Abstract
Background and purpose:
Nitric oxide (NO) production through the inducible nitric oxide synthase (iNOS) pathway is increased in response to pro-inflammatory cytokines and bacterial products. In inflammation, NO has pro-inflammatory and regulatory effects. Peroxisome proliferator-activated receptors (PPARs), members of the nuclear steroid receptor superfamily, regulate not only metabolic but also inflammatory processes. The aim of the present study was to investigate the role of PPARα in the regulation of NO production and iNOS expression in activated macrophages.
Experimental approach:
The effects of PPARα agonists were investigated on iNOS mRNA and protein expression, on NO production and on the activation of transcription factors NF-κB and STAT1 in J774 murine macrophages exposed to bacterial lipopolysaccharide (LPS).
Key results:
PPARα agonists GW7647 and WY14643 reduced LPS-induced NO production in a dose-dependent manner as measured by the accumulation of nitrite into the culture medium. However, PPARα agonists did not alter LPS-induced iNOS mRNA expression or activation of NF-κB or STAT1 which are important transcription factors for iNOS. Nevertheless, iNOS protein levels were reduced by PPARα agonists in a time-dependent manner. The reduction was markedly greater after 24 h incubation than after 8 h incubation. Treatment with the proteasome inhibitors, lactacystin or MG132, reversed the decrease in iNOS protein levels caused by PPARα agonists.
Conclusions and implications:
The results suggest that PPARα agonists reduce LPS-induced iNOS expression and NO production in macrophages by enhancing iNOS protein degradation through the proteasome pathway. The results offer an additional mechanism underlying the anti-inflammatory effects of PPARα agonists.
Keywords: iNOS, macrophages, nitric oxide, PPAR, protein degradation, proteasome
Introduction
Nitric oxide (NO) is an important modulator of immune response in human tissues. It has cytotoxic and cytostatic effects, which are beneficial in host defence against pathogenic microbes. In inflammatory diseases, the regulatory, pro-inflammatory and destructive effects of NO modulate the responses also in host tissues (Moilanen et al., 1999; Abramson et al., 2001; Korhonen et al., 2005) and inhibitors of iNOS have been found to be beneficial in various models of inflammatory diseases (Vallance and Leiper, 2002). High amounts of NO are produced through the inducible nitric oxide synthase (iNOS) pathway in response to proinflammatory cytokines and bacterial products. Expression of iNOS has been shown to be regulated both at transcriptional and post-translational levels in activated macrophages, but many of the mechanisms are still unknown (MacMicking et al., 1997; Alderton et al., 2001; Kleinert et al., 2003; Aktan, 2004; Korhonen et al., 2005).
Peroxisome proliferator-activated receptors (PPARs) belong to the nuclear steroid receptor superfamily. Three members of the family have been identified: PPARα, PPARβ/δ and PPARγ. Originally, the receptors were found to be involved in the regulation of the oxidation of fatty acids, but recently other functions of PPARs have been described (Berger and Moller, 2002; Kota et al., 2005). For example, they regulate the transcription of genes that are involved in lipid and glucose metabolism and play a role in adipocyte differentiation and apoptosis (Delerive et al., 2001; Moore et al., 2001a; Kota et al., 2005). Furthermore, recent observations suggest that the PPARs, especially PPARα and PPARγ, are involved in the regulation of the immune and inflammatory responses. Although both anti-inflammatory and pro-inflammatory effects of PPARs have been reported (Delerive et al., 2001; Moore et al., 2001a; Cabrero et al., 2002; Clark, 2002; Zhang and Young, 2002; Genolet et al., 2004) the role of PPARs in inflammation is not clear.
PPARγ agonists have been shown to decrease interferon γ- or lipopolysaccharide (LPS)-induced NO production (Ricote et al., 1998; Alleva et al., 2002; Chen et al., 2003) and iNOS expression (Castrillo et al., 2000; Chen et al., 2003). iNOS expression was shown to be modulated at the transcriptional level. PPARγ agonists were proposed to inhibit the action of inflammatory transcription factors nuclear factor κappa B (NF-κB), activator protein 1 and signal transducer and activator of transcription 1 (STAT1) (Ricote et al., 1998; Chen et al., 2003). The effects of PPARα agonists on NO production and iNOS expression in macrophages have been less studied (Colville-Nash et al., 1998; Cernuda-Morollón et al., 2002). The results of the two studies were contradictory and the mechanisms of action were not investigated in detail.
