Abstract
We have identified a spontaneous mutation in mice, which we term HD for “helper T cell deficient.” This mouse is distinguished by the virtual absence of peripheral T cells of the CD4+8− major histocompatibility complex (MHC) class II-restricted T helper subset due to a specific block in thymic development. The developmental defect is selective for CD4+8− cells; the maturation of CD4−8+ and γδ T cells is normal. The autosomal recessive mutation underlying the HD phenotype is unrelated to MHC class II, since it segregates independently of the MHC class II locus. Moreover, the HD phenotype is not caused by a defect of the CD4 gene. Bone marrow transfer experiments demonstrate that the defect is intrinsic to cells of the hematopoietic lineage, i.e., most likely to developing thymocytes themselves. The frequency of CD4+8low intermediate cells is markedly increased in HD mice, suggesting that class II-restricted thymocytes are arrested at this stage. This is the first genetic defect of its kind to be described in the mouse and may prove highly informative in understanding the molecular pathways underlying lineage commitment.
Developing αβ T cells mature through three major stages characterized by differential expression of the coreceptor molecules CD4 and CD8. Initially, thymocytes express neither coreceptor (CD4−CD8−: double negative or DN), then up-regulate both molecules (CD4+CD8+: double positive or DP), and finally selectively down-modulate CD4 or CD8 (CD4+CD8− or CD4−CD8+: single positive or SP). The vast majority of thymocytes belong to the DP stage. It is at this stage that complete αβ T cell antigen receptor (TCR) is first expressed at the cell surface and thymocytes undergo selection on the basis of their antigen receptor specificity. Thymocytes are selected to undergo alternate developmental fates depending largely on the relative affinities of their clonotypic TCRs for thymic-selecting ligands. Thymocytes interacting too strongly with thymic ligands undergo negative selection, i.e., suicide by apoptosis, whereas those interacting with intermediate affinity undergo positive selection, i.e., maturation to the SP stage.
Thymocytes that undergo positive selection mature into either of two cell types, SP CD4+ helper T cells or SP CD8+ cytotoxic T cells. The decision to enter one or the other developmental pathway is referred to as lineage commitment. Mature cells of the CD4 and CD8 lineages express TCRs restricted to major histocompatibility complex (MHC) class II or class I ligands, respectively. Several alternative models have been proposed to explain how this correlation is achieved: In the instructive model, TCR specificity directs coreceptor down-modulation, i.e., when a TCR recognizes a class I ligand, a specific signal is sent to turn off CD4 expression and vice versa (1). The stochastic model, on the other hand, postulates that positive selection leads to random coreceptor down-modulation; thymocytes that down-modulate the wrong coreceptor are unable to respond to subsequent TCR-mediated stimuli and die (2, 3). In the asymmetric model, only differentiation to the CD8 lineage requires an instructive signal, whereas development to the CD4 lineage occurs by default (4). The signal strength model, finally, proposes that lineage commitment is determined by the overall intensity of signaling through the TCR complex and appropriate coreceptor, such that stronger and weaker signals promote development to the CD4 and CD8 lineages, respectively (5).
Recent studies have shown that progression from the DP to the SP stage is marked by passage through a series of transitional stages characterized by intermediate levels of CD4 and CD8 expression (4, 6–8). An important transitional stage in this progression is the CD4+8low stage, originally identified in both normal and class II-deficient mice (9, 10). Differentiation to the CD4+8low stage indicates that thymocytes have received a primary positive selection signal (4, 6–8). CD4+8low cells in normal mice include progenitors of both the CD4 and CD8 lineages (4, 6–8) and can either give rise directly to CD4+8− thymocytes or, via two further intermediate stages (CD4low8low and CD4low8+), to CD4−8+ thymocytes (4, 8). There is compelling evidence that the DP to SP transition is a multistep process requiring repeated or continual stimuli (11, 12). In particular, it has been proposed that CD4+8low cells must receive a distinct secondary selective signal to undergo final maturation to the SP stage (2).
The differences in signaling processes that mediate alternate maturation to either CD4 or CD8 lineages have yet to be clarified. Although there is abundant evidence that CD4 and CD8 molecules are intimately involved in signal transduction and influence lineage commitment (13–20), they are not essential for this process (21, 22). There is some evidence that additional receptors, specifically homologues of the Drosophila cell fate regulator Notch, can influence lineage choice of developing thymocytes (23). The functionally important outcome of the lineage commitment process is the initiation of distinct patterns of gene expression consistent with the thymocyte’s specificity toward class I or II MHC. The silencing of one or the other coreceptor molecule represents the most obvious example of this divergence in gene expression. In the case of the CD4 gene, a cis-acting negative regulatory element has been defined that is required for silencing of CD4 in cells committed to the CD8 lineage (24). In the case of the CD8α and β genes, an enhancer has recently been defined that directs expression of CD8 only in mature SP CD8+ not DP cells (25, 26). The transcription factors that bind to these regulatory elements remain unknown.
The mutant helper T cell-deficient (HD) mouse that we describe here is characterized by a striking deficiency in the generation of peripheral CD4+ T cells. No mouse model with a similar phenotype has been described previously, with the exception of induced mutants lacking expression of CD4 or MHC class II products (10, 27, 28). We provide evidence that the HD phenotype is not caused by mutations of the CD4 or class II loci. Instead, we speculate that HD mice may bear a defect in a critical signaling pathway initiated either by the TCR/coreceptor complex or some other receptor expressed on thymocytes, e.g., a Notch family member or a cytokine receptor. The eventual identification of the specific gene defect in HD mice should provide significant insights into the mechanisms underlying lineage commitment.
MATERIALS AND METHODS
Animals.
The HD mouse arose as a spontaneous mutant in our animal colony at the Fox Chase Cancer Center. HD mice are derived from an intercross between CD3δ-deficient and human CD3δ transgenic mice which have been described previously (29). RAG2-deficient mice were provided by F. Alt (Howard Hughes Medical Institute, Children’s Hospital, Boston, MA). I-Aβ−/− (MHC class II-deficient) and CD4−/− mice were obtained, respectively, from Taconic Farms and The Jackson Laboratory. C57BL/6 and BALB/c mice were obtained from the Fox Chase Cancer Center Laboratory Animal Facility. Progeny of (HD × BALB/c) intercrosses were H-2 typed by staining of peripheral blood lymphocytes (PBLs) with specific antibodies against MHC class I products Kb and Kd.
Cell Preparation and Flow Cytometry.
PBLs were obtained by retro-orbital bleeding and purified by density gradient centrifugation over Lympholyte M (Cedarlane, ON). PBLs (105) or thymocytes were incubated with the relevant combinations of fluorescently labeled antibodies for 15 min at 4°C and analyzed using a Becton Dickinson FACStar Plus or FACScan. Fluorescent-labeled antibodies against CD4, CD8, αβTCR, γδTCR, I-Ab, H-2 Kb, H-2 Kd, CD69, HSA, and Mel14 were obtained from PharMingen.
Radiation Chimeras.
Bone marrow was isolated from the rear leg bones of a HD mouse and depleted of αβTCR+ cells by fluorescence-activated cell sorting. Five to 10 × 105 T cell-depleted bone marrow cells in 0.2 ml of RPMI 1640 were injected i.v. into 10 RAG2-deficient H-2b recipients that had been sublethally irradiated (900 rads) 24 hr previously. Peripheral blood and thymus samples were obtained 5 wk after bone marrow transfer. Seven of 10 animals showed reconstitution of both B and T cell compartments.
RESULTS
HD Mice Lack Peripheral SP CD4+ T Cells.
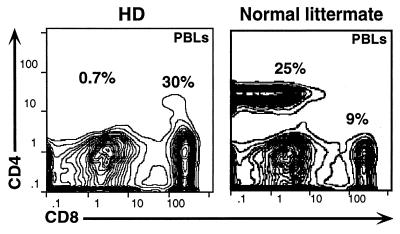
We have identified a cohort of mice in our colony that essentially lack peripheral CD4+ T cells and instead possess elevated numbers of CD8+ αβTCR+ T cells (Fig. 1). These mice are provisionally designated as HD mice. Although SP CD4+ T cells are lacking in HD mice, the total number of peripheral αβTCR+ cells is roughly the same as for wild-type (wt) littermates [i.e., in peripheral blood 24.5 ± 5.4% (n = 6) for HD and 26.7 ± 7.9% (n = 9) for wt littermates]. The HD phenotype is highly consistent (no animals display intermediate reductions of CD4 T cells) and does not undergo significant variation with age up to 8 mo (the oldest animals examined). HD mice are healthy under specific pathogen-free conditions.
Figure 1.
HD mice specifically lack peripheral CD4+ T lymphocytes. PBLs from a HD mouse or a normal littermate were stained with antibodies specific for CD4, CD8, and αβTCR and then analyzed by flow cytometry.
The HD Defect Is Due to an Autosomal Recessive Mutation.
To determine whether the HD phenotype has a genetic basis and, if so, whether it is due to a single dominant or recessive mutation, we have backcrossed HD mice either to other HD mice or to wt C57BL/6, animals. All (HD × HD) progeny bear the HD phenotype, consistent with a heritable defect, whereas all (HD × wt) progeny are normal in phenotype, i.e., possess normal numbers of peripheral CD4+ T cells, indicating that the mutation is not dominant. Finally, among F2s, i.e., progeny of an (HD × wt)F1 intercross, 5 of 24 animals show the HD phenotype (21%), in close agreement with the expected frequency of 6 of 24 (25%) predicted for a single recessive gene (Table 1). Interestingly, (HD × HD) intercrosses consistently produce small litters of two to four pups. This is also true of intercrosses between wt males and HD females, but not HD males and wt females, suggesting that female fertility is impaired in HD mice.
Table 1.
Transmission of HD phenotype
| Cross | No. of progeny | HD phenotype (%) |
|---|---|---|
| (HD × HD)* | 14 | 14 (100%) |
| (HD × C57BL/6)† F1 | 12 | 0 (0%) |
| (F1 × F1)‡ | 24 | 5 (21%) |
A total of 14 progeny were obtained from four separate matings.
All 12 progeny are from a single mating.
A total of 24 progeny were obtained from three separate matings between 6 of the 12 F1 progeny.
Originally, seven HD mice were discovered during routine screening by PBL analysis of 34 mixed F1 and F2 progeny derived from a cross between a knockout line lacking the CD3δ component of the TCR/CD3 complex and a transgenic strain expressing the human CD3δ homologue, neither of which lacks peripheral SP CD4+ T cells (29). No HD mice have arisen from any other crosses between individuals of these two strains, indicating that the HD phenotype is due to a spontaneous mutation in one of the parents in this particular mating. In backcrosses with wt mice, the HD phenotype segregates independently of both knockout and transgenic loci (data not shown).
The HD Phenotype Is Not Due to a Mutation in MHC Class II.
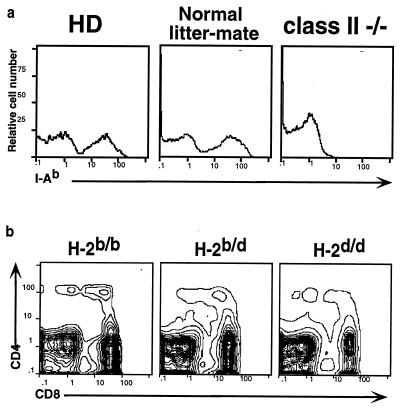
The HD phenotype is superficially similar to that of engineered mutants lacking expression of class II MHC. Hence, it was important to exclude a structural defect in class II, leading to impaired antigen presentation, as the cause of the HD phenotype. All original HD animals are homozygous for the H-2b haplotype and would be expected to express a single class II product, I-Ab. Indeed, flow cytometric analysis of PBLs from HD mice shows normal expression of the I-Ab molecule on peripheral blood B cells and macrophages (Fig. 2a). The possibility of genetic linkage between the HD phenotype and the MHC region was directly tested by backcrossing HD mice to BALB/c mice, which express a different MHC haplotype, H-2d. If the HD phenotype is unlinked to the MHC, it would segregate independently of the H-2b haplotype, i.e., F2 mice would arise bearing the HD phenotype, as defined by the lack of peripheral SP CD4+ T cells, on both heterozygous b/d and homozygous d/d H-2 backgrounds. In fact, several such progeny were identified (Fig. 2b). Thus, we can exclude mutations in the coding regions as well as in cis-acting regulatory elements of class II genes as the cause of the HD phenotype.
Figure 2.
HD phenotype is not caused by the absence of MHC class II products and is genetically unlinked to the class II locus. (a) PBLs from HD mice, normal littermates, and MHC class II-deficient mice were stained with antibody specific for the MHC class II I-Ab product. (b) F2 progeny from the intercross between HD and BALB/c mice were stained with antibodies against CD4 and CD8. Data are shown for one mouse each of the HD phenotype that typed as H-2 b/b, b/d, or d/d.
HD Mice Are Not CD4 Deficient.
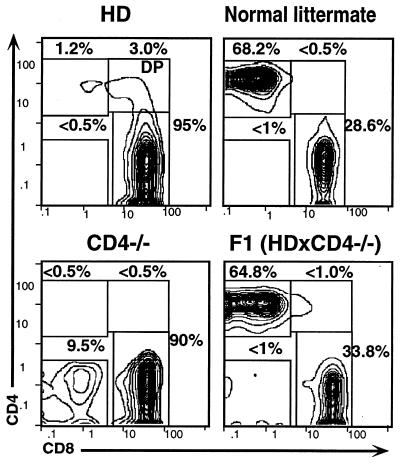
To determine whether the deficit in peripheral CD4+ T cells in HD mice reflects inactivation of the CD4 gene, thymocytes were isolated from 6- to 8-wk-old mice previously identified as HD by PBL analysis and compared with their normal littermates. HD mice displayed normal thymic cellularity (1.5–2 × 108). Most significantly, CD4+CD8+ (DP) thymocytes were present at a normal frequency (80–85%), demonstrating that the HD phenotype is not caused by a null mutation at the CD4 locus (Fig. 3 a and b). TCR and CD4 levels on DP thymocytes have been shown to be regulated by the interaction between CD4 and class II ligands, such that when this interaction is disrupted TCR and CD4 levels increase (30, 31). In HD mice, both CD4 and TCR levels on DP thymocytes are normal, indicating that the ability of CD4 to interact with class II MHC is unimpaired (Fig. 3k: note the difference in TCR levels between HD mice and class II−/− mice, in which the interaction with CD4 is abrogated). In this context, it should be noted that a small but reproducible fraction of peripheral αβTCR+ cells also express cell surface CD4, mostly in combination with CD8 giving rise to a DP phenotype (Fig. 4; note the absence of DP cells in the normal littermate control). In induced mutants lacking expression of CD4, a distinct population of CD4−CD8− (DN) T cells arises in the periphery at relatively high frequency (10–15% of total αβTCR+ T cells) (Fig. 4). These cells are class II-restricted and presumed to belong to the CD4 lineage, although CD4 expression is blocked due to the induced structural mutation (28, 32). The fact that such cells do not arise in HD mice provides further confirmation that the HD phenotype is not due to a defect in CD4. Finally, we have crossed HD and CD4−/− mice to test for genetic linkage. The fact that F1 progeny are phenotypically normal, formally demonstrates that the HD defect cannot lie in the CD4 protein or in cis-acting transcriptional regulatory elements that control its expression (Fig. 4, Right).
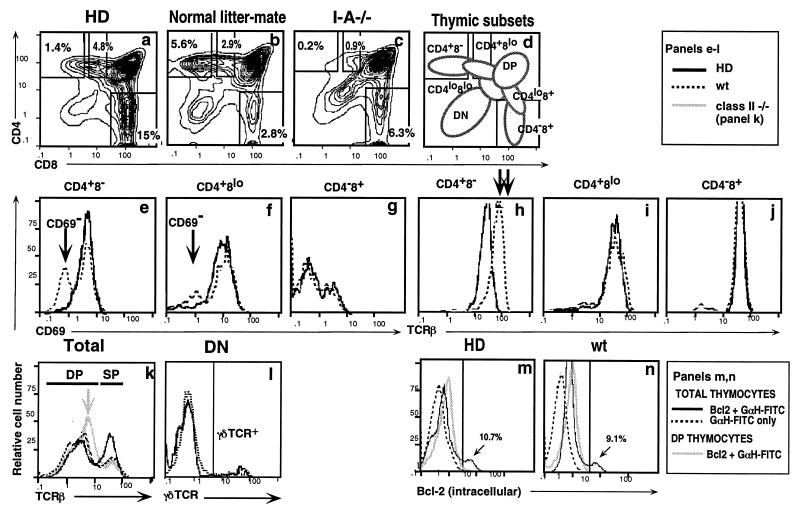
Figure 3.
Thymic development of CD4 lineage cells is abrogated in HD mice. Thymocytes from HD mice, normal littermates, or MHC class II-deficient controls were stained with antibodies specific for CD4, CD8, αβTCR, and CD69 and analyzed by flow cytometry. Histograms represent total thymocytes (a–c and k) or thymic subsets, as specified. In e–l, solid and dashed dark lines correspond to HD and wt mice, respectively, whereas the solid light line in k represents class II−/− mice. In m and n, solid and dashed dark lines correspond to total thymocytes stained either with anti-Bcl-2 and fluoresceinated goat anti-hamster fluorescein isothiocyanate (GαH–FITC) or only GαH–FITC, respectively, whereas the solid light lines represent DP thymocytes stained with anti-Bcl-2 and GαH-FITC. Bcl-2high populations correspond to SP thymocytes. Thymic subset designations are defined in d. Gates used to define CD4+8−, CD4+8low, and CD4−8+ subsets are shown in a–c. Arrows point out key differences between HD mice and wt or class II−/− controls as follows: In e and f, arrows identify the mature CD69− population that is present in wt but absent in HD mice. In h, arrows highlight the 2-fold difference in TCR expression between CD4+8− cells from HD versus wt mice. In k, the arrow highlights the higher TCR expression levels seen on DP thymocytes from class II−/− but not HD or wt mice.
Figure 4.
HD mice lack the αβTCR+ peripheral T cell subset found in CD4−/− mice. PBLs from a HD mouse, a wt littermate, or a CD4-deficient mouse were stained with antibodies against CD4, CD8, and αβTCR and analyzed by flow cytometry. Plots are gated to show CD4:CD8 expression profiles for αβTCR+ cells.
Thymic Development of CD4 Cells Is Specifically Abrogated in HD Mice.
Although generation of DP thymocytes was normal in HD mice, progression to the SP stage was altered in several respects (Fig. 3): (i) The ratio of SP CD8+ to SP CD4+ (CD4+8− + CD4+8low) thymocytes is reversed from 1:3 to 2:1. (ii) The great majority of SP CD4+ thymocytes from HD mice are actually immature CD4+8low rather than CD4+8− cells (Fig. 3 a and b; note that only 1.4% of HD thymocytes fall within the CD4+8− gate). Furthermore, all SP CD4+ thymocytes from HD mice express the lower TCR surface levels that are characteristic of CD4+8low cells (4, 8) (Fig. 3 h and i). (iii) CD4+ SP thymocytes from HD mice are uniformly CD69+, specifically lacking the CD69− subpopulation found in normal mice (Fig. 3 e and f). The absence of CD69− SP CD4+ thymocytes in HD mice is highly significant, because these comprise the most mature thymocytes that are ready to emigrate to the periphery (33). In contrast, CD8+ SP thymocytes from HD mice show the typical biphasic CD69 staining pattern (Fig. 3g), indicative of normal maturation. Development of γδ T lineage cells, which diverge from the αβ lineage at the DN stage, is also unaffected in HD mice, as judged by the normal frequency with which they arise from the DN compartment (Fig. 3l). Finally, since it has been shown that expression of Bcl-2 at the DP stage can cause skewing to the CD8 lineage (34), we investigated whether Bcl-2 is abnormally up-regulated in DP thymocytes from HD mice. Intracellular staining with specific antibody showed that there is no difference in Bcl-2 expression between DP thymocytes of normal and HD mice (Fig. 3 m and n).
The HD Defect Is Intrinsic to the Hematopoietic Lineage.
In principle, the HD defect in CD4 development could reflect a defect in gene expression intrinsic to the developing thymocytes themselves or intrinsic to another thymic cell type that provides an essential developmental signal. Radiation chimeras can be used to dissect the role of radioresistant epithelial cells from that of radiosensitive hematopoietic cells, including thymocytes, macrophages, and dendritic cells. Thus, we isolated bone marrow cells from a HD mouse, depleted mature T cells by fluorescence-activated cell sorting, and transferred them into sublethally irradiated RAG2-deficient hosts. Five weeks later the chimeras were typed for reconstitution of their peripheral immune system. A total of seven chimeras with reconstituted immune systems were identified. All of these bore an abundance of peripheral CD8+ T cells but virtually lacked CD4+ cells, thereby recapitulating the original HD phenotype (Fig. 5e). As would be predicted, analysis of thymocytes from these chimeras confirmed that the specific defect in CD4 development had also been transferred (Fig. 5 f–h). Again we noted that the ratio of CD4:CD8 SP thymocytes was reversed and that SP CD4+ cells were predominantly replaced by CD4+8lo cells (Fig. 5f). A more detailed four-color fluorescence-activated cell sorting analysis of these CD4+8lo cells was carried out using a panel of antibodies specific for the differentiation markers CD69, HSA, and l-selectin. In wt mice fully mature cells that are competent to exit the thymus comprise a distinct and readily detectable subset of SP CD4+ thymocytes that type as HSA− CD69− l-selectin+ (Fig. 5, populations in insets in c and d) (33). In contrast, in HD chimeras these mature cells are essentially lacking (Fig. 5 g and h).
Figure 5.
The HD defect maps to the hematopoietic compartment. Sublethally irradiated RAG2−/− recipients were reconstituted with T cell-depleted bone marrow from wt (a–d) or HD (e–h) mice. PBLs and thymocytes were collected after 4 wk and stained with antibodies against CD4, CD8, HSA, CD69, and l-selectin. Staining profiles are shown for total PBLs (a and e), total thymocytes (b and f), and SP CD4+ thymocytes (c, d, g, and h). Insets in c, d, g, and h indicate the position of the most mature SP CD4+ subpopulation which is present in wt but not in HD mice.
DISCUSSION
We present the initial phenotypic characterization of a novel immunodeficient mouse mutant, the HD mouse, which bears a striking defect in T lymphoid development. We show that these animals are characterized by the inability to support thymic development of mature SP CD4+ thymocytes and a virtual absence of peripheral CD4+ T cells. Two types of mouse models have been described previously with phenotypes superficially similar to that of HD mice, i.e., engineered mutants lacking MHC class II or CD4 (10, 27, 28). We provide compelling evidence that neither of these genes is defective in HD mice.
Involvement of MHC class II is excluded because: (i) the HD phenotype segregates independently of the MHC locus; (ii) peripheral B cells and macrophages express normal surface levels of class II products; and (iii) intrathymic interactions between CD4 and class II appear unperturbed as evidenced by normal TCR surface expression levels on DP thymocytes. A defect in CD4 function or expression is excluded because: (i) CD4 expression is normal on immature DP and intermediate CD4+8low thymocytes; (ii) peripheral T cells committed to the CD4 lineage fail to arise in HD mice, although these cells can develop in induced mutant mice in the complete absence of CD4; and (iii) F1 progeny of an intercross between HD and CD4−/− mice are phenotypically normal. We conclude that the HD mutation affects expression of a novel gene or, at least, a gene whose critical role in lineage commitment is unappreciated.
Using bone marrow transfer experiments, we demonstrate that the HD defect is intrinsic to the hematopoietic lineage rather than the epithelial lineage. Coreconstitution experiments demonstrate that wt thymocytes mature efficiently to the CD4 lineage, whereas HD-derived thymocytes present in the same thymus give rise only to SP CD8 thymocytes (V.P.D. and D.J.K., unpublished work). Hence, the HD defect does not affect thymocytes that do not themselves carry the HD mutation. It has previously been shown that positive selection and lineage commitment of CD4+ cells requires expression of class II, and hence the capacity for antigen presentation, only on cortical epithelial cells and specifically not on cells of hematopoietic origin (35). Thus, our data demonstrate that the HD defect cannot lie in antigen presentation or costimulatory function provided by epithelial cells. The most likely possibility is that the defect is intrinsic to developing thymocytes.
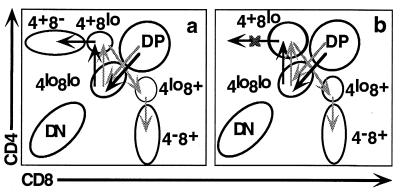
We have shown that the lack of peripheral CD4+ T cells in HD mice reflects a specific block in thymic development of the CD4 lineage. We postulate that the blockade does not affect positive selection of class II-restricted thymocytes but rather a subsequent lineage-commitment step. As mentioned above the transition from the DP to the SP stages is not a simple one-step process, but rather involves progression through a number of intermediate stages defined by different relative levels of surface expression of the CD4 and CD8 molecules (4, 6–8; see Fig. 6a). The CD4+8low population may represent a key stage in this process, since it is the last intermediate common to both the CD4 and CD8 maturation pathways. CD4+8low cells have undergone positive selection as judged by their increased levels of CD69 and TCR surface expression (4, 6–8). It has been proposed that further maturation is not automatic for these cells, but rather requires a specific selective signal (2). HD mice show a striking accumulation of these CD4+8low cells, as compared with wt and particularly class II-deficient mice (Fig. 6b). In class II-deficient mice, positive selection of class II-restricted thymocytes is blocked and CD4+8low cells should consist exclusively of class I-restricted transitional cells on their way to the SP CD8+ compartment. HD mice possess 6-fold more CD4+8low cells than class II-deficient mice. We postulate that the additional cells in HD mice comprise class II-restricted thymocytes that have successfully undergone positive selection but are blocked in a subsequent maturation stage (Fig. 6b). However, since CD4+8low cells can also give rise to SP CD8+ thymocytes, it is alternatively possible that in HD mice these intermediate cells are all CD8 committed and that development of class II-restricted thymocytes is actually arrested earlier, e.g., at the DP stage. The increased number of CD4+8low cells could then reflect enhanced maturation to the CD8 lineage, as also suggested by the increased number of mature SP CD8+ thymocytes. It has been reported that the LEC rat strain also bears an unknown defect in generation of SP CD4+ thymocytes (36). The defect is likely to be different, since LEC rats lack substantial numbers of CD4+8low intermediate thymocytes. Another difference is that LEC rats actually bear substantial numbers of peripheral SP CD4+ T cells, particularly in the lymph nodes where they reach almost normal levels by 12 mo of age (37).
Figure 6.
Absence of mature SP CD4+ thymocytes in HD mice reflects a blockade of development at the CD4+8low stage. (a) Developmental routes postulated to be followed by class I- (light arrows) and class II-restricted (dark arrows) thymocytes in wt mice. (b) Model of HD defect as a blockade in CD4 development at the CD4+8low stage.
The nature of the genetic defect in HD mice remains unclear. However, we think that it is unlikely to affect proximal components of the TCR-mediated signaling pathway, since induced mutants of such components, i.e., CD3ζ, p56lck, and ZAP-70, affect both lineages and, in the case of CD3ζ and p56lck, already block the DN to DP transition (38–42). In addition, we have preliminary data indicating that early events in TCR/coreceptor-mediated signaling, i.e., phosphorylation of CD3ζ and ZAP-70, are normal in HD mice. Rather we suspect that the defect lies in a different process critical for lineage commitment. The impairment in female fertility in HD mice, a feature not usually associated with immunodeficiency, suggests that the effect of the mutation is not lymphoid specific.
In conclusion, we have established that the HD mouse bears a novel mutation of a gene that is specifically required for CD4 lineage commitment. The defect appears to be intrinsic to developing thymocytes themselves. Given that lineage commitment remains very poorly understood at the molecular level, the identification of the genetic defect responsible for the HD phenotype should provide important new insights into our understanding of this phenomenon.
Acknowledgments
We thank D. L. Wiest for critical reading of the manuscript. This work was supported by National Institutes of Health Grants CA74620 and AI34472, Institutional Grant CA06927 from the National Institutes of Health, and also by an appropriation from the Commonwealth of Pennsylvania.
ABBREVIATIONS
- MHC
major histocompatibility complex
- DN
double negative (CD4−8−)
- DP
double positive (CD4+8+)
- SP
single positive (CD4+8− or CD4−8+)
- TCR
T cell antigen receptor
- PBL
peripheral blood lymphocyte
References
- 1.Robey E A, Fowlkes B J, Gordon J W, Kioussis D, von Boehmer H, Ramsdell F, Axel R. Cell. 1991;64:99–107. doi: 10.1016/0092-8674(91)90212-h. [DOI] [PubMed] [Google Scholar]
- 2.Chan S H, Cosgrove D, Waltzinger C, Benoist C, Mathis D. Cell. 1993;73:225–236. doi: 10.1016/0092-8674(93)90225-f. [DOI] [PubMed] [Google Scholar]
- 3.Davis C B, Killeen N, Crooks M E C, Raulet D, Littman D R. Cell. 1993;73:237–247. doi: 10.1016/0092-8674(93)90226-g. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki H, Punt J A, Granger L G, Singer A. Immunity. 1995;2:413–425. doi: 10.1016/1074-7613(95)90149-3. [DOI] [PubMed] [Google Scholar]
- 5.Matechak E O, Killeen N, Hedrick S M, Fowlkes B J. Immunity. 1996;4:337–347. doi: 10.1016/s1074-7613(00)80247-2. [DOI] [PubMed] [Google Scholar]
- 6.Kydd R, Lundberg K, Vremec D, Harris A W, Shortman K. J Immunol. 1995;155:3806–3814. [PubMed] [Google Scholar]
- 7.Lundberg K, Heath W, Köntgen F, Carbone F R, Shortman K. J Exp Med. 1995;181:1643–1651. doi: 10.1084/jem.181.5.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucas B, Germain R N. Immunity. 1996;5:461–477. doi: 10.1016/s1074-7613(00)80502-6. [DOI] [PubMed] [Google Scholar]
- 9.Guidos C J, Danska J S, Fathman C G, Weissman I L. J Exp Med. 1990;172:835–845. doi: 10.1084/jem.172.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosgrove D, Gray D, Dierich A, Kaufman J, Lemeur M, Benoist C, Mathis D. Cell. 1991;66:1051–1066. doi: 10.1016/0092-8674(91)90448-8. [DOI] [PubMed] [Google Scholar]
- 11.Kisielow P, Miazek A. J Exp Med. 1995;181:1975–1984. doi: 10.1084/jem.181.6.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrie H T, Strasser A, Harris A W, Hugo P, Shortman K. J Immunol. 1993;151:1273–1279. [PubMed] [Google Scholar]
- 13.Veillette A, Bookman M A, Horak E M, Bolen J B. Cell. 1988;55:301–308. doi: 10.1016/0092-8674(88)90053-0. [DOI] [PubMed] [Google Scholar]
- 14.Ravichandran K S, Burakoff S J. J Exp Med. 1994;179:727–732. doi: 10.1084/jem.179.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Killeen N, Littman D R. Nature (London) 1993;364:729–732. doi: 10.1038/364729a0. [DOI] [PubMed] [Google Scholar]
- 16.Fung-Leung W P, Louis M C, Limmer A, Ohashi P S, Ngo K, Chen L, Kawai K, Lacy E, Loh D Y, Mak T W. Eur J Immunol. 1993;23:2834–2840. doi: 10.1002/eji.1830231117. [DOI] [PubMed] [Google Scholar]
- 17.Norment A M, Forbush K A, Nguyen N, Malissen M, Perlmutter R M. J Exp Med. 1997;185:121–130. doi: 10.1084/jem.185.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crooks M E C, Littman D R. Immunity. 1994;1:277–285. doi: 10.1016/1074-7613(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 19.Nakayama K, Nakayama K, Negishi I, Kuida K, Louie M C, Kanagawa O, Nakauchi H, Loh D Y. Science. 1994;263:1131–1133. doi: 10.1126/science.8108731. [DOI] [PubMed] [Google Scholar]
- 20.Itano A, Cado D, Chan F K M, Robey E. Immunity. 1994;1:287–290. doi: 10.1016/1074-7613(94)90080-9. [DOI] [PubMed] [Google Scholar]
- 21.Goldrath A W, Hogquist K A, Bevan M J. Immunity. 1997;6:633–642. doi: 10.1016/s1074-7613(00)80351-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sebzda E, Choi M, Fung-Leung W P, Mak T W, Ohashi P S. Immunity. 1997;6:643–653. doi: 10.1016/s1074-7613(00)80352-0. [DOI] [PubMed] [Google Scholar]
- 23.Robey E, Chang D, Itano I, Cado D, Alezander H, Lans D, Weinmaster G, Salmon P. Cell. 1996;87:483–492. doi: 10.1016/s0092-8674(00)81368-9. [DOI] [PubMed] [Google Scholar]
- 24.Sawada S, Scarborough J D, Killeen N, Littman D R. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 25.Ellmeier W, Sunshine M J, Losos K, Hatam F, Littman D R. Immunity. 1997;7:537–547. doi: 10.1016/s1074-7613(00)80375-1. [DOI] [PubMed] [Google Scholar]
- 26.Hostert A, Tolaini M, Roderick K, Harker N, Norton T, Kioussis D. Immunity. 1997;7:525–536. doi: 10.1016/s1074-7613(00)80374-x. [DOI] [PubMed] [Google Scholar]
- 27.Grusby M J, Johnson R S, Papaioannou V E, Glimcher L H. Science. 1991;253:1417–1420. doi: 10.1126/science.1910207. [DOI] [PubMed] [Google Scholar]
- 28.Rahemtulla A, Fung-Leung W P, Schilham M W, Kuendig T M, Samhara S R, Narendran A, Arabian A, Wakeham A, Paige C J, Zinkernagel R M, Miller R G, Mak T W. Nature (London) 1991;353:180–184. doi: 10.1038/353180a0. [DOI] [PubMed] [Google Scholar]
- 29.Dave V P, Cao Z, Browne C, Alarcon B, Fernandez-Miguel G, Lafaille J, de la Hera A, Tonegawa S, Kappes D J. EMBO J. 1997;16:1360–1370. doi: 10.1093/emboj/16.6.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakayama T, June C H, Munitz T I, Sheard M, McCarthy S A, Sharrow S O, Samelson L E, Singer A. Science. 1990;249:1558–1561. doi: 10.1126/science.2120773. [DOI] [PubMed] [Google Scholar]
- 31.McCarthy S A, Kruisbeek A M, Uppenkamp I K, Sharrow S O, Singer A. Nature (London) 1988;336:76–79. doi: 10.1038/336076a0. [DOI] [PubMed] [Google Scholar]
- 32.Killeen N, Sawada S, Littman D R. EMBO J. 1993;12:1547–1553. doi: 10.1002/j.1460-2075.1993.tb05798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabor M J, Godfrey D I, Scollay R. Eur J Immunol. 1997;27:2019–2015. doi: 10.1002/eji.1830271135. [DOI] [PubMed] [Google Scholar]
- 34.Linette G P, Grusby M J, Hedrick S M, Hansen T H, Glimcher L H, Korsmeyer S J. Immunity. 1994;1:197–205. doi: 10.1016/1074-7613(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 35.Laufer T M, DeKoning J, Markowitz J S, Lo D, Glimcher L H. Nature (London) 1996;383:81–85. doi: 10.1038/383081a0. [DOI] [PubMed] [Google Scholar]
- 36.Agui T, Oka M, Yamada T, Sakai T, Izumi K, Ishida Y, Himeno K, Matsumoto K. J Exp Med. 1990;172:1615–1624. doi: 10.1084/jem.172.6.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamada T, Natori T, Izumi K, Sakai T, Agui T, Matsumoto K. Immunogenetics. 1991;33:216–219. doi: 10.1007/BF01719246. [DOI] [PubMed] [Google Scholar]
- 38.Molina T J, Kishihara K, Siderovski D P, van Ewijk W, Narendran A, Timms E, Wakeham A, Paige C J, Hartmann K-U, Veillette A, Davidson D, Mak T W. Nature (London) 1992;357:161–164. doi: 10.1038/357161a0. [DOI] [PubMed] [Google Scholar]
- 39.Ohno H, Aoe T, Taki S, Kitamura D, Ishida Y, Rajewsky K, Saito T. EMBO J. 1993;12:4357–4366. doi: 10.1002/j.1460-2075.1993.tb06120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malissen M, Gillet A, Rocha B, Trucy J, Vivier E, Boyer C, Koentgen F, Brun N, Mazza G, Spanopoulou E, Guy-Grand D, Malissen B. EMBO J. 1993;12:4347–4355. doi: 10.1002/j.1460-2075.1993.tb06119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Negishi I, Motoyama N, Nakayama K-i, Nakayama K, Senju S, Hatakeyama S, Zhang Q, Chan A C, Loh D H. Nature (London) 1995;376:435–438. doi: 10.1038/376435a0. [DOI] [PubMed] [Google Scholar]
- 42.Wiest D L, Ashe J M, Howcraft K, Lee H-M, Kemper D M, Negishi I, Singer A, Abe R. Immunity. 1997;6:663–671. doi: 10.1016/s1074-7613(00)80442-2. [DOI] [PubMed] [Google Scholar]