Abstract
Background
In summer 2003, a respiratory outbreak was investigated in British Columbia, during which nucleic acid tests and serology unexpectedly indicated reactivity for severe acute respiratory syndrome coronavirus (SARS-CoV).
Methods
Cases at a care facility were epidemiologically characterized and sequentially investigated for conventional agents of respiratory infection, SARS-CoV and other human CoVs. Serological cross-reactivity between SARS-CoV and human CoV-OC43 (HCoV-OC43) was investigated by peptide spot assay.
Results
Ninety-five of 142 residents (67%) and 53 of 160 staff members (33%) experienced symptoms of respiratory infection. Symptomatic residents experienced cough (66%), fever (21%) and pneumonia (12%). Eight residents died, six with pneumonia. No staff members developed pneumonia. Findings on reverse transcriptase-polymerase chain reaction assays for SARS-CoV at a national reference laboratory were suspected to represent false positives, but this was confounded by concurrent identification of antibody to N protein on serology. Subsequent testing by reverse transcriptase-polymerase chain reaction confirmed HCoV-OC43 infection. Convalescent serology ruled out SARS. Notably, sera demonstrated cross-reactivity against nucleocapsid peptide sequences common to HCoV-OC43 and SARS-CoV.
Conclusions
These findings underscore the virulence of human CoV-OC43 in elderly populations and confirm that cross-reactivity to antibody against nucleocapsid proteins from these viruses must be considered when interpreting serological tests for SARS-CoV.
Key Words: Coronavirus, Human coronavirus OC43, Outbreak, Respiratory infection, SARS-CoV, SARS coronavirus, Severe acute respiratory syndrome
On July 5, 2003, the World Health Organization (WHO) announced that human transmission of severe acute respiratory syndrome coronavirus (SARS-CoV) had ceased (1). Many drew the inference that the virus, whose causal role for SARS had been established (2-5), had been driven back into nature. From its emergence in November 2002, SARS was characterized as a lower respiratory syndrome involving pneumonia and/or acute respiratory distress syndrome (6,7). Transmission to others occurred most easily from severely affected patients, and was rare without fever (8).
Hence, a report of reactive tests for SARS-CoV on respiratory specimens submitted during an outbreak of generally mild respiratory infection accompanied by limited mortality in the elderly residents of a British Columbia long-term care facility during the summer of 2003 was unexpected (9). In the present article, the outbreak is described, the initial and subsequent laboratory findings are summarized, and the probable basis of serological cross-reactivity responsible for early findings is elucidated.
Methods
Epidemiological Methods
A case definition was established as “any resident or staff member who experienced symptoms of respiratory infection – rhinitis, sore throat, fever, cough or pneumonia – after July 1, 2003”. Assessment of the extent and timing of symptoms was performed by chart review for residents of the facility and for patients transferred to hospital, and by interview of health care professionals. Records of staff absenteeism were examined, and interviews with ill workers were conducted. The accumulated findings were entered into a database (Microsoft Access, Microsoft Corporation, USA). Tabulation and construction of epidemic curves were accomplished in Microsoft Excel (Microsoft Corporation, USA).
Specimen Collection
Nasopharyngeal, nasal and lower respiratory specimens were collected from a subset of symptomatic residents and transported to the University of British Columbia Centre for Disease Control (BCCDC) laboratories within 24 h. Specimens from the lower respiratory tract, stools and autopsy materials were obtained where available. Sera were collected from a subset of symptomatic residents in various stages of presentation. Sera collected before the 21st day following symptom onset were considered acute; after the 28th day, they were considered convalescent.
Microbiological Testing
Respiratory specimens were tested for adenovirus, parainfluenza virus, respiratory syncytial virus, enterovirus and influenza virus by immunofluorescence microscopy and isolation in cell culture (10). Serological testing was carried out for Mycoplasma pneumoniae, Chlamydophila pneumoniae and Chlamydophila psittaci. Respiratory specimens were cultured for bacterial pathogens and, together with urine specimens, were evaluated for Legionella antigen using standard methods (10).
Nucleic Acid-based Testing
The reverse transcriptase-polymerase chain reaction (RT-PCR) assays for SARS-CoV were conducted at the National Microbiology Laboratory (NML) in Winnipeg, Manitoba, the BCCDC and the British Columbia Cancer Agency’s Genome Sciences Centre (Vancouver, British Columbia); the results are summarized in Table 1. A detailed description of assays is available as a technical appendix through personal communication with the authors. Of note, the ‘pancoronavirus’ PCR (BCCDC) targeted a 220-base pair region of the polymerase 1b gene that is highly conserved among CoVs (11). Amplicons from this assay were sequenced for phylogenetic analysis. RT-PCR was also performed for human metapneumovirus (hMPV) at the NML (12).
TABLE 1.
Listing of diagnostic methods and key study findings
| PCR assay | Laboratory | Gene target(s) | Results |
|---|---|---|---|
| RT-nested PCR for SARS-CoV | NML | Membrane glycoprotein and polymerase | Specimens from 9 of 40 patients were positive (two on repeat extraction) |
| TaqMan RT-PCR (Applied Biosystems, USA) for SARS-CoV | NML | Polymerase and nucleocapsid | 3 of 9 specimens were positive |
| RT-PCR for SARS-CoV | BCCDC | Nucleocapsid | 0 of 66 specimens from 39 residents were positive |
| Roche LightCycler PCR (Roche Diagnostics, USA) for SARS-CoV | BCCDC | Polymerase 1b | 0 of 40 specimens from 25 residents were positive |
| ‘Pancoronavirus’ PCR | BCCDC | Conserved region of polymerase 1b | 14 of 39 residents positive in one or more specimen |
| RT-PCR for HCoV-OC43 | BCGSC | Nucleocapsid and matrix | 3 of 3 specimens positive |
| RT-PCR for HCoV-OC43 | NML | Polymerase and nucleocapsid | 4 specimens positive |
| Serological assay | Laboratory | Antigen/method | Results |
| ELISA for SARS-CoV antibody | NML | Infected cell lysate, baculovirus expressed N protein | 6 of 35 acute specimens reactive for antibody to SARS-CoV by ELISA, neutralizing antibody or both. Three others with weak N band by WB. No positives by neutralizing antibody on convalescent testing |
| IFA for SARS-CoV antibody | NML | Infected cells | |
| Neutralizing antibody to SARS-CoV | NML | Plaque reduction on Vero E6 cells | |
| WB | NML | Baculovirus expressed N protein* | |
| Euroimmun indirect immunofluorescence (Medizinische Labordiagnostika, Germany) | BCCDC | Escherichia coli expressed HIS-tagged SARS-CoV nucleocapsid protein | 0 of 35 specimens positive. Indeterminate NA assay of one in eight found in six patients. No confirmed positives with convalescent serology |
| WB | BCCDC | Infected cell lysate and recombinant N protein | |
| NA to SARS-CoV | BCCDC | Microneutralization test on Vero E6 cells |
Data from references 3 and 7. BCCDC British Columbia Centre for Disease Control; BCGSC British Columbia Cancer Agency’s Genome Sciences Centre; HCoV Human coronavirus; IFA Immunofluorescent antibody; NA Neutralizing antibody; NML National Microbiology Laboratory; RT-PCR Real-time polymerase chain reaction; SARS-CoV Severe acute respiratory syndrome CoV; WB Western blot
Serological Methods
Methods used at the NML and BCCDC are summarized in Table 1. The BCCDC diagnostic algorithm required that specimens reactive by ELISA be confirmed using the neutralizing antibody test.
Assessing the Potential for Cross-reactivity
Multiple 10 amino acid peptides incorporating all the sequences in the SARS-CoV and human CoV (HCoV)-OC43 nucleocapsid proteins were prepared on derivatized cellulose membranes using a robotic peptide synthesizer (Autospot ASP 222, Intavis Bioanalytical Instruments, Germany) as previously described (13). The peptides overlapped by eight residues, with each subsequent peptide on a membrane being shifted by two amino acids toward the C-terminal end. There were 220 peptide spots prepared from the sequence of the HCoV-OC43 nucleocapsid protein and 207 spots from the corresponding SARS-CoV protein.
The membranes were blocked overnight at 4°C with 5% skim milk. They were then incubated with serum samples diluted 100-fold in buffer for 1 h at 37°C. After washing, membranes were treated with horseradish peroxidase-conjugated goat antihuman immunoglobulin G (Sigma, USA). The membranes were developed with enhanced chemiluminescence reagents and the positive spots were visualized on a Bio-Rad Fluor-S Multi-Imager (Bio-Rad Laboratories, USA).
Results
Epidemiology
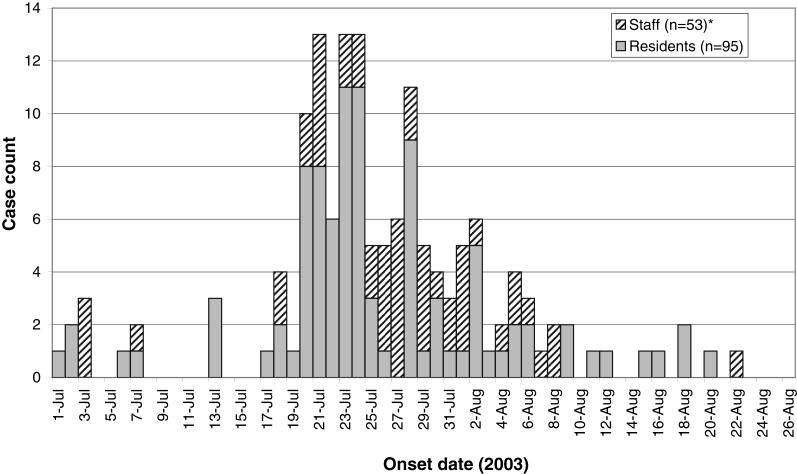
The outbreak occurred in a long-term care facility located in British Columbia. Residents were elderly or disabled adults in need of intermediate or extended care. From July 1 to August 22, 2003, 95 of 142 residents (67%) and 53 of 160 staff members (33%) experienced respiratory tract infections. The median age of affected residents was 83.7 years, with a range of 43.6 to 98.7 years. The epidemic curve is depicted in Figure 1. Peak incidence occurred between July 20 and 28, and the outbreak had tapered off by August 22, 2003. A floor-by-floor pattern of spread was not discernible, but a common area on the ground floor was used by most residents. Symptoms, findings and outcomes in residents and staff are summarized in Table 2. Most had mild clinical findings. Of the residents who met the case definition, 66% experienced cough, 21% fever, 66% rhinitis and 12% pneumonia. Twelve residents were admitted to hospital, eight for pneumonia. There were eight deaths among patients with respiratory symptoms. During the preceding six months, there was a mean of three deaths per month. None of the 53 affected staff members died, was hospitalized or developed pneumonia.
Figure 1.
Epidemic curve: Onset of respiratory illness in a long-term care facility. *Onset dates for two staff members are missing
TABLE 2.
Spectrum of symptoms in affected residents and staff
| Symptom or finding | Residents (n=95) | Staff (n=53) | ||
|---|---|---|---|---|
| n | % | n | % | |
| Fever | 20 | 21.1 | 11 | 20.8 |
| Dyspnea | 8 | 8.4 | 4 | 7.5 |
| Cough | 63 | 66.3 | 18 | 34.0 |
| Sore throat | 23 | 24.2 | 34 | 64.2 |
| Runny nose | 63 | 66.3 | 36 | 67.9 |
| Myalgia | 9 | 9.5 | 20 | 37.7 |
| Headache | 2 | 2.1 | 22 | 41.5 |
| Vomiting | 2 | 2.1 | 2 | 3.8 |
| Diarrhea | 6 | 6.3 | 8 | 15.1 |
| Rash | 0 | 0.0 | 1 | 1.9 |
| Fatigue | 9 | 9.5 | 21 | 39.6 |
| Nausea | 2 | 2.1 | 6 | 11.3 |
| Chills | 1 | 1.1 | 2 | 3.8 |
| Pneumonia | 12 | 12.6 | 0 | 0.0 |
| Hospitalized (x-ray confirmed) | 8 | 8.4 | 0 | 0.0 |
| Clinically diagnosed | 4 | 4.2 | 0 | 0.0 |
| Hospitalized without pneumonia* | 4 | 4.2 | 0 | 0.0 |
| Deaths† | 8 | 8.4 | 0 | 0.0 |
Reasons for hospitalization among those admitted without pneumonia included gastric bleed (n=1), pulmonary embolus (n=1) and exacerbation of chronic obstructive pulmonary disease (n=1);
Two residents with congestive heart failure/pneumonia, two with pneumonia, one with chronic obstructive pulmonary disease/pneumonia, one with pulmonary embolism, one with ruptured aortic aneurism, and one admitted for palliative care
Microbiology Findings
No pathogenic organisms were identified on routine testing from any of the residents. The exception was the finding of hMPV in respiratory specimens from four of 17 residents tested by RT-PCR. Molecular or serological testing for SARS-CoV was performed on specimens from 51 of 95 affected residents, the results of which are summarized in the final column of Table 1.
Findings from Nucleic Acid Testing for SARS-CoV
Specimens from nine patients had a signal on one or more RT-PCR assays at the NML and were suspected of being false positives. These included two of the patients who had tested positive for hMPV by PCR at the NML. Analysis of the sequences of amplicons from three suspected false-positive PCR specimens at the NML (lung, stool and respiratory) showed homology to sequences of SARS-CoV (14,15). No specimens were positive for SARS-CoV by RT-PCR at the BCCDC.
Findings from Nucleic Acid Tests for Other CoVs
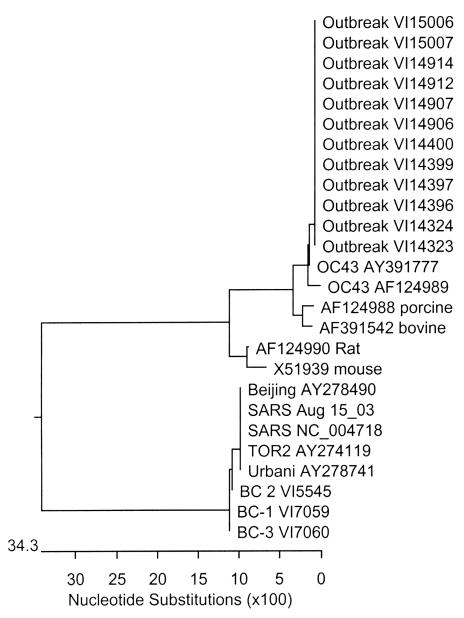
Table 1 summarizes the findings from nucleic acid tests for other CoVs. RT-PCR amplicons from specimens of nine of the 14 patients obtained by the ‘pancoronavirus’ assay were of concentrations adequate for sequencing. The following 163 base pair sequence internal to the primers was obtained from 12 specimens from these nine patients (including three who had tested positive for hMPV by PCR at the NML): TCGTGCTATGCCAAACCTACTACGTATTGTTAGTAGTTTGGTATTAGCCCGAAAACATGAGACATGTTGCTCGCAAAGCGATAGGTTTTATCGACTTGCGAATGAATGCGCACAAGTTTTGAGTGAAATTGTTATGTGTGGTGGCTGTTATTATGTTAAGCCT. This sequence has a homology of over 99% (162 of 163) to the corresponding region from HCoV-OC43, but of only 51.2% to the corresponding region of SARS-CoV (Figure 2) (16). Three of these specimens were independently tested by RT-PCR targeted to the matrix and nucleocapsid genes at the British Columbia Cancer Agency’s Genome Sciences Centre, and the amplicons were sequenced. Analysis of the sequences (GenBank accession number AY382775-7) showed that they were 98.5% homologous to the corresponding sequences of the HCoV-OC43 genes (16,17). HCoV-OC43 sequences in specimens from four patients were also identified by the NML.
Figure 2.
Phylogenetic tree from sequencing of the pancoronavirus polymerase 1b gene region. Image reproduced with permission from the University of British Columbia Centre for Disease Control Molecular Services
Findings from Serological Testing
Acute and convalescent sera were collected from 35 affected residents (Table 1). For all nine patients for whom antibody to SARS-CoV were detected on preliminary testing of acute specimens by any of the above methods, no significant neutralizing antibody titres were detected in the acute or convalescent sera by microneutralization test at the BCCDC or by repeat plaque reduction neutralization assay at the NML. In six patients, neutralizing activity was detected in the serum at a dilution of 1:8, but this was not repeatable. HCoV-OC43-like virus was detected in specimens from three of these six cases. (In this test, neutralizing activity at a dilution of 1:8 has been detected sporadically in healthy controls. In contrast, activity in convalescent sera from SARS patients ranges between dilutions of 1:32 and 1:1024). There was no evidence of seroconversion when acute and convalescent sera were tested in parallel by the virus neutralization test.
Correlation of Clinical and Laboratory Findings
Clinical and laboratory findings for all 18 patients whose specimens on initial or subsequent testing by any method had evidence of SARS-CoV are displayed in Table 3. Residents in whom SARS-CoV was potentially indicated by initial nucleic acid or serological tests expressed a range of symptoms, from mild disease through to lobar pneumonia, which were not consistent with the known profile of SARS.
TABLE 3.
Correlation of laboratory and clinical findings for 18 patients with an initial assay positive for severe acute respiratory syndrome coronavirus (SARS-CoV)
| Patient number | NML SARS-CoV PCR | BCCDC HCoV-OC43 PCR | NML SARS-CoV serology acute/conv | BCCDC SARS-CoV serology acute/conv | hMPV byPCR | Fever | URIsymptoms | Pneumonia | Disposition |
|---|---|---|---|---|---|---|---|---|---|
| 12 | + Nasal | – Nasal | ND/– | ND/Ind | + | + | + | + | Hospitalized |
| 13 | + Stool | – NP | +/– | Ind/Ind | + | Recovered | |||
| 14 | + Nasal | – Nasal | ND/– | –/– | + | Recovered | |||
| 17 | + NP | + NP | –/ND | –/ND | + | + | Died | ||
| 18 | ND | ND | –/– | Ind/Ind | + | Recovered | |||
| 19 | ND | – Stool | WB+/– | –/– | + | Recovered | |||
| 21 | + Nasal | + Nasal | –/– | –/– | + | Recovered | |||
| 26 | + Autopsy | + Autopsy | ND | ND | + | + | + | Died | |
| 27 | + Nasal | + Nasal | –/– | –/– | + | Recovered | |||
| 29 | ND | ND | +/– | –/– | + | + | Recovered | ||
| 30 | ND | ND | WB+/– | –/– | + | Recovered | |||
| 32 | + Nasal | + NP | +/– | –/– | + | Recovered | |||
| 35 | – Nasal | + Nasal | +/– | –/– | + | + | Recovered | ||
| 36 | – Stool | – Stool | –/– | Ind/Ind | + | + | Recovered | ||
| 44 | ND | ND | +/– | Ind/Ind | + | Recovered | |||
| 46 | + Nasal | + Nasal | –/– | –/– | + | Recovered | |||
| 47 | ND | ND | WB+/– | –/– | + | Recovered | |||
| 50 | ND | ND | +/– | –/– | + | Recovered |
NML serology was denoted as ‘+’ if positive by ELISA, neutralizing antibody assay or both. BCCDC British Columbia Centre for Disease Control; Conv Convalescent; hMPV Human metapneumovirus; Ind Indeterminate (screening ELISA weakly positive but confirmatory neutralization assay negative); ND Not done; NML National Microbiology Laboratory; NP Nasopharyngeal specimen; PCR Polymerase chain reaction; URI Upper respiratory infection; WB+ Serology positive only by Western blot (ELISA and indirect fluorescent antibody negative on same specimen)
Findings from Peptide Assay for Cross-reactivity
Eight patient sera from the outbreak were analyzed (Table 4). Five of these had tested weakly reactive by the recombinant nucleocapsid protein ELISA for SARS-CoV at the BCCDC, but had no detectable neutralizing antibody activity (patients 12, 13, 18, 36 and 44). Two of these and one other (patients 13, 44 and 50) were found to have antibody to SARS-CoV by the NML, and two sera that were nonreactive for SARS-CoV antibody on retesting in any laboratory were further analyzed for reactivity to the peptides of the N proteins of SARS-CoV and CoV-OC43.
TABLE 4.
Reactivity of sera with severe acute respiratory syndrome coronavirus (SARS-CoV) and human CoV-OC43 peptides
| Case number | S2 | 43-2 | S3 | 43-3 | S4 | 43-4 | S5 | 43-5 | SARS-CoVELISA-ODBCCDC | SARS-CoVELISA NML |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | + | + | – | – | + | – | + | + | 0.060 | neg |
| 12 | + | + | – | + | + | + | – | + | 0.825 | neg |
| 13 | + | + | + | + | + | + | + | + | 0.255 | 1:800 |
| 18 | + | + | + | – | + | + | + | + | 0.243 | neg |
| 36 | + | – | + | – | + | + | – | + | 0.435 | neg |
| 44 | + | + | + | + | + | + | + | + | 0.320 | neg |
| 48 | – | – | – | – | – | – | – | + | 0.029 | neg |
| 50 | + | + | + | + | + | + | + | + | 0.065 | 1:3200 |
| For SARS nucleocapsid protein, | For OC43 nucleocapsid protein, | |||||||||
| the sequences recognized are: | the sequences recognized are: | |||||||||
| S2=GLPNNTASWFTA | 43-2=GNVVPYYSWFSG | |||||||||
| S3=KMKELSPRWYFYYLGT | 43-3=NQRQLLPRWYFYYLGT | |||||||||
| S4=SQASSRSSSRSRGNSR | 43-4=RSAPNSRSTSRTSSRA | |||||||||
| S5=SAFFGMSRIGME | 43-5=APTAGAFFFGSRLELA | |||||||||
| S3 homology with 43-3 | Abbreviations: | |||||||||
| S4 homology with 43-4 | ELISA NML: ELISA report from the | |||||||||
| S5 homology with 43-5 | National Microbiology Laboratory | |||||||||
| S2 homology with 43-2 | ELISA-OD: Optical density reading | |||||||||
| of ELISA at the British Columbia | ||||||||||
| ‘+’ indicates patient serum reactive | Centre for Disease Control (usual | |||||||||
| to the indicated peptide spot. | cut-off is 0.20) | |||||||||
Four of the sites recognized by the sera shared homology between SARS-CoV and HCoV-OC43, and these have been designated S2 to S5 and 43-2 to 43-5, respectively. Where two or three peptides in a row were recognized, the overall sequence was documented (ie, 12 amino acids were listed in the case of two in a row; 14 for three in a row). Serum from patient 48 that was not reactive by SARS-CoV nucleocapsid protein ELISA was only reactive for peptide site 43-5. Conversely, the sera from patients that were weakly reactive by the BCCDC or NML ELISA (patients 13, 44 and 50) were reactive to all peptides expressed by both SARS-CoV and HCoV-OC43. Sera from the other patients were reactive for one to three of the four homologous peptides. These findings show that there are epitopes on the nucleocapsid protein at which antibodies to SARS-CoV and HCoV-OC43 may cross-react.
Public Health Measures
Once the outbreak was reported, routine influenza-control measures were instituted. After findings suggesting possible involvement of SARS-CoV were reported, these measures were enhanced, and SARS-type respiratory and contact precautions were exercised around symptomatic cases (18). This was to assure that measures shown to limit the spread of SARS-CoV (19,20) were in place until or unless it could be demonstrated that the agent responsible for the facility outbreak was not SARS-CoV.
Discussion
We have characterized an outbreak of respiratory illness due to HCoV-OC43. The observed attack rate of 67% and case fatality rate of 8% underscore the pathogenic potential of HCoVs in frail populations. This adds to other observations underscoring that CoVs other than SARS-CoV may be responsible for a broader spectrum of disease than coryza alone (21-23).
The fact that no staff members were severely affected was clearly incongruent with tests indicating the possible presence of SARS-CoV. British Columbia had experienced three importations and one occupational transmission of SARS during the original global outbreak (24). None of the four SARS patients were epidemiologically connected to the facility where this outbreak took place.
While initial RT-PCR findings at the NML were suspected to be false-positive because they did not fit the epidemiological picture, early signals on serological testing meant that SARS precautions were prudent as the investigation progressed. The outbreak was observed for 60 days, during which no severe illness among staff and no change toward a pattern reminiscent of SARS occurred. Subsequent molecular and serological investigations conducted at three laboratories ruled against a role for SARS-CoV and in favour of HCoV-OC43 as the etiological agent for this outbreak. Consultation among the BCCDC, Health Canada and the WHO concluded that the outbreak was not SARS, and was unlikely to have involved SARS-CoV.
In 2003, the SARS diagnosis was in its infancy. Several reports have emphasized the increased risk of false-positive SARS tests in the postoutbreak period because of very low pretest likelihood and discussed the potential for cross-reactivity (25,26). The WHO called for further standardization of SARS testing. Since then, several reports (27-31) have made it clear that cross-reactivity on serological testing may occur, especially when the target is the nucleocapsid protein. Such cross-reactivity has been demonstrated to nucleocapsid proteins of other CoVs but also to human interleukin-11. These studies confirm that antibodies to spike protein are more specific and generally recommend the use of more than one antigen in confirmatory testing or the use of a virus neutralization test.
In the present study, most sera evaluated for cross-reactivity recognized homologous peptide sequences on HCoV-OC43 and SARS-CoV nucleocapsid protein, establishing that these cases were indeed producing cross-reacting antibodies. The immune response to the peptides was most likely a broad-spectrum response that the patients had evolved as a result of repeat infections by different CoVs throughout their lives. Hence, serological tests based on reactions to the SARS-CoV nucleocapsid protein may inadvertently be interpreted as reactive due to cross-reactivity with antibodies from a current or preceding CoV infection, and it is not surprising that Western blot and ELISA assays based on that moiety should have given false-positive results. Because the epitopes to which neutralizing antibodies are directed are believed to be on the spike protein, such cross-reactions may be less likely during the neutralizing antibody test.
The issue of serological cross-reactivity is also compelling when interpreting reports on the seroprevalence of SARS-CoV in animal and human hosts (32,33) and reports of apparent sporadic SARS-CoV infection (34).
With respect to the nucleic acid-based tests used by the NML in these investigations, false-positive results driven by intrinsic test performance or amplicon contamination remain most likely. The reference laboratory had processed hundreds of specimens during the preceding SARS outbreak. Depletion of original specimens made it impossible to completely confirm this assumption. The NML has maintained rigorous quality control since then, with no evidence of similar findings. Since 2003, there has been some effort to refine RT-PCR testing for SARS-CoV generally, and most experts now caution against the use of nested methods for routine diagnostic testing (35).
The present investigation underscores the fact that laboratory testing is but one way to form inferences on the etiology of outbreaks, and cannot replace scrupulous clinical and epidemiological observation. When these different approaches lead to inconsistent observations, it is important to remain open to all possible explanations.
Acknowledgments
The authors wish to acknowledge the following individuals: Dr Murray Fyfe and Ms Cheryl McIntyre for assistance with epidemiology, interviews and organization of materials; Ms Lisan Kwindt and Ms Natalie Gaudun for assistance with manuscript preparation; Hayley Rees-Howlett for assistance during record review at the long-term care facility; Lisa Chu, who organized the public health nursing response for contact investigation and follow-up; L Liu, E Grudeski, M Traykova-Andonova, J Krishnan, C Ouellette and D Dick (National Microbiology Laboratory) and Kathy Adie, Loretta Janz, A Mak and R Chow (British Columbia Centre for Disease Control) for many hours contributed to resolving diagnostic test findings; and William Campbell for assistance with the peptide spot assay. The authors also express their appreciation for the cooperation of residents and families of the affected facility. Investigations were performed as a component of an outbreak investigation and did not involve human experimentation. The authors have no relevant conflict of interest to declare. Financial support was provided by the home institutions as listed on the title page.
References
- 1.World Health Organization. Update 96 - Taiwan, China: SARS transmission interrupted in last outbreak area <www.who.int/csr/ don/2003_07_05/en> (Version current at November 6, 2006).
- 2.Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003;361:1319-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003;348:1953-66. [DOI] [PubMed] [Google Scholar]
- 4.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003;348:1967-76. [DOI] [PubMed] [Google Scholar]
- 5.Fouchier RA, Kuiken T, Schutten M, et al. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature 2003;423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drazen JM. SARS - looking back over the first 100 days. N Engl J Med 2003;349:319-20. [DOI] [PubMed] [Google Scholar]
- 7.Poutanen SM, Low DE, Henry B, et al. Identification of severe acute respiratory syndrome in Canada. N Engl J Med 2003;348:1995-2005. [DOI] [PubMed] [Google Scholar]
- 8.Varia M, Wilson S, Sarwal S, et al. Investigation of a nosocomial outbreak of severe acute respiratory syndrome (SARS) in Toronto, Canada. CMAJ 2003;169:285-92. [PMC free article] [PubMed] [Google Scholar]
- 9.Enserink M. Infectious diseases. Unexplained false alarm may hold lessons. Science 2003;302:767. [DOI] [PubMed] [Google Scholar]
- 10.Patrick R, Murray P, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH. Manual of Clinical Microbiology. Washington: ASM Press, 2003. [Google Scholar]
- 11.Adachi D, Johnson G, Draker R, et al. Comprehensive detection and identification of human coronaviruses, including the SARS-associated coronavirus, with a single RT-PCR assay. J Virol Methods 2004;122:29-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bastien N, Normand S, Taylor T, et al. Sequence analysis of the N, P, M and F genes of Canadian human metapneumovirus strains. Virus Res 2003;93:51-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo JP, Petric M, Campbell W, McGeer PL. SARS corona virus peptides recognized by antibodies in the sera of convalescent cases. Virology 2004;324:251-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gene sequence of the SARS Coronavirus (Frankfurt Strain). GenBank accession number AY291315. <www.ncbi.nlm.nih.gov/ entrez/viewer.fcgi?db=nucleotide&val=31581502> (Version current at November 6, 2006).
- 15.Marra MA, Jones SJ, Astell CR, et al. The Genome sequence of the SARS-associated coronavirus. Science 2003;300:1399-404. [DOI] [PubMed] [Google Scholar]
- 16.Gene sequence of human coronavirus OC43. GenBank accession number AY391777. <http://www.ncbi.nlm.nih.gov/entrez/ viewer.fcgi?db=nucleotide&val=37702815> (Version current at November 6, 2006).
- 17.Kamahora T, Soe LH, Lai MM. Sequence analysis of nucleocapsid gene and leader RNA of human coronavirus OC43. Virus Res 1989;12:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Health Canada. Public health management of SARS cases and contacts: Interim guidelines. <http://www.phac-aspc.gc.ca/ sars-sras/pdf/phmanagementofcases12-17_e.pdf> (Version current at November 12, 2006).
- 19.Fisher DA, Chew MH, Lim YT, Tambyah PA. Preventing local transmission of SARS: Lessons from Singapore. Med J Aust 2003;178:555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riley S, Fraser C, Donnelly CA, et al. Transmission dynamics of the etiological agent of SARS in Hong Kong: Impact of public health interventions. Science 2003;300:1961-6. [DOI] [PubMed] [Google Scholar]
- 21.Birch CJ, Clothier HJ, Seccull A, et al. Human coronavirus OC43 causes influenza-like illness in residents and staff of aged-care facilities in Melbourne, Australia. Epidemiol Infect 2005;133:273-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Sahly HM, Atmar RL, Glezen WP, Greenberg SB. Spectrum of clinical illness in hospitalized patients with “common cold” virus infections. Clin Infect Dis 2000;31:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vabret A, Mourez T, Gouarin S, Petitjean J, Freymuth F. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin Infect Dis 2003;36:985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skowronski DM, Petric M, Daly P, et al. Coordinated response to SARS, Vancouver, Canada. Emerg Infect Dis 2006;12:155-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.World Health Organization. Summary of the discussion and recommendations of the SARS Laboratory Workshop, 22 October 2003. <www.who.int/csr/sars/guidelines/en/SARSLabmeeting.pdf> (Version current at November 6, 2006).
- 26.Peiris JS, Yuen KY, Osterhaus AD, Stohr K. The severe acute respiratory syndrome. N Engl J Med 2003;349:2431-41. [DOI] [PubMed] [Google Scholar]
- 27.Chan KH, Cheng VC, Woo PC, et al. Serological responses in patients with severe acute respiratory syndrome coronavirus infection and cross-reactivity with human coronaviruses 229E, OC43, and NL63. Clin Diagn Lab Immunol 2005;12:1317-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Che XY, Qiu LW, Liao ZY, et al. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J Infect Dis 2005;191:2033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng M, Chan CW, Cheung RC, et al. Cross-reactivity of antibody against SARS-coronavirus nucleocapsid protein with IL-11. Biochem Biophys Res Commun 2005;338:1654-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maache M, Komurian-Pradel F, Rajoharison A, et al. False-positive results in a recombinant severe acute respiratory syndrome-associated coronavirus (SARS-CoV) nucleocapsid-based Western blot assay were rectified by the use of two subunits (S1 and S2) of spike for detection of antibody to SARS-CoV. Clin Vaccine Immunol 2006;13:409-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Chang Z, Ouyang J, et al. Profiles of IgG antibodies to nucleocapsid and spike proteins of the SARS-associated coronavirus in SARS patients. DNA Cell Biol 2005;24:521-7. [DOI] [PubMed] [Google Scholar]
- 32.Centers for Disease Control and Prevention (CDC). Prevalence of IgG antibody to SARS-associated coronavirus in animal traders - Guangdong Province, China, 2003. MMWR Morb Mortal Wkly Rep 2003;52:986-7. [PubMed] [Google Scholar]
- 33.Zhong NS, Zheng BJ, Li YM, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 2003;362:1353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Health Organization. New case of laboratory-confirmed SARS in Guangdong, China - update 5. <www.who.int/csr/don/ 2004_01_31/en> (Version current at November 6, 2006).
- 35.Chui L, Drebot M, Andonov A, Petrich A, Glushek M, Mahony J. Comparison of 9 different PCR primers for the rapid detection of severe acute respiratory syndrome coronavirus using 2 RNA extraction methods. Diagn Microbiol Infect Dis 2005;53:47-55. [DOI] [PMC free article] [PubMed] [Google Scholar]