The aim of the present study was to investigate the effects of PPARα agonists on LPS-induced NO production and iNOS expression in macrophages. The results suggest that PPARα agonists suppress LPS-induced NO production and iNOS expression by enhancing the degradation of iNOS protein through the proteasome pathway.
Methods
Cell culture
J774 macrophages (American Type Culture Collection) were cultured at 37 °C in 5% CO2 atmosphere in Dulbecco's modified Eagle's medium with Ultraglutamine 1 (Cambrex BioScience, Verviers, Belgium), supplemented with 10% heat-inactivated fetal bovine serum (Cambrex BioScience), 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and 250 ng ml−1 amphotericin B (Gibco, Paisley, UK) and harvested with trypsin-EDTA (Gibco). Cells were seeded on 24-well plates (0.2 × 106 cells per well) for nitrite measurements and real-time PCR assays, on six-well plates (0.9 × 106 cells per well) for preparation of cell lysates for iNOS and PPARα Western blot analysis, on 10 cm dishes (4 × 106 cells per dish) for preparation of nuclear extracts and cell lysates for ubiquitin western blotting, and on 96-well plates (4 × 104 cells per well) for cell viability assays. Confluent cells were exposed to fresh culture medium containing the compounds of interest. PPAR agonists were added together with LPS (10 ng ml−1) in all experiments.
Nitrite determination
Measurement of nitrite accumulation into the culture medium was used to determine NO production. The culture medium was collected at indicated time points and nitrite was measured by the Griess reaction (Green et al., 1982). The concentration of nitrite was calculated by using sodium nitrite added to the culture medium (including supplements) as a standard. A selective iNOS inhibitor 1400W was used to differentiate nitrite derived from other biochemical pathways and cellular sources.
Cell viability assays
Cell viability was tested using Cell Proliferation Kit II (Roche Diagnostics, Indianapolis, IN, USA). Cells were incubated with the tested compounds for 20 h before addition of sodium 3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis (4-methoxy-6-nitro) benzene sulphonic acid hydrate (final concentration 0.3 mg ml−1) and N-methyl dibenzopyrazine methyl sulphate (final concentration 1.25 mM). Then the cells were further incubated for 3 h and the amount of formazan accumulated into the growth medium was assessed spectrophotometrically. Triton X-100-treated cells were used as a positive control. A direct cytotoxicity of the tested compounds was evaluated by Trypan blue staining. Triton X-100-treatment was used as a positive control in the cytotoxicity tests.
Preparation of cell lysates for iNOS, PPARα and ubiquitin western blotting
At indicated time points, the cells were rapidly washed with ice-cold phosphate-buffered saline and solubilized in cold lysis buffer containing 10 mM Tris-base, pH 7.4, 5 mM EDTA, 50 mM NaCl, 1% Triton X-100, 0.5 mM phenylmethylsulphonyl fluoride, 1 mM sodium orthovanadate, 20 μg ml−1 leupeptin, 50 μg ml−1 aprotinin, 5 mM NaF, 2 mM sodium pyrophosphate and 10 μM n-octyl-β-D-glucopyranoside. When preparing cell lysates for ubiquitin western blotting, lysis buffer contained also 20 μg ml−1 ubiquitin aldehyde and 25 μM MG132. After incubation on ice for 15 min, lysates were centrifuged (13 400 g, 4 °C, 10 min), supernatants were collected and mixed 3:1 with SDS sample buffer (62.5 mM Tris–HCl, pH 6.8, 10% glycerol, 2% SDS, 0.025% bromophenol blue and 5% β-mercaptoethanol). An aliquot of the supernatant was used to determine protein concentration by the Coomassie blue method (Bradford, 1976).
Preparation of nuclear extracts for STAT1α, NF-κB and PPARγ western blotting
At indicated time points, the cells were rapidly washed with ice-cold phosphate-buffered saline and solubilized in hypotonic buffer A (10 mM HEPES-KOH, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulphonyl fluoride, 1 mM sodium orthovanadate, 10 μg ml−1 leupeptin, 25 μg ml−1 aprotinin, 1 mM NaF and 0.1 mM EGTA). After incubation for 10 min on ice, the cells were vortexed for 30 s and the nuclei were separated by centrifugation at 4 °C, 21 000 g for 10 s. Nuclei were resuspended in buffer C (20 mM HEPES-KOH, pH 7.9, 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulphonyl fluoride, 1 mM sodium orthovanadate, 10 μg ml−1 leupeptin, 25 μg ml−1 aprotinin, 1 mM NaF and 0.1 mM EGTA) and incubated for 20 min on ice. Nuclei were vortexed for 30 s and nuclear extracts were obtained by centrifugation at 4 °C, 21 000 g for 2 min. Supernatants were collected and mixed 3:1 with SDS sample buffer. Coomassie blue was used to measure the protein content of the samples (Bradford, 1976).
Western blotting
Prior to western blotting, samples were boiled for 10 min and 20 μg (240 μg in ubiquitin western blotting) of protein was loaded per lane on 5% (ubiquitin), 8% (iNOS, STAT1α), 10% (PPARα, PPARγ) or 12% (NF-κB p65) SDS-polyacrylamide gel. After electrophoresis, the proteins were transferred to Hybond ECL nitrocellulose membrane (Amersham Biosciences UK Ltd, Little Chalfont, Buckinghamshire, UK). The membrane was blocked in TBS/T (20 mM Tris-base pH 7.6, 150 mM NaCl, 0.1% Tween-20) containing 5% of non-fat dry milk for 1 h at room temperature and incubated with primary antibody in the blocking solution at 4 °C overnight. Thereafter, the membrane was washed with TBS/T, incubated with secondary antibody in the blocking solution for 30 min at room temperature and washed. Bound antibody was detected using SuperSignal West Pico, Dura or Femto chemiluminescent substrate (Pierce, Rockford, IL, USA) and FluorChem 8800 imaging system (Alpha Innotech Corporation, San Leandro, CA, USA). Actin or lamin A was used as a loading control.
RNA extraction and quantitative real-time PCR
Cell homogenization, RNA extraction, reverse transcription of RNA to cDNA and PCR of iNOS were performed as described previously (Lahti et al., 2003). Glyceraldehyde-3-phosphate dehydrogenase was used as a control gene.
Statistics
Results are expressed as mean±s.e.m. When indicated, statistical significance was calculated by analysis of variance followed by Dunnett's multiple comparisons test. Differences were considered significant at P<0.05.
Materials
Reagents were obtained as follows: GW7647 and MG132 from Tocris Cookson Ltd. (Bristol, UK), 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2) from Calbiochem (San Diego, CA, USA), ubiquitin aldehyde from Boston Biochem (Cambridge, MA, USA), LPS (Escherichia coli 0111:B4, product no. L-4391) from Sigma Chemical Co. (St Louis, MO, USA), rabbit polyclonal actin, lamin A/C, iNOS, NF-κB subunit p65, PPARγ and STAT1α p91 antibodies and goat anti-rabbit polyclonal HRP-conjugated antibody from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA), rabbit polyclonal PPARα antibody from Alexis Biochemicals (Lausen, Switzerland), mouse monoclonal ubiquitin antibody from Zymed (San Fransisco, CA, USA) and anti-mouse polyclonal HRP-conjugated antibody from Pierce (Cheshire, UK). 1400W was a kind gift from Dr Richard Knowles (GlaxoSmithKline, Stevenage, UK). All other reagents were from Sigma Chemical Co.
Results
Effects of PPARα agonists on LPS-induced NO production
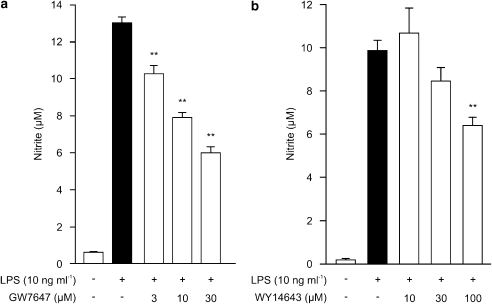
J774 macrophages were found to express PPARα and PPARγ as detected by western blot and LPS treatment for 24 h did not alter their expression levels (data not shown). Resting cells did not produce detectable amounts of NO (measured as nitrite accumulated in the culture medium), but LPS induced NO production and iNOS expression in J774 macrophages. To test the effect of PPARα activation on LPS-induced NO production, we measured NO production in the presence of a selective PPARα agonist GW7647 or WY14643. GW7647 and WY14643 inhibited LPS-induced NO production in a dose-dependent manner, GW7647 being more potent than WY14643 (Figures 1a and b). GW7647 and WY14643 did not affect cell viability when determined by sodium 3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis (4-methoxy-6-nitro) benzene sulphonic acid hydrate test or Trypan blue staining.
Figure 1.
Effects of PPARα agonists on NO production in J774 macrophages. Cells were stimulated by LPS (10 ng ml−1) and treated with increasing concentrations of GW7647 (a) or WY14643 (b). After 24 h incubation nitrite accumulated in the culture medium was measured by Griess reaction, as a marker of NO production. Results are expressed as mean±s.e.m. (n=6). **P<0.01 as compared to cells treated with LPS alone.
Effects of PPARα agonists on iNOS mRNA levels and activation of transcription factors NF-κB and STAT1
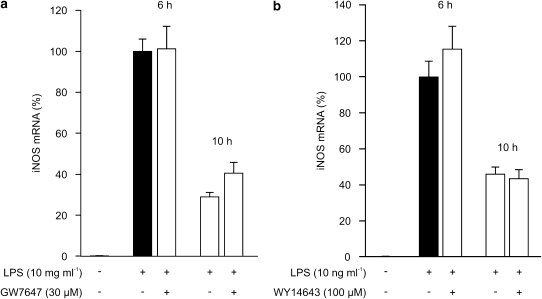
To measure the effects of PPARα agonists on iNOS mRNA expression, the LPS-induced iNOS mRNA levels in the presence and absence of PPARα agonists were determined by quantitative RT-PCR. Neither GW7647 nor WY14643 had any effect on iNOS mRNA expression when measured 6h or 10 h after addition of LPS (Figures 2a and b).
Figure 2.
Effects of PPARα agonists on iNOS mRNA expression in J774 macrophages. Cells were incubated with LPS (10 ng ml−1) and GW7647 (30 μM) (a) or WY14643 (100 μM) (b). Total RNA was extracted at the indicated time points and iNOS mRNA was measured by RT-PCR. The results were normalized against GAPDH mRNA. Levels of iNOS mRNA are expressed relative to that induced by LPS at 6 h (set to 100%). Results are expressed as mean±s.e.m. (n=3).
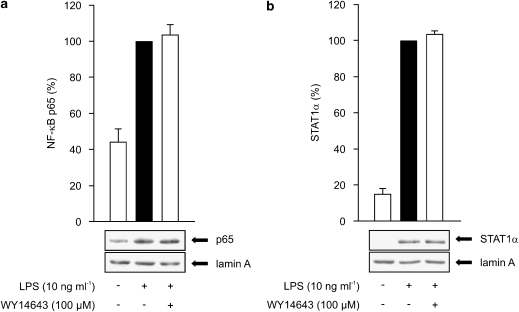
We tested also the effect of WY14643 on the activation of NF-κB and STAT1, which are important transcription factors for iNOS expression. The activation was examined by measuring the translocation of NF-κB (as measured by an antibody against p65 subunit) or STAT1α to the nuclei by western blot. LPS increased the translocation of NF-κB, which peaked at 30 min and decreased thereafter, and that of STAT1, which increased up to 6 h after LPS. WY14643 did not alter LPS-induced NF-κB or STAT1 translocation (Figures 3a and b).
Figure 3.
(a) Effects of PPARα agonists on NF-κB activity in J774 macrophages. Cells were stimulated by LPS (10 ng ml−1) and treated with WY14643 (100 μM) for 30 min. Nuclear extracts were prepared and the p65 subunit of NF-κB was measured by western blot. (b) Effects of PPARα agonists on STAT1α activity in J774 macrophages. Cells were stimulated by LPS (10 ng ml−1) and treated with WY14643 (100 μM) for 6 h. Nuclear extracts were prepared and STAT1α was measured by western blot. Protein levels are expressed relative to that in LPS-treated cells (set to 100%). Lamin A was used as a loading control. Results are expressed as mean±s.e.m. (n=3).
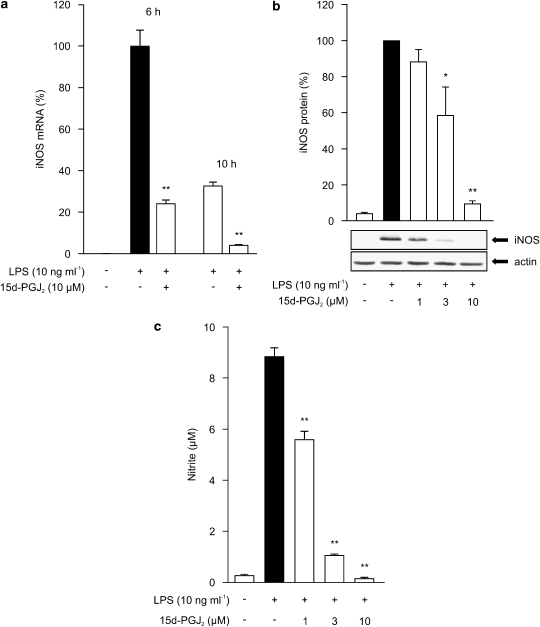
Since PPARγ agonists have been previously reported to inhibit LPS-induced iNOS mRNA expression in macrophages (Ricote et al., 1998; Castrillo et al., 2000; Chen et al., 2003), we wanted to compare the effects of PPARα agonists to those of PPARγ agonists. Although PPARα agonists had no effect on iNOS mRNA expression, we saw a marked reduction in LPS-induced iNOS mRNA levels after treatment with 15d-PGJ2, a natural ligand of PPARγ (Figure 4a). 15d-PGJ2 reduced also LPS-induced iNOS protein expression and NO production (Figures 4b and c) as reported previously (Ricote et al., 1998; Petrova et al., 1999). These results suggest that the mechanism of the inhibitory effect of PPARα agonists on NO production is different from that of PPARγ agonists.
Figure 4.
(a) Effect of PPARγ agonist 15d-PGJ2 on iNOS mRNA expression in J774 macrophages. Cells were incubated with LPS (10 ng ml−1) and 15d-PGJ2 (10 μM). Total RNA was extracted at the indicated time points and iNOS mRNA was measured by real-time PCR. The results were normalized against GAPDH mRNA. Levels of iNOS mRNA are expressed relative to that induced by LPS at 6 h (set to 100%). Results are expressed as mean±s.e.m. (n=3). **P<0.01 as compared to cells treated with LPS alone. (b) Effect of PPARγ agonist 15d-PGJ2 on iNOS protein expression in J774 macrophages. Cells were stimulated by LPS (10 ng ml−1) and treated with increasing concentrations of 15d-PGJ2. After 24 h incubations, proteins were extracted and iNOS protein was measured by western blot. Protein levels are expressed relative to that in LPS-treated cells (set to 100%). Actin was used as a loading control. Results are expressed as mean±s.e.m. (n=3). *P<0.05 and **P<0.01 as compared to cells treated with LPS alone. (c) Effect of PPARγ agonist 15d-PGJ2 on NO production in J774 macrophages. Cells were stimulated by LPS (10 ng ml−1) and treated with increasing concentrations of 15d-PGJ2. After 24 h incubation, nitrite accumulated into the culture medium was measured by Griess reaction as a marker of NO production. Results are expressed as mean±s.e.m. (n=6). **P<0.01 as compared to cells treated with LPS alone.
Effects of PPARα agonists on iNOS protein levels
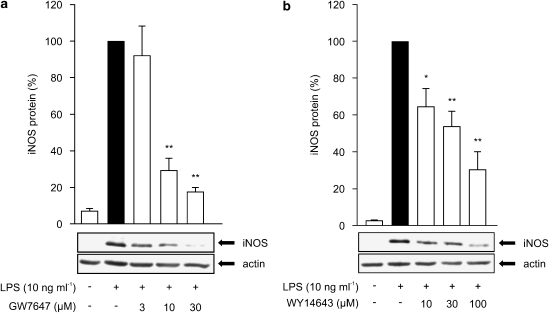
In further studies, we determined the effects of PPARα agonists on iNOS protein expression by western blot analysis. LPS-induced iNOS expression was reduced by PPARα agonists in a dose-dependent manner (Figures 5a and b). After 24 h incubation, the reduction of iNOS expression was about 70% (WY14643) and 80% (GW7647) at the highest agonist concentrations used, thus showing a greater reduction on iNOS protein levels than on NO production (Figure 1).
Figure 5.
Dose-dependent effects of PPARα agonists on iNOS protein expression in J774 macrophages. Cells were stimulated by LPS (10 ng ml−1) and treated with increasing concentrations of GW7647 (a) or WY14643 (b). After 24 h incubations, proteins were extracted and iNOS protein was measured by Western blot. iNOS protein levels are expressed relative to that in LPS-treated cells (set to 100%). Actin was used as a loading control. Results are expressed as mean±s.e.m. (n=3). *P<0.05 and **P<0.01 as compared to cells treated with LPS alone.
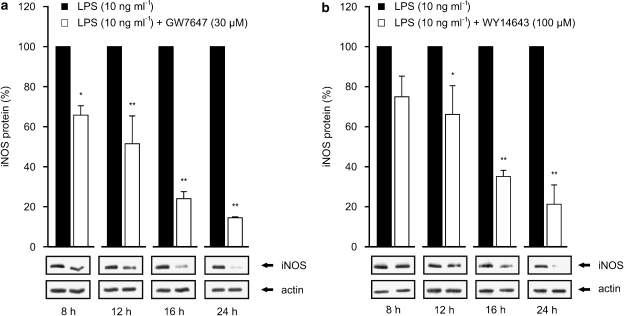
Since PPARα agonists reduced the expression of iNOS protein, but had no effect on iNOS mRNA levels, we hypothesized that PPARα agonists could enhance iNOS degradation. Therefore, we measured the effects of PPARα agonists on LPS-induced iNOS protein levels by western blot after different incubation times (Figures 6a and b). After 8 h incubation, the level of iNOS protein expression was 20–30% lower in cells treated with combinations of LPS and GW7647 or LPS and WY14643 than in cells treated with LPS alone. In contrast, when measured after 12, 16 and 24 h incubations, iNOS protein levels were 50, 75 and 85% lower, respectively in (LPS + GW7647)-treated cells than in cells treated with LPS alone (Figure 6a). A similar pattern of reduction was seen in cells treated with LPS+WY14643 as compared to cells treated with LPS only (Figure 6b).
Figure 6.
Time-dependent effects of PPARα agonists on iNOS protein expression in J774 macrophages. Cells were stimulated by LPS (10 ng ml−1) and treated with GW7647 (30 μM) (a) or WY14643 (100 μM) (b). Proteins were extracted at indicated time points and iNOS protein was measured by western blot. At each time point, iNOS protein levels are expressed relative to that in LPS-treated cells (set to 100%). Actin was used as a loading control. Results are expressed as mean±s.e.m. (n=3). *P<0.05 and **P<0.01 as compared to cells treated with LPS alone.
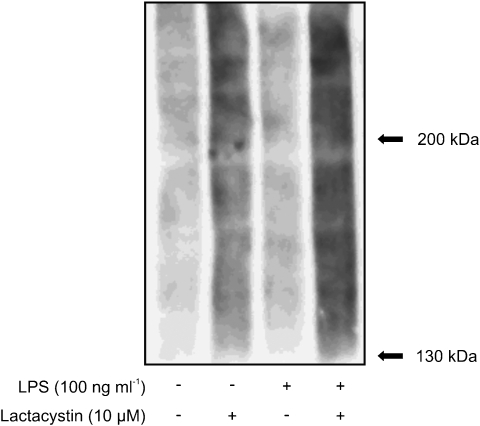
iNOS protein has been reported to be degraded through the proteasome pathway (Felley-Bosco et al., 2000; Musial and Eissa, 2001). Therefore we investigated the role of proteasomes in the suppressive effect of GW7647 and WY14643 on iNOS protein levels. For this purpose, we used two proteasome inhibitors, lactacystin and MG132. To ensure that the proteasome pathway was blocked by these proteasome inhibitors, we first assessed the effect of lactacystin on ubiquitinated protein levels. As detected by western blot, lactacystin increased the ubiquitinated protein levels both in cells incubated with and without LPS (Figure 7).
Figure 7.
Effect of lactacystin on ubiquitinated protein levels in J774 macrophages. When indicated, LPS 10 ng ml−1 was added 8 h prior to lactacystin (10 μM). Proteins were extracted 16 h after the addition of lactacystin and ubiquitinated protein levels were analysed by Western blot. A representative gel is shown, from three experiments with similar results.
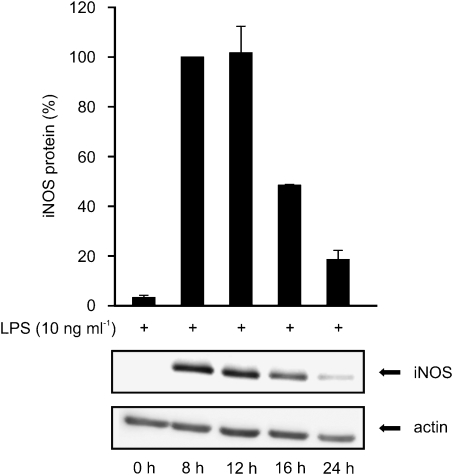
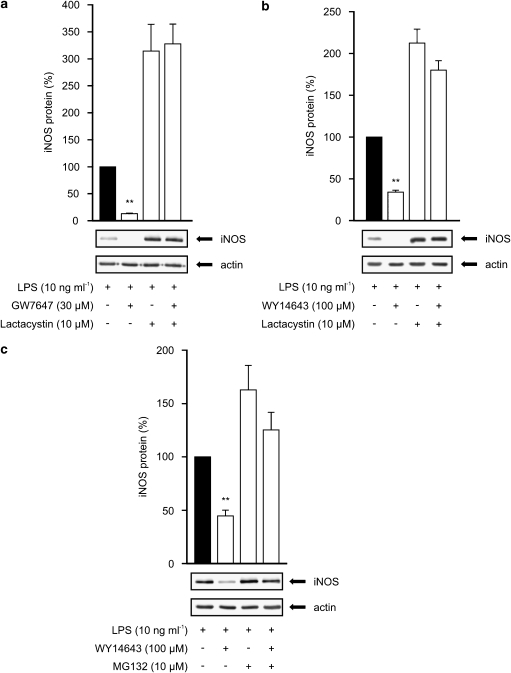
In subsequent studies, lactacystin (10 μM) or MG132 (10 μM) was added to the cells 8 hours after the commencement of the incubation with LPS or LPS and a PPARα agonist, and the cells were harvested after 24 h incubation. As a response to LPS, J774 macrophages expressed iNOS protein reaching maximum between 8 and 12 h after stimulation and decreasing thereafter (Figure 8). In the (LPS+lactacystin)-treated cells, iNOS protein levels were higher after 24 h incubation than in LPS-treated cells (Figures 9a and b) supporting the idea that lactacystin inhibits iNOS degradation. In addition, GW7647 and WY14643 had practically no effect on iNOS levels in the presence of lactacystin while they reduced iNOS protein levels by more than 65% in the absence of lactacystin (Figures 9a and b). Another proteasome inhibitor, MG132, also reduced the inhibitory effect of WY14643 on LPS-induced iNOS protein expression (Figure 9c). Similarly, proteasome inhibitors reversed the inhibitory effects of WY14643 on NO production as measured by nitrite accumulation in the culture medium (data not shown). These results suggest that treatment with proteasome inhibitors reversed the degradation of iNOS protein induced by PPARα agonists GW7647 and WY14643.
Figure 8.
The effect of LPS on iNOS protein expression in J774 macrophages. J774 macrophages were stimulated by LPS (10 ng ml−1). Proteins were extracted at indicated time points and iNOS protein was measured by western blot. iNOS protein levels are expressed relative to that in LPS-treated cells at 8 h (set to 100%). Actin was used as a loading control. Values expressed are mean±s.e.m. (n=3).
Figure 9.
Effects of proteasome inhibitors on iNOS protein expression in J774 macrophages. Cells were stimulated by LPS (10 ng ml−1) with or without GW7647 (a) or WY14643 (b, c). After 8 h incubation a proteasome inhibitor lactacystin (a, b) or MG132 (c) was added into the culture medium. Proteins were extracted after 24 h incubation and iNOS protein was analysed by western blot. iNOS protein levels are expressed relative to that in LPS-treated cells (set to 100%). Actin was used as a loading control. Results are expressed as mean±s.e.m. (n=3). **P<0.01 as compared to cells treated with LPS alone.
Discussion
In the present study, we have shown that PPARα agonists GW7647 and WY14643 reduce LPS-induced iNOS expression and NO production in macrophages. Our results suggest that this effect is mediated through enhanced degradation of iNOS protein via the proteasome pathway. Because the proteasome pathway is involved in the degradation of several inflammatory factors, the present findings may well provide an explanation for the anti-inflammatory effects of PPARα agonists.
In several studies, activation of PPARα has been reported to have anti-inflammatory effects in vivo. A clear evidence of the immunomodulating effects of PPARα was unveiled in 1996, when PPARα-null mice were shown to present a prolonged inflammatory reaction in response to leukotriene B4 as compared to wild-type animals (Devchand et al., 1996). Later, fibrates, which act as PPARα ligands, have been shown to decrease plasma levels of interleukin-6 (IL-6), interferon-γ, tumour necrosis factor-α, fibrinogen