also lead to propagation and lysis (the lytic pathway), this is not always the case. If the phage lytic functions become repressed, the virus genome coexists in a stable form (ie, the prophage) within its host. In the latter case, the phage genome is either integrated into the bacterial chromosome (most common) or remains separate, as a ‘plasmid’ (lysogenic pathway).
Figure 1.
The lytic and lysogenic pathways of bacteriophages. In the lysogenic pathway, the phage genome (thick line) is shown integrated into the bacterial genome. Bacterial genome projects have revealed that, on average, there are 2.5 phage genomes per bacterium. In the case of Streptococcus pyogenes (154-157) and Escherichia coli O157:H7 (158,159), the bacterial genomes contain multiple phage genomes. The reversion to the lytic state (‘induction’) occurs spontaneously but can be enhanced by certain chemical and antibiotics (160,161)
Temperate phages are seldom used in phage therapy because, first, they do not kill 100% of the infected bacteria, and second, in certain cases, they contain genes that render the bacterium more virulent – a phenomenon known as ‘lysogenic conversion’. This has been observed with many human pathogens, including Vibrio cholerae (18-20), Escherichia coli (21,22), Salmonella typhimurium (23-25), Pseudomonas aeruginosa (26,27), Staphylococcus aureus (28), Streptococcus pyogenes (29,30), Clostridium botulinum (31,32) and Corynebacterium diphtheriae (33,34).
Life Cycle of Lytic Phages
Phages, like other viruses, can be characterized on the basis of their host range. While most phages probably possess a narrow host range – that is, they lyse relatively few bacterial strains – others have a broader spectrum of hosts. Some, like phages Felix O1, lyse almost all Salmonella serotypes (35-37), while φS1 lyses a broad range of fluorescent pseudomonads (38). From a practical perspective, those possessing a broader host range offer advantages as therapeutic agents.
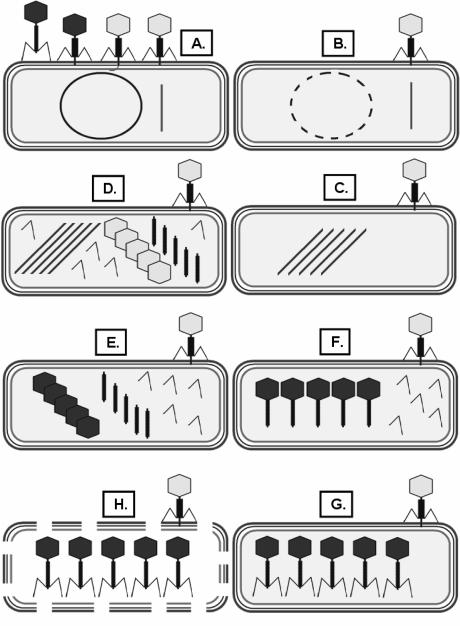
Diffusion brings the phage and potential host into contact. Adsorption occurs through interaction of the distal ends of the phage tails with (usually one of) a plethora of cell-surface components, including pili, flagella, capsules, proteins, lipopolysaccharides (LPSs) and teichoic acids (39). Strong binding then leads to injection of the phage DNA – contained within the capsid – into the host cell (Figure 2A); the mechanisms by which this happens are just beginning to be understood. Once inside the host, the viruses take over the cell’s machinery, subverting it to make new phage particles. This process involves degradation of the host genome and the reutilization of the nucleotides in phage DNA replication (Figure 2B and Figure 2C). Electron microscopy and the analysis of mutants have revealed that, in the subsequent step, phage precursors (such as proheads) appear within the infected cell (Figure 2D). The DNA is packaged into the proheads (Figure 2E), thereby making them competent for tail assembly (Figure 2F). This ultimately leads to the appearance of complete phage particles in the infected cell (Figure 2G).
Figure 2.
The life cycle of lytic bacteriophages. A Phage adsorption and DNA injection; B Host genome degradation; C Phage DNA replication; D Appearance of morphogenesis intermediates, including empty heads (proheads); E Packaging of phage DNA into capsids; F, G Phage assembly; H Lysis and release of progeny phage. The time to lysis varies with the phage host system but can be as little as 10 min
Release of these intracellular viruses requires two phage products, commonly referred to as ‘holins’ and ‘lysins’. The former are small, pore-forming proteins that permit the cytoplasmic lysins access to the peptidoglycan layer in the periplasm. The degradation of this layer leads to osmotic lysis of the cell and the release of tens to hundreds of progeny virus particles that can, in turn, infect and lyse remaining host cells (Figure 2H).
Importance of Phages
Phages are important ecologically as agents in the recycling of organic matter, including cells, and as tools in molecular biology and epidemiology. Almost all natural environments have high concentrations of phage-like particles (40-45). It is conservatively estimated that the prevalence of phages worldwide is in excess of 1031 (46) – equivalent to approximately 109 metric tons – making viruses the most abundant life form on earth.
Epidemiological Fingerprinting of Bacterial Isolates (phage typing)
In epidemiological work, it is necessary to track individual bacterial isolates from clinical specimens to their source (47). This is accomplished by fingerprinting the strains using a wide variety of phenotype- or genotype-based typing methods. One of the classical procedures is phage typing, which is still used in Canada at the Laboratory for Foodborne Zoonoses (Guelph, Ontario) and the National Microbiology Laboratory (Winnipeg, Manitoba), for Salmonella and E coli strains. This procedure involves exposing the bacterial isolate to a battery of ‘typing’ phages, and recording the pattern and degree of lysis. This approach offers import advantages, including the incredible specificity of phage, and a high degree of typability and reproducibility (48,49). Furthermore, in contrast to serotyping and pulsed field gel electrophoresis analysis, phage typing is relatively inexpensive.
Use of Phages As Tools in Molecular Biology
Phage research has had a pivotal impact on molecular biology. In their book Phage and the Origins of Molecular Biology, Cairns et al (50) show how phages have contributed not only to our understanding of vital cellular processes, but also to the development of a considerable number of important genetic and biochemical tools. For example, the realization that viable bacteriophage lambda particles could be constructed with a significant portion of their genome deleted led to the development of insertional and replacement vectors, as well as cosmids and integrative plasmids. Phage serine integrases, particularly those of Streptomyces phageφC31, have been exploited by Michele P Calos (Stanford University, USA) to integrate foreign DNA into mammalian cells (51-53) and Drosophila (54), with the goal of producing transgenic animals or curing biochemical defects (55-57). In addition, phage packaging signals, promoters and terminators, together with a great variety of enzymes, are used in today’s molecular biology laboratory – including polynucleotide kinases, DNA ligases, DNA polymerases, RNA polymerases, recombinases, single-stranded DNA binding proteins, endo- and exonucleases, and even methylases and restriction endonucleases (58).
Use of Bacteriophages to Express Peptides and Proteins (phage Display)
Several systems have been developed to create peptide or protein fusions on capsid proteins of bacteriophages of coliphages lambda (59,60), M13 (61), T7 (62-64) and T4 (65). The M13 and T7 systems have been commercialized. These extremely elegant molecular tools have been used to identify antibody binding epitopes (66-68), amino acid residues involved in protein-protein interactions (69-71), peptides that mimic nonpeptide ligands (72), enzyme substrates and inhibitors, and have even been used to express proteins. One major advantage of this system over standard protein chemistry is that the sequence of the peptide insert can be rapidly and inexpensively determined by DNA sequencing.
A fascinating application of phage display technology is the production of M13 derivatives that express specific antibodies (73). This was pioneered by Cambridge Antibody Technology Ltd (United Kingdom) and is commercially available through GE Healthcare (formerly Amersham Biosciences) as the Recombinant Phage Antibody System. This technology has been the subject of numerous recent reviews (74-77).
New Phage Diagnostic Tools
In addition to the classical uses of phages in molecular biology and diagnostic microbiology, new tools have been developed. The phage amplification assay, for example, is a simple yet elegant way to identify the presence of specific pathogens in food products. The intracellular replication of phage and concomitant lysis of the susceptible bacteria leads to an increase in free phage, which can be easily measured (78,79).
Another new tool involves phage-luciferase fusions. Phages active against a wide variety of bacteria have been tagged with luciferase cassettes (created from either Vibrio luxAB genes or Aequorea green fluorescent protein), which, when expressed, result in the emission of visible light. The major advantages of this system are that light production is absolutely dependent on phage infection of sensitive cells and can easily be measured with exquisite sensitivity.
Phage-luciferase fusions have been used to identify E coli O157 (80-82), Listeria (83), Salmonella (84) and Mycobacterium (85-88). Lastly, bioluminescent phages for the latter bacterium have been used to rapidly determine the antibiotic susceptibility properties of the host cells; that is, in the presence of an effective antimicrobial, light production is inhibited (89-91).
Animal Studies
The earliest research on the therapeutic efficacy of phages was conducted by Felix d’Herelle, who carried out field studies on fowl typhoid (Salmonella enterica subspecies enterica serovar Gallinarum) and laboratory studies on dysentery in rabbits (Shigella dysenteriae) (92). Subsequent animal studies have shown that phage therapy works and is, at least in theory, a practical therapeutic modality.
Unfortunately, one has to wade through a number of studies that, while offering tantalizing evidence of the efficacy of phages (see, for example, the chicken studies by WE Huff and colleagues [93-98]), were either poorly conceived or used impractical treatment regimens with a low probability of being used in agriculture. Examples include treatments that are ineffective once the disease symptoms are expressed, the coadministration of high doses of phage preparations with the pathogen and treatment delivery protocols that would be too costly on a large scale. In addition, a wide variety of animal types and infection strategies have been used, and the phages, which have generally been isolated on a needs basis, are all poorly characterized. This makes comparisons across studies difficult at best.
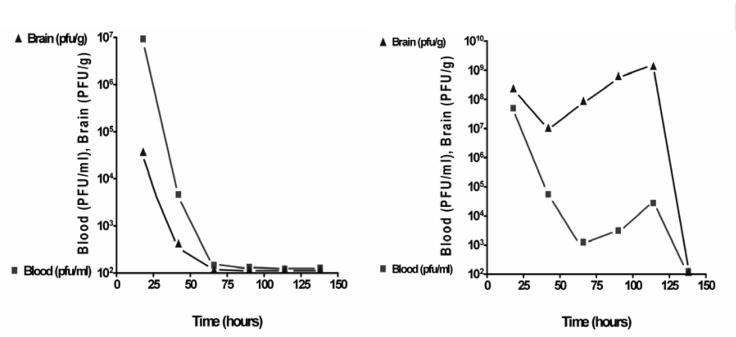
Nevertheless, there do exist well-controlled animal studies that indicate that phages can be used therapeutically in animals and have some advantages over antibiotics. In 1943, René Dubos and colleagues (99) at Harvard University (USA) demonstrated that anti-S dysenteriae phage injected intraperitoneally in mice appeared in the blood stream (and even crossed the brain-blood barrier) but were rapidly cleared (by the reticuloendothetial system, particularly the spleen) (Figure 3, left). When approximately 2×107 colony-forming units of the bacterium were injected intracerebrally, more than 95% of the control mice died within four days; this rate was reduced to 28% with the intraperitoneal injection of bacteriophage (109 plaque-forming units). In contrast, sterile, uninoculated broth, bacterial filtrates and heated phage preparations (60°C) were ineffectual in rescuing the mice. Of particular note was the observation that in the presence of susceptible bacteria (Figure 3, right), the level of phage actually increased and remained high for longer. In the words of Ryland Young (Texas A&M University, USA; personal communication), phages are “the only medicine that multiplies”.
Figure 3.
Left panel Fate of 109 plaque-forming units (PFU) of Shigella dysenteriae-specific phage injected intraperitoneally into a mouse. Right panel Change in titre of the phage as a result of simultaneous injection of phage (intraperitoneally) and S dysenteriae (approximately 2×107 colony-forming units) intracranially
Substantially similar results were obtained in chickens inoculated intramuscularly or intracranially with E coli O18:K1 by Paul Barrow and colleagues (Institute for Animal Health, United Kingdom) (100).
For the most promising animal studies, one must turn to those conducted in the 1980s by H Williams Smith and colleagues at the Institute for Animal Disease Research (United Kingdom) (101-104); these still stand up to scientific scrutiny and indicate clearly the potential for phage therapy in animals. Six- to 12-hour-old calves were fed a mixture of enteropathogenic E coli strains (at a dose of 109), nonpathogenic E coli (1010) and Lactobacillus species (1010). Ninety-seven per cent of the treated animals developed diarrhea within 12 h to 46 h, and 80% died. Microbiological examinations showed the highest counts of bacteria (109.3-9.8/g) in the anterior of the small intestinal and rectal area.
In studies with E coli O101:K30, the addition of 105 K-antigen specific phage B41/1 orally at the onset of diarrhea resulted in no deaths. Other studies showed that the in vitro virulence of the phage did not always correlate with its effect in vivo; phage could have a protective effect if introduced up to 6 h before and 12 h postinfection; and phage could be administered in milk, or picked up from sprayed stall litter or even from contaminated stool material.
Earlier mouse studies by Smith and Huggins (101) using E coli demonstrated that single doses of phage were just as effective as a single administration of streptomycin in eliminating infections, but delaying the treatment regimens negatively affected the outcome for both bacterial viruses and antibiotics. This was re-examined by JJ Bull and coworkers (105), who showed that delaying the treatment in mice by 8 h had little effect on the efficacy of the phage, but markedly reduced the percentage of survivors in the antibiotic-treated group.
Lastly, James Soothill and colleagues (Great Ormond Street Hospital, United Kingdom) (106) demonstrated the potential of using phages against otitis externa caused by P aeruginosa. Like humans, dogs are susceptible to Pseudomonas ear infections. The animals in the study responded well to a single dose of a polyvalent phage preparation (Biovet-PA).
Human Studies
Safety
The first safety trials were conducted by d’Herelle (3), whose methods would not pass bioethics approval today: he tested the safety of phage preparations (taken orally or by injection) on himself, his family and colleagues. Neither they nor subsequent test subjects (patients) experienced any ill effects (107). To these early studies, we can add a wide range of anecdotal evidence, as well as results of actual human experimentation (though not always deliberate), that support the conclusion that exposure to phages poses minimal, if any, health risk. It is impossible to avoid ingesting phages. Both the human oral cavity (108,109) and fecal matter contain phages (41,110), and they are present in municipal drinking water (111), food substances such as yogourt (112) and salami (113), and have even been contaminants in live polio vaccine preparations (114).
Extensive safety trials were undertaken on Staphage Lysate by Delmont Laboratories (USA). This product, which contains high concentrations of antistaphylococcal phages (15,16), was administered to humans intranasally, topically, orally, subcutaneously and intravenously. In over 12 years of use in humans, only minor side effects were observed (16). Lastly, double-blind studies by Anne Bruttin and Harald Brüssow (Nestle Research Center, Switzerland) in 2005 showed that the oral administration of coliphage T4 had no ill effects on human volunteers (115). It is worth reiterating the words of Andrzej Górski et al (116) (Institute of Immunology and Experimental Therapy, Poland), whose institute has 60 years of experience with phage therapy:
“While our past studies did not formally meet current strict criteria of controlled clinical research, they still strongly suggest a high efficacy of phage treatment, its safety, and virtual lack of side effects. Our more recent studies also suggest that phages can migrate to organs that are usually not readily accessible to drugs (prostate gland, bone)”.
Successes
As with the earliest animal experiments, the first studies in humans were conducted successfully by d’Herelle, who used them on patients suffering with dysentery and bubonic plague (3,92). This work led d’Herelle and the Georgian scientist, Giorgi Eliava, to establish an institute, which now bears his name, in Tbilisi, Georgia (1923), for the investigation of practical uses of phage in therapy. In one study (of many) conducted by the Institute in their 83-year history, over 31,000 children younger than seven years of age participated in a 16-week clinical trial of the efficacy of anti-Shigella phage tablets. One-half of the children (from one side of the streets) received weekly tablets containing the phages, while those on the opposite side of the streets received a placebo. The incidence of clinically verified shigellosis in the subjects receiving the treatment was reduced 2.6-fold (5).
The most clearly documented phage therapeutic research – and the most relevant to the global problem of antimicrobial resistance – was conducted at the Institute of Immunology and Experimental Therapy (also known as the ‘Hirszfeld Institute’) in Wroclaw, Poland. Their experience with phages over the past thirty years has been the subject of numerous publications (many of which are accessible from their Web site at http://www.iitd.pan.wroc.pl/about_en.html). In all, almost 2000 patients infected with a variety of life-threatening (predominantly antibiotic-resistant) infections have been treated with phages. The overall success rate is from 60% to 90% (117,118). These studies have been described as the “most detailed studies published in English on the use of phages in clinical settings” (14) and “probably the most important data published in the English literature” (119).
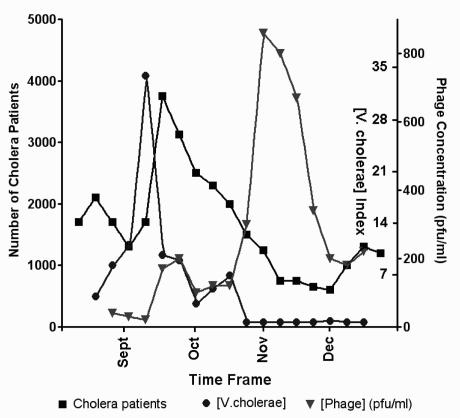
One study that deserves mention did not involve active intervention by the physician, nor was it a clinical study of a therapeutic agent. Rather, Shah M Faruque and colleagues from the International Centre for Diarrhoeal Disease Research, Bangladesh, examined the self-limiting nature of seasonal cholera epidemics and the role of host-mediated amplification of phage (110,120). The team found that the number of cholera patients spikes in mid-September, just after the level of V cholerae in water reaches its maximum, and then decreases in subsequent months (Figure 4). This was found to be a natural consequence of enrichment for lytic phages in the intestinal tracts of infected patients. These phages then enter (‘contaminate’) the community water sources, leading to further decreases in the levels of this pathogen. These studies suggest that the early phage studies by d’Herelle on the ecological control of cholera in India bear re-examination with a less skeptical eye (16).
Figure 4.
Changes in the concentration of Vibrio cholerae in water over the summer and autumn months in Bangladesh, as well as its impact on the number of cholera patients. The decrease in phage patients and the pathogen (measured in plaque-forming units [pfu]) are due to enrichment in specific cholera phages within the patients and in water. Data from reference 120
Problems
There are some major problems with the reintroduction of phage therapy into North America, but the alarming increase in antibiotic-resistant bacteria and the move against the use of antimicrobials in food production in Europe is forcing us to look more favourably at phages, either for treating infections in humans or (more likely) to reduce the spread of zoonotic bacterial diseases. The problems arise from phage biology, from proteomics and from difficulties in obtaining regulatory approval. Some of these challenges have been addressed experimentally as outlined below.
The behaviour of phages under anaerobic conditions (ie, in the gut) or in starved cells has been addressed by relatively few studies (121-126) and requires further study. Sandra Chibani-Chennoufi and colleagues (6) showed that E coli isolated from normal mice was generally susceptible to a cocktail of four broad host range coliphages in the laboratory, but oral ingestion of these phages had little impact on the resident flora, suggesting that the bacteria were somehow protected or that the phages behaved differently under anaerobic conditions. Phage Rb33, for example, is a broad host range, Teven-like virus that displays oxygen-dependent growth on ECOR4 (127). The scientists also noted extensive lysis inhibition under anaerobic conditions. They hypothesized that, under anaerobic conditions, different phage receptors were expressed, a theory that echoes the observations of Wegrzyn and Thomas (125). It is also possible that gut carbohydrates and bile salts chelate divalent ions, which are required for the adsorption and replication of many phages.
Mutation of bacteria to phage resistance is often listed among the problems with phage therapy. Indeed, in the laboratory, the famous Luria and Delbrück fluctuation test (128) showed that the rate of mutation of E coli to phage T1 resistance was 1.4×10–8 to 4.1×10–8. In phage-treated animals, bacteria resistant to the therapeutic phage have been observed to arise. This has been modelled in the laboratory in studies on predator-prey successions. But in vitro predator-prey studies do not accurately reflect in vivo conditions. If one is dealing with K1 or LPS-specific phages, the most likely resistant mutants are capsule or LPS defective. In both cases, the mutant bacteria are less virulent. In a recent in vitro study of E coli, S typhimurium and coliphages T5 and T7, William R Harmcombe and James J Bull (University of Texas at Austin, USA) (129) showed that resistant mutant outgrowth occurred in pure E coli cultures, but in mixed culture conditions, the phage-resistant coliforms were outcompeted by Salmonella. It is also worthy noting that therapeutic phage cocktails may contain viruses with different receptor specificities; this would reduce the likelihood of the outgrowth of virulent, phage-resistant mutants.
Bacteriophages are antigenic and are rapidly cleared by the reticuloendothelial system. The former problem can only be dealt with by substituting another phage, if proven necessary. In their mice studies at the National Institutes of Health (USA), Carl Merril et al (130) were able to select “long-circulating” or “Argo” mutants from virulent derivatives of the well-studied coliphage lambda and Salmonella phage P22 (λvir and P22vir). These possess changes in their capsid proteins (131).
Relatively little is known about the proteomes of the large, virulent phages. In silico analysis has failed to reveal ‘toxin’ genes in virulent coliphages, and the studies by Bruttin and Brüssow (115) revealed no human toxicity, but we have identified the functions of at most 50% of proteins of coliphage T4 – one of the best-studied viruses. We know far less about many of the other potential therapeutic phages; indeed, most have not been sequenced.
Regulatory approval is the major hurdle for standard mono- or polyvalent phage therapy. In the words of Harald Brüssow, “phagetherapy … challenges current pharmacokinetic concepts”, in large part because this is “the only medicine that multiplies”. The Georgian approach – to isolate new phages when the existing cocktail does not work – is unacceptable to North American regulators.
In addition, phage therapy is decidedly ‘low technology’. It is questionable whether drug companies would be willing to invest in the development of phage-based products without considerable confidence that they could make a profit. And the patenting of viral cocktails would be venturing into new legal territory.
New Alternative Therapies
The newest therapies involving phages can be arbitrarily divided in two, based on whether the approach is simply an outgrowth of traditional phage therapy or is truly unique. In the former group, we have the immobilization of phages in or on membranes, as in PhagoBioDerm (Phage International, USA), a biodegradable, polymer-based membrane that contains phages, ciprofloxacin, benzocaine and alpha-chymotrypsin, which was developed at the Eliava Institute (132-134). The phages (PyoPhage, BioPharm-L, Georgia) target a variety of Gram-positive and Gram-negative bacteria. The preparation has been successfully used (70%) to treat recurrent leg ulcers and infections in burn victims. Janice Spencer and colleagues (University of Strathclyde, United Kingdom) (135) developed a variant of this procedure. They immobilized phages on nylon strips and found that the preparation was “effective against most of the major epidemic methicillin-resistant Staphylococcus aureus strains”. They have proposed that this could be used “in different forms, including strips, sutures and beads” (see http://www.news-medical.net/?id=8938).
The potential exists for at least two types of novel phage therapies: recombinant therapeutic phages and phage-derived lysins. In the first therapy, highly lytic phages could be selected or engineered for different receptor specificities (136,137). This has yet to be attempted, but its proof of concept is the isolation of spontaneous phage host range mutants and the existence of recombinant phages with different receptor specificities (138). Diane L Schaak (Rowland Institute, Harvard University, USA) (139) has suggested developing what she calls “toxin-phage bacteriocides” or “trojan silver bullets” – that is, phages whose genomes are supplemented with host lethal toxins to enhance cell killing.
As proofs of concept, both temperate and several nonlytic phages have been loaded with toxic protein genes. Steven Hagens et al (Max F Perutz Laboratories, Austria) (140) created a nonreplicating mutant of P aeruginosa phage Pf3 that carries the restriction endonuclease BglII. This virus was more effective than the pilus-specific virulent phage Pt1 in animal protection studies and had the added benefit of abrogating the release of LPS as a result of cell lysis, thus decreasing the danger of endotoxic shock. Similar studies have been carried out in E coli by Caroline Westwater et al using phage M13 carrying the addiction toxins Gef and ChpBK (141). Very recently, Michelle L Embleton (Eastman Dental Institute, University College, United Kingdom) (142) labelled the S aureus serogroup F typing phage 75 with the photosensitizer tin (IV) chlorin e6 and noted its enhanced activity against this bacterium. This targeted photosensitizer was even effective in significantly reducing the viability of strains that were apparently not susceptible to this bacteriophage, suggesting that phages may still bind to bacteria that they cannot productively infect.
The construction of lysis-deficient phage mutants and their use therapeutically is the subject of patents issued to Gangagen (USA) (143,144). If effective, one can envisage their immediate application to bacteria containing intracellular toxins, such as Clostridium difficile.
One of the most exciting new technologies is based on the activity of phage lysins and is derived from the pioneering work of WM Mullan and RJ Crawford (145). This approach has been used by Vincent Fischetti (Rockefeller University, USA) (146) and others (147,148). The advantages of lysin-based therapy are numerous: they can be prepared with high purity and possess high specific activity; they exhibit rapid lethal action; they are nontoxic; and apparently, antibodies that form against these proteins do not neutralize their lytic activity. Lastly, no bacterial resistance develops to these proteins, probably because they possess multiple domains for cell wall binding and hydrolysis.
Experimental studies have indicated that lysins possess considerable potential for decontaminating foods (149) or treating infections. For example, lysin PlyGBS was found to be effective in preventing group B streptococcal colonization of the mouse vagina and oropharynx (150), the C(1) lysin prevents group A streptococcal colonization of the mouse upper respiratory tract (151); and Cpl-1 lysin prevented Streptococcus pneumoniae endocarditis in a rat model (152). Likewise, LysK cloned from broad host range staphylococcal phage K may have therapeutic efficacy against methicillin-resistant S aureus (153). (This therapeutic modality is only effective against Gram-positive bacteria because of the outer membrane permeability barrier in Gram-negative cells.)
Where Next in Canada?
The Public Health Agency of Canada’s Laboratory for Foodborne Zoonoses and its director, Mohammed Karmali, are intent on it becoming the research centre in Canada investigating the use of phages to limit the animal carriage and transmission of zoonotic bacteria (Salmonella, E coli O157 and Campylobacter).
They propose becoming the organizational home of a network of Canadian scientists and clinicians interested in all aspects of phage therapy. Interested clinicians should contact Mohamed Karmali (Mohamed_Karmali@phac-aspc.gc.ca), while basic scientists should contact the author (Andrew_Kropinski@phac-aspc.gc.ca).
Acknowledgments
The author thanks Betty Kutter for many fruitful discussions on phage therapy, as well as Peggy A Pritchard (Editor extraordinaire of Success Strategies for Women in Science: A Portable Mentor), without whose help this manuscript would not have been written.
Resources
Two basic resources on phage therapy are the Web sites of Elizabeth Kutter (Evergreen State College, USA) at <http://www.evergreen.edu/phage/> and Stephen Abedon (Ohio State University, USA) at <http://www.mansfield.ohio.edu/~sabedon/>. The Appendix lists some of the companies involved in phage research or phage therapy. The Intralytix Web site contains Alexander Sulakvelidze’s detailed history of phagotherapy.
APPENDIX 1.
Companies involved in phage and phage therapy research
| Company | Location | Web site address |
|---|---|---|
| Biophage | Canada | http://www.biophage.com/ |
| Pharma Inc | ||
| Exponential | USA | http://www.expobio.com/ |
| Biotherapies, Inc | ||
| Gangagen Inc | USA | http://www.gangagen |
| Hexal Genentech | Germany | http://www.hexal-gentech.de/ |
| InnoPhage | Portugal | http://www.innophage.com/ |
| Intralytix, Inc | USA | http://www.intralytix.com/ |
| New Horizons | USA | http://www.nhdiag.com/index.htm |
| Diagnostics Inc | ||
| Novolytics Ltd | United Kingdom | http://www.novolytics.co.uk/ |
| about_us.html | ||
| Phage | Israel | http://www.phage-biotech.com/ |
| Biotech Ltd | ||
| Phage | USA | http://www.phageinternational.com/ |
| International, Inc | ||
| Phage Therapy | Georgia | http://www.phagetherapycenter.com/ |
| Targanta | Canada | http://www.targanta.com/ |
| Therapeutics Inc | ||
| Biochimpharm | Georgia | http://www.biochimpharm.ge/ |
USA United States of America
References
- 1.Twort FW. An investigation on the nature of the ultramicroscopic viruses. Lancet 1915;189:1241-3. [Google Scholar]
- 2.d’Herelle F. Sur un microbe invisible antagoniste des bacilles dysentériques. C R Acad Sci (Paris) 1917;165:373-5. [Google Scholar]
- 3.d’Herelle F. The Bacteriophage and its Behavior. Baltimore: The Williams&Wilkins Company, 1926. [Google Scholar]
- 4.Borysowski J, Weber-Dabrowska B, Gorski A. The potential use of bacteriophages in view of the current antibiotic therapy crisis. Pol Arch Med Wewn 2005;113:73-8. [PubMed] [Google Scholar]
- 5.Brussow H. Phage therapy: The Escherichia coli experience. Microbiology 2005;151:2133-40. [DOI] [PubMed] [Google Scholar]
- 6.Chibani-Chennoufi S, Sidoti J, Bruttin A, Kutter E, Sarker S, Brussow H. In vitro and in vivo bacteriolytic activities of Escherichia coli phages: Implications for phage therapy. Antimicrob Agents Chemother 2004;48:2558-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischetti VA. Phage antibacterials make a comeback. Nat Biotechnol 2001;19:734-5. [DOI] [PubMed] [Google Scholar]
- 8.Fischetti VA. Novel method to control pathogenic bacteria on human mucous membranes. Ann NY Acad Sci 2003;987:207-14. [DOI] [PubMed] [Google Scholar]
- 9.Gorski A, Weber-Dabrowska B. The potential role of endogenous bacteriophages in controlling invading pathogens. Cell Mol Life Sci 2005;62:511-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inal JM. Phage therapy: A reappraisal of bacteriophages as antibiotics. Arch Immunol Ther Exp (Warsz) 2003;51:237-44. [PubMed] [Google Scholar]
- 11.Matsuzaki S, Rashel M, Uchiyama J, et al. Bacteriophage therapy: A revitalized therapy against bacterial infectious diseases. J Infect Chemother 2005;11:211-9. [DOI] [PubMed] [Google Scholar]
- 12.Payne RJ, Jansen VA. Pharmacokinetic principles of bacteriophage therapy. Clin Pharmacokinet 2003;42:315-25. [DOI] [PubMed] [Google Scholar]
- 13.Payne RJ, Phil D, Jansen VA. Phage therapy: The peculiar kinetics of self-replicating pharmaceuticals. Clin Pharmacol Ther 2000;68:225-30. [DOI] [PubMed] [Google Scholar]
- 14.Sulakvelidze A, Alavidze Z, Morris JG Jr. Bacteriophage therapy. Antimicrob Agents Chemother 2001;45:649-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sulakvelidze A, Barrow P. Phage therapy in animals and agribusiness. In: Kutter R, Sulakvelidze A, eds. Bacteriophages: Biology and Application. Boca Raton: CRC Press, 2005:335-80. [Google Scholar]
- 16.Sulakvelidze A, Kutter E. Bacteriophage therapy in humans. In: Kutter E, Sulakvelidze A, eds. Bacteriophages: Biology and Application. Boca Raton: CRC Press, 2005:381-436. [Google Scholar]
- 17.Ackermann H-W. Tailed bacteriophages: The order Caudovirales. Adv Virus Res 1999;51:135-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyd EF, Davis BM, Hochhut B. Bacteriophage-bacteriophage interactions in the evolution of pathogenic bacteria. Trends Microbiol 2001;9:137-44. [DOI] [PubMed] [Google Scholar]
- 19.O’Shea YA, Boyd EF. Mobilization of the Vibrio pathogenicity island between Vibrio cholerae isolates mediated by CP-T1 generalized transduction. FEMS Microbiol Lett 2002;214:153-7. [DOI] [PubMed] [Google Scholar]
- 20.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 1996;272:1910-4. [DOI] [PubMed] [Google Scholar]
- 21.Miyamoto H, Nakai W, Yajima N, et al. Sequence analysis of Stx2-converting phage VT2-Sa shows a great divergence in early regulation and replication regions. DNA Res 1999;6:235-40. [DOI] [PubMed] [Google Scholar]
- 22.Plunkett G 3, Rose DJ, Durfee TJ, Blattner FR. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J Bacteriol 1999;181:1767-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Figueroa-Bossi N, Uzzau S, Maloriol D, Bossi L. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol Microbiol 2001;39:260-71. [DOI] [PubMed] [Google Scholar]
- 24.Ho TD, Figueroa-Bossi N, Wang M, Uzzau S, Bossi L, Slauch JM. Identification of GtgE, a novel virulence factor encoded on the Gifsy-2 bacteriophage of Salmonella enterica serovar Typhimurium. J Bacteriol 2002;184:5234-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mirold S, Rabsch W, Rohde M, et al. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc Natl Acad Sci USA 1999;96:9845-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi T, Terawaki Y. Molecular approach to Pseudomonas aeruginosa cytotoxin: Structure, activation mechanism and phage conversion. Antibiot Chemother 1991;44:48-53. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama K, Kanaya S, Ohnishi M, Terawaki Y, Hayashi T. The complete nucleotide sequence of fCTX, a cytotoxin-converting phage of Pseudomonas aeruginosa: Implications for phage evolution and horizontal gene transfer via bacteriophages. Mol Microbiol 1999;31:399-419. [DOI] [PubMed] [Google Scholar]
- 28.Coleman DC, Sullivan DJ, Russell RJ, Arbuthnott JP, Carey BF, Pomeroy HM. Staphylococcus aureus bacteriophages mediating the simultaneous lysogenic conversion of beta-lysin, staphylokinase and enterotoxin A: Molecular mechanism of triple conversion. J Gen Microbiol 1989;135:1679-97. [DOI] [PubMed] [Google Scholar]
- 29.Broudy TB, Fischetti VA. In vivo lysogenic conversion of Tox(-) Streptococcus pyogenes to Tox(+) with Lysogenic Streptococci or free phage. Infect Immun 2003;71:3782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canchaya C, Desiere F, McShan WM, Ferretti JJ, Parkhill J, Brüssow H. Genome analysis of an inducible prophage and prophage remnants integrated in the Streptococcus pyogenes strain SF370. Virology 2002;302:245-58. [DOI] [PubMed] [Google Scholar]
- 31.Hauser D, Eklund MW, Boquet P, Popoff MR. Organization of the botulinum neurotoxin C1 gene and its associated non-toxic protein genes in Clostridium botulinum C 468. Mol Gen Genet 1994;243:631-40. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Y, Sugiyama H, Johnson EA. Transfer of neurotoxigenicity from Clostridium butyricum to a nontoxigenic Clostridium botulinum type E-like strain. Appl Environ Microbiol 1993;59:3825-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buck GA, Cross RE, Wong TP, Loera J, Groman N. DNA relationships among some tox-bearing corynebacteriophages. Infect Immun 1985;49:679-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Groman NB. Conversion by corynephages and its role in the natural history of diphtheria. J Hyg (Lond) 1984;93:405-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirsh DC, Martin LD. Rapid detection of Salmonella spp. by using Felix-O1 bacteriophage and high-performance liquid chromatography. Appl Environ Microbiol 1983;45:260-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kallings LO. Sensitivity of various salmonella strains to felix 0-1 phage. Acta Pathol Microbiol Scand 1967;70:446-54. [DOI] [PubMed] [Google Scholar]
- 37.van der Walt ML, Steyn HC. The biochemical differentiation between Salmonella and Citrobacter. Onderstepoort J Vet Res 1989;56:263-9. [PubMed] [Google Scholar]
- 38.Kelln RA, Warren RA. Isolation and properties of a bacteriophage lytic for a wide range of pseudomonads. Can J Microbiol 1971;17:677-82. [DOI] [PubMed] [Google Scholar]
- 39.Lindberg AA. Bacteriophage receptors. Annu Rev Microbiol 1973;27:205-41. [DOI] [PubMed] [Google Scholar]
- 40.Breitbart M, Felts B, Kelley S, et al. Diversity and population structure of a near-shore marine-sediment viral community. Proc Biol Sci 2004;271:565-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breitbart M, Hewson I, Felts B, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol 2003;185:6220-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breitbart M, Salamon P, Andresen B, et al. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA 2002;99:14250-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Breitbart M, Wegley L, Leeds S, Schoenfeld T, Rohwer F. Phage community dynamics in hot springs. Appl Environ Microbiol 2004;70:1633-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frederickson CM, Short SM, Suttle CA. The physical environment affects cyanophage communities in British Columbia inlets. Microb Ecol 2003;46:348-57. [DOI] [PubMed] [Google Scholar]
- 45.Kepner RL Jr, Wharton RA Jr, Suttle CA. Viruses in Antarctic lakes. Limnol Oceanogr 1998;43:1754-61. [DOI] [PubMed] [Google Scholar]
- 46.Rohwer F. Global phage diversity. Cell 2003;113:141. [DOI] [PubMed] [Google Scholar]
- 47.Pitt TL, Gaston MA. Bacteriophage typing. Methods Mol Biol 1995;46:15-26. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen JP, Rosdahl VT. Development and epidemiological applications of a bacteriophage typing system for typing Pasteurella multocida. J Clin Microbiol 1990;28:103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weischer M, Kolmos HJ, Kaufmann ME, Rosdahl VT. Biotyping, phage typing, and O-serotyping of clinical isolates of Enterobacter cloacae. APMIS 1993;101:838-44. [DOI] [PubMed] [Google Scholar]
- 50.Cairns J, Stent GS, Watson JD. Phage and the Origins of Molecular Biology. Cold Spring Harbor: Cold Spring Harbor Laboratory Press, 1966. [Google Scholar]
- 51.Chalberg TW, Genise HL, Vollrath D, Calos MP. φC31 integrase confers genomic integration and long-term transgene expression in rat retina. Invest Ophthalmol Vis Sci 2005;46:2140-6. [DOI] [PubMed] [Google Scholar]
- 52.Hollis RP, Stoll SM, Sclimenti CR, Lin J, Chen-Tsai Y, Calos MP. Phage integrases for the construction and manipulation of transgenic mammals. Reprod Biol Endocrinol 2003;1:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP. Site-specific genomic integration in mammalian cells mediated by phage φC31 integrase. Mol Cell Biol 2001;21:3926-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage φC31. Genetics 2004;166:1775-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ginsburg DS, Calos MP. Site-specific integration with φC31 integrase for prolonged expression of therapeutic genes. Adv Genet 2005;54:179-87. [DOI] [PubMed] [Google Scholar]
- 56.Groth AC, Calos MP. Phage integrases: Biology and applications. J Mol Biol 2004;335:667-78. [DOI] [PubMed] [Google Scholar]
- 57.Held PK, Olivares EC, Aguilar CP, Finegold M, Calos MP, Grompe M. In vivo correction of murine hereditary tyrosinemia type I by fC31 integrase-mediated gene delivery. Mol Ther 2005;11:399-408. [DOI] [PubMed] [Google Scholar]
- 58.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE: Restriction enzymes and methyltransferases. Nucleic Acids Res 2003;31:418-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maruyama IN, Maruyama HI, Brenner S. λfoo: A λ phage vector for the expression of foreign proteins. Proc Natl Acad Sci USA 1994;91:8273-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sternberg N, Hoess RH. Display of peptides and proteins on the surface of bacteriophage λ. Proc Natl Acad Sci USA 1995;92:1609-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Makowski L. Phage display: Structure, assembly and engineering of filamentous bacteriophage M13. Curr Biol 1994;4:225-30. [Google Scholar]
- 62.Danner S, Belasco JG. T7 phage display: A novel genetic selection system for cloning RNA-binding proteins from cDNA libraries. Proc Natl Acad Sci USA 2001;98:12954-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takakusagi Y, Kobayashi S, Sugawara F. Camptothecin binds to a synthetic peptide identified by a T7 phage display screen. Bioorg Med Chem Lett 2005;15:4850-3. [DOI] [PubMed] [Google Scholar]
- 64.Xie B, Tassi E, Swift MR, et al. Identification of the fibroblast growth factor (FGF)-interacting domain in a secreted FGF-binding protein by phage display. J Biol Chem 2006;281:1137-44. [DOI] [PubMed] [Google Scholar]
- 65.Ren ZJ, Lewis GK, Wingfield PT, Locke EG, Steven AC, Black LW. Phage display of intact domains at high copy number: A system based on SOC, the small outer capsid protein of bacteriophage T4. Protein Sci 1996;5:1833-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bradbury A, Cattaneo A. The use of phage display in neurobiology. Trends Neurosci 1995;18:243-9. [PubMed] [Google Scholar]
- 67.Cortese R, Felici F, Galfre G, Luzzago A, Monaci P, Nicosia A. Epitope discovery using peptide libraries displayed on phage. Trends Biotechnol 1994;12:262-7. [DOI] [PubMed] [Google Scholar]
- 68.Scott JS. Discovering peptide ligands using epitope libraries. Trends Biochem Sci 1992;17:241-5. [DOI] [PubMed] [Google Scholar]
- 69.Allen JB, Walberg MW, Edwards MC, Elledge SJ. Finding prospective partners in the library: The two-hybrid system and phage display find a match. Trends Biochem Sci 1995;20:511-6. [DOI] [PubMed] [Google Scholar]
- 70.Cortese R, Monaci P, Nicosia A, et al. Identification of biologically active peptides using random libraries displayed on phage. Curr Opin Biotechnol 1995;6:73-80. [DOI] [PubMed] [Google Scholar]
- 71.Sidhu SS, Fairbrother WJ, Deshayes K. Exploring protein-protein interactions with phage display. Chembiochem 2003;4:14-25. [DOI] [PubMed] [Google Scholar]
- 72.O’Neil KT, DeGrado WF, Mousa SA, Ramachandran N, Hoess RH. Identification of recognition sequences of adhesion molecules using phage display technology. Methods Enzymol 1994;245:370-86. [DOI] [PubMed] [Google Scholar]
- 73.Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu Rev Immunol 1994;12:433-55. [DOI] [PubMed] [Google Scholar]
- 74.Bradbury AR, Marks JD. Antibodies from phage antibody libraries. J Immunol Methods 2004;290:29-49. [DOI] [PubMed] [Google Scholar]
- 75.Conrad U, Scheller J. Considerations on antibody-phage display methodology. Comb Chem High Throughput Screen 2005;8:117-26. [DOI] [PubMed] [Google Scholar]
- 76.Hoogenboom HR, de Bruine AP, Hufton SE, Hoet RM, Arends JW, Roovers RC. Antibody phage display technology and its applications. Immunotechnology 1998;4:1-20. [DOI] [PubMed] [Google Scholar]
- 77.Mancini N, Carletti S, Perotti M, et al. Phage display for the production of human monoclonal antibodies against human pathogens. New Microbiol 2004;27:315-28. [PubMed] [Google Scholar]
- 78.Badrinath P, Sundkvist T, Mahgoub H, Kent R. An outbreak of Salmonella enteritidis phage type 34a infection associated with a Chinese restaurant in Suffolk, United Kingdom. BMC Public Health 2004;4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Isaacs S, Aramini J, Ciebin B, et al; Salmonella Enteritidis PT30 Outbreak Investigation Working Group. An international outbreak of salmonellosis associated with raw almonds contaminated with a rare phage type of Salmonella enteritidis. J Food Prot 2005;68:191-8. [DOI] [PubMed] [Google Scholar]
- 80.Funatsu T, Taniyama T, Tajima T, Tadakuma H, Namiki H. Rapid and sensitive detection method of a bacterium by using a GFP reporter phage. Microbiol Immunol 2002;46:365-9. [DOI] [PubMed] [Google Scholar]
- 81.Oda M, Morita M, Unno H, Tanji Y. Rapid detection of Escherichia coli O157:H7 by using green fluorescent protein-labeled PP01 bacteriophage. Appl Environ Microbiol 2004;70:527-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Waddell TE, Poppe C. Construction of mini-Tn10luxABcam/Ptac-ATS and its use for developing a bacteriophage that transduces bioluminescence to Escherichia coli O157:H7. FEMS Microbiol Lett 2000;182:285-9. [DOI] [PubMed] [Google Scholar]
- 83.Loessner MJ, Rees CE, Stewart GS, Scherer S. Construction of luciferase reporter bacteriophage A511::luxAB for rapid and sensitive detection of viable Listeria cells. Appl Environ Microbiol 1996;62:1133-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuhn J, Suissa M, Wyse J, et al. Detection of bacteria using foreign DNA: The development of a bacteriophage reagent for Salmonella. Int J Food Microbiol 2002;74:229-38. [DOI] [PubMed] [Google Scholar]
- 85.Fullner KJ, Hatfull GF. Mycobacteriophage L5 infection of Mycobacterium bovis BCG: Implications for phage genetics in the slow-growing mycobacteria. Mol Microbiol 1997;26:755-66. [DOI] [PubMed] [Google Scholar]
- 86.Riska PF, Jacobs WR Jr, Bloom BR, McKitrick J, Chan J. Specific identification of Mycobacterium tuberculosis with the luciferase reporter mycobacteriophage: Use of p-nitro-alpha-acetylamino-beta-hydroxy propiophenone. J Clin Microbiol 1997;35:3225-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Riska PF, Su Y, Bardarov S, et al. Rapid film-based determination of antibiotic susceptibilities of Mycobacterium tuberculosis strains by using a luciferase reporter phage and the Bronx Box. J Clin Microbiol 1999;37:1144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sasahara KC, Gray MJ, Shin SJ, Boor KJ. Detection of viable Mycobacterium avium subsp. paratuberculosis using luciferase reporter systems. Foodborne Pathog Dis 2004;1:258-66. [DOI] [PubMed] [Google Scholar]
- 89.Banaiee N, Bobadilla-del-Valle M, Riska PF, et al. Rapid identification and susceptibility testing of Mycobacterium tuberculosis from MGIT cultures with luciferase reporter mycobacteriophages. J Med Microbiol 2003;52:557-61. [DOI] [PubMed] [Google Scholar]
- 90.Bardarov S Jr, Dou H, Eisenach K, et al. Detection and drug-susceptibility testing of M. tuberculosis from sputum samples using luciferase reporter phage: Comparison with the Mycobacteria Growth Indicator Tube (MGIT) system. Diagn Microbiol Infect Dis 2003;45:53-61. [DOI] [PubMed] [Google Scholar]
- 91.Hazbon MH, Guarin N, Ferro BE, et al. Photographic and luminometric detection of luciferase reporter phages for drug susceptibility testing of clinical Mycobacterium tuberculosis isolates. J Clin Microbiol 2003;41:4865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Summers WC. Bacteriophage therapy. Annu Rev Microbiol 2001;55:437-51. [DOI] [PubMed] [Google Scholar]
- 93.Mendez J, Audicana A, Cancer M, et al. Assessment of drinking water quality using indicator bacteria and bacteriophages. J Water Health 2004;2:201-14. [PubMed] [Google Scholar]
- 94.Huff WE, Huff GR, Rath NC, Balog JM, Donoghue AM. Bacteriophage treatment of a severe Escherichia coli respiratory infection in broiler chickens. Avian Dis 2003;47:1399-405. [DOI] [PubMed] [Google Scholar]
- 95.Milch H, Fornosi F. Bacteriophage contamination in live poliovirus vaccine. J Biol Stand 1975;3:307-10. [DOI] [PubMed] [Google Scholar]
- 96.Huff WE, Huff GR, Rath NC, Balog JM, Donoghue AM. Therapeutic efficacy of bacteriophage and Baytril (enrofloxacin) individually and in combination to treat colibacillosis in broilers. Poult Sci 2004;83:1944-7. [DOI] [PubMed] [Google Scholar]
- 97.Huff WE, Huff GR, Rath NC, Balog JM, Donoghue AM. Alternatives to antibiotics: Utilization of bacteriophage to treat colibacillosis and prevent foodborne pathogens. Poult Sci 2005;84:655-9. [DOI] [PubMed] [Google Scholar]
- 98.Huff WE, Huff GR, Rath NC, et al. Prevention of Escherichia coli respiratory infection in broiler chickens with bacteriophage (SPR02). Poult Sci 2002;81:437-41. [DOI] [PubMed] [Google Scholar]
- 99.Dubos RJ, Straus JH, Pierce C. The multiplication of bacteriophage in vivo and its protective effect against an experimental infection with Shigella dysenteriae. J Exp Med 1943;20:161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barrow P, Lovell M, Berchieri A Jr. Use of lytic bacteriophage for control of experimental Escherichia coli septicemia and meningitis in chickens and calves. Clin Diagn Lab Immunol 1998;5:294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smith HW, Huggins MB. Successful treatment of experimental Escherichia coli infections in mice using phage: Its general superiority over antibiotics. J Gen Microbiol 1982;128:2-18. [DOI] [PubMed] [Google Scholar]
- 102.Smith HW, Huggins MB. Effectiveness of phages in treating experimental Escherichia coli diarrhoea in calves, piglets and lambs. J Gen Microbiol 1983;129:2659-75. [DOI] [PubMed] [Google Scholar]
- 103.Smith HW, Huggins MB, Shaw KM. Factors influencing the survival and multiplication of bacteriophages in calves and in their environment. J Gen Microbiol 1987;133:1127-35. [DOI] [PubMed] [Google Scholar]
- 104.Smith HW, Huggins MB, Shaw KM. The control of experimental Escherichia coli diarrhoea in calves by means of bacteriophages. J Gen Microbiol 1987;133:1111-26. [DOI] [PubMed] [Google Scholar]
- 105.Bull JJ, Levin BR, DeRouin T, Walker N, Bloch CA. Dynamics of success and failure in phage and antibiotic therapy in experimental infections. BMC Microbiol 2002;2:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Soothill J, Hawkins C, Anggard E, Harper D. Therapeutic use of bacteriophages. Lancet Infect Dis 2004;4:544-5. [DOI] [PubMed] [Google Scholar]
- 107.Summers WC. Félix d’Herelle and the Origins of Molecular Biology. New Haven: Yale University Press, 1999. [Google Scholar]
- 108.Bachrach G, Leizerovici-Zigmond M, Zlotkin A, Naor R, Steinberg D. Bacteriophage isolation from human saliva. Lett Appl Microbiol 2003;36:50-3. [DOI] [PubMed] [Google Scholar]
- 109.Hitch G, Pratten J, Taylor PW. Isolation of bacteriophages from the oral cavity. Lett Appl Microbiol 2004;39:215-9. [DOI] [PubMed] [Google Scholar]
- 110.Faruque SM, Islam MJ, Ahmad QS, et al. Self-limiting nature of seasonal cholera epidemics: Role of host-mediated amplification of phage. Proc Natl Acad Sci USA 2005;102:6119-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huff WE, Huff GR, Rath NC, Balog JM, Donoghue AM. Prevention of Escherichia coli infection in broiler chickens with a bacteriophage aerosol spray. Poult Sci 2002;81:1486-91. [DOI] [PubMed] [Google Scholar]
- 112.Brussow H, Bruttin A, Desiere F, Lucchini S, Foley S. Molecular ecology and evolution of Streptococcus thermophilus bacteriophages - A review. Virus Genes 1998;16:95-109. [DOI] [PubMed] [Google Scholar]
- 113.Sozzi T, Maret R, Cerise L. Isolation and some characteristics of two Micrococcus phages from Italian salami, type Varzi. Arch Mikrobiol 1973;92:313-20. [DOI] [PubMed] [Google Scholar]
- 114.Huff WE, Huff GR, Rath NC, Balog JM, Donoghue AM. Evaluation of aerosol spray and intramuscular injection of bacteriophage to treat an Escherichia coli respiratory infection. Poult Science 2003;82:1108-12. [DOI] [PubMed] [Google Scholar]
- 115.Bruttin A, Brussow H. Human volunteers receiving Escherichia coli phage T4 orally: A safety test of phage therapy. Antimicrob Agents Chemother 2005;49:2874-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gorski A, Weber-Dabrowska B, Dabrowska K, et al. Clinical phage therapy: Present and future. America Society for Microbiology Conference on the New Phage Biology. Key Biscayne, 2004. [Google Scholar]
- 117.Slopek S, Weber-Dabrowska B, Dabrowski M, Kucharewicz-Krukowska A. Results of bacteriophage treatment of suppurative bacterial infections in the years 1981-1986. Arch Immunol Ther Exp (Warsz) 1987;35:569-83. [PubMed] [Google Scholar]
- 118.Weber-Dabrowska B, Mulczyk M, Gorski A. Bacteriophage therapy of bacterial infections: An update of our institute’s experience. Arch Immunol Ther Exp (Warsz) 2000;48:547-51. [PubMed] [Google Scholar]
- 119.Stone R. Stalin’s forgotten cure. Science 2002;298:728-31. [DOI] [PubMed] [Google Scholar]
- 120.Faruque SM, Naser IB, Islam MJ, et al. Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc Natl Acad Sci USA 2005;102:1702-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brussow H. Phage-host interaction: An ecological perspective. J Bacteriol 2004;186:3677-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hadas H, Einav M, Fishov I, Zaritsky A. Bacteriophage T4 development depends on the physiology of its host Escherichia coli. Microbiology 1997;143:179-85. [DOI] [PubMed] [Google Scholar]
- 123.Rabinovitch A, Fishov I, Hadas H, Einav M, Zaritsky A. Bacteriophage T4 development in Escherichia coli is growth rate dependent. J Theor Biol 2002;216:1-4. [DOI] [PubMed] [Google Scholar]
- 124.Rabinovitch A, Zaritsky A, Fishov I, Einav M, Hadas H. Bacterial lysis by phage - A theoretical model. J Theor Biol 1999;201:209-13. [DOI] [PubMed] [Google Scholar]
- 125.Wegrzyn G, Thomas MS. Modulation of the susceptibility of intestinal bacteria to bacteriophages in response to Ag43 phase variation - A hypothesis. Med Sci Monit 2002;8:HY15-8. [PubMed] [Google Scholar]
- 126.You L, Suthers PF, Yin J. Effects of Escherichia coli physiology on growth of phage T7 in vivo and in silico. J Bacteriol 2002;184:1888-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Varey P, Alione C, Robison M, et al. Single-step growth curves involving T-even like phages in anaerobic respiration conditions. <http://www.evergreen.edu/phage/research/petecostanzoabstract.htm> (Version current at September 29, 2006).
- 128.Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 2006;28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Harcombe WR, Bull JJ. Impact of phages on two-species bacterial communities. Appl Environ Microbiol 2005;71:5254-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Merril CR, Biswas B, Carlton R, et al. Long-circulating bacteriophage as antibacterial agents. Proc Natl Acad Sci USA 1996;93:3188-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vitiello CL, Merril CR, Adhya S. An amino acid substitution in a capsid protein enhances phage survival in mouse circulatory system more than a 1000-fold. Virus Res 2005;114:101-3. [DOI] [PubMed] [Google Scholar]
- 132.Jikia D, Chkhaidze N, Imedashvili E, et al. The use of a novel biodegradable preparation capable of the sustained release of bacteriophages and ciprofloxacin, in the complex treatment of multidrug-resistant Staphylococcus aureus-infected local radiation injuries caused by exposure to Sr90. Clin Exp Dermatol 2005;30:23-6. [DOI] [PubMed] [Google Scholar]
- 133.Markoishvili K, Tsitlanadze G, Katsarava R, Morris JG Jr, Sulakvelidze A. A novel sustained-release matrix based on biodegradable poly(ester amide)s and impregnated with bacteriophages and an antibiotic shows promise in management of infected venous stasis ulcers and other poorly healing wounds. Int J Dermatol 2002;41:453-8. [DOI] [PubMed] [Google Scholar]
- 134.Shaishmelashvili G. Adaptation of standard method of experimental burn injury for phagobioderm investigation. Georgian Med News 2005:51-3. [PubMed] [Google Scholar]
- 135.Spencer J, McColm F, Mattey M. Treatment of methicillin resistant Staphylococcus aureus (MRSA) infection by immobilized bacteriophage. 156th Annual Meeting of the Society for General Microbiology. Edinburgh, April 4 to 7, 2005. (Abst) [Google Scholar]
- 136.Scholl D, Adhya S, Merril C. Escherichia coli K1’s capsule is a barrier to bacteriophage T7. Appl Environ Microbiol 2005;71:4872-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Scholl D, Merril C. The genome of bacteriophage K1F, a T7-like phage that has acquired the ability to replicate on K1 strains of Escherichia coli. J Bacteriol 2005;187:8499-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yamamoto N. Genetic evolution of bacteriophage. I. Hybrids between unrelated bacteriophages P22 and Fels 2. Proc Natl Acad Sci USA 1969;62:63-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schaak D. Toxin-phage bacteriocide as an antibiotic therapy. Rowland Institute of Harvard, 2002. <http://www.rowland.harvard.edu/organization/past_research/bacteriophage/bacteriophage.html> (Version current at October 10, 2006).
- 140.Hagens S, Habel A, von AU, von GA, Bläsi U. Therapy of experimental pseudomonas infections with a nonreplicating genetically modified phage. Antimicrob Agents Chemother 2004;48:3817-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Westwater C, Schofield DA, Schmidt MG, Norris JS, Dolan JW. Development of a P1 phagemid system for the delivery of DNA into Gram-negative bacteria. Microbiology 2002;148:943-50. [DOI] [PubMed] [Google Scholar]
- 142.Embleton ML, Nair SP, Heywood W, Menon DC, Cookson BD, Wilson M. Development of a novel targeting system for lethal photosensitization of antibiotic-resistant strains of Staphylococcus aureus. Antimicrob Agents Chemother 2005;49:3690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ramachandran J, Padmanabhan S, Sriram B. Incapacitated whole-cell immunogenic bacterial compositions. US patent 2002;6,913,753 B2. [Google Scholar]
- 144.Ramachandran J, Padmanabhan S, Sriram B. Lysin-deficient bacteriophages having reduced immunogenicity. US patent 2002;6,896,882 B2. [Google Scholar]
- 145.Mullan WM, Crawford RJ. Partial purification and some properties of φC2(W) lysin, a lytic enzyme produced by phage-infected cells of Streptococcus lactis C2. J Dairy Res 1985;52:123-38. [DOI] [PubMed] [Google Scholar]
- 146.Fischetti VA. Bacteriophage lytic enzymes: Novel anti-infectives. Trends Microbiol 2005;13:491-6. [DOI] [PubMed] [Google Scholar]
- 147.Loessner MJ. Bacteriophage endolysins - Current state of research and applications. Curr Opin Microbiol 2005;8:480-7. [DOI] [PubMed] [Google Scholar]
- 148.Schuch R, Nelson D, Fischetti VA. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 2002;418:884-9. [DOI] [PubMed] [Google Scholar]
- 149.Carlton RM, Noordman WH, Biswas B, de Meester ED, Loessner MJ. Bacteriophage P100 for control of Listeria monocytogenes in foods: Genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul Toxicol Pharmacol 2005;43:301-12. [DOI] [PubMed] [Google Scholar]
- 150.Cheng Q, Nelson D, Zhu S, Fischetti VA. Removal of group B streptococci colonizing the vagina and oropharynx of mice with a bacteriophage lytic enzyme. Antimicrob Agents Chemother 2005;49:111-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nelson D, Schuch R, Zhu S, Tscherne DM, Fischetti VA. Genomic sequence of C1, the first streptococcal phage. J Bacteriol 2003;185:3325-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Entenza JM, Loeffler JM, Grandgirard D, Fischetti VA, Moreillon P. Therapeutic effects of bacteriophage Cpl-1 lysin against Streptococcus pneumoniae endocarditis in rats. Antimicrob Agents Chemother 2005;49:4789-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.O’Flaherty S, Coffey A, Meaney W, Fitzgerald GF, Ross RP. The recombinant phage lysin LysK has a broad spectrum of lytic activity against clinically relevant staphylococci, including methicillin-resistant Staphylococcus aureus. J Bacteriol 2005;187:7161-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Aziz RK, Edwards RA, Taylor WW, Low DE, McGeer A, Kotb M. Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J Bacteriol 2005;187:3311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Banks DJ, Beres SB, Musser JM. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol 2002;10:515-21. [DOI] [PubMed] [Google Scholar]
- 156.Nakagawa I, Kurokawa K, Yamashita A, et al. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res 2003;13:1042-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sumby P, Porcella SF, Madrigal AG, et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis 2005;192:771-82. [DOI] [PubMed] [Google Scholar]
- 158.Hayashi T, Makino K, Ohnishi M, et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 2001;8:11-22. (Erratum in 2001;8:96). [DOI] [PubMed] [Google Scholar]
- 159.Perna NT, Plunkett G III, Burland V, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 2001;409:529-33. (Erratum in 2001;410:240). [DOI] [PubMed] [Google Scholar]
- 160.Herold S, Siebert J, Huber A, Schmidt H. Global expression of prophage genes in Escherichia coli O157:H7 strain EDL933 in response to norfloxacin. Antimicrob Agents Chemother 2005;49:931-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ingrey KT, Ren J, Prescott JF. A fluoroquinolone induces a novel mitogen-encoding bacteriophage in Streptococcus canis. Infect Immun 2003;71:3028-33. [DOI] [PMC free article] [PubMed] [Google Scholar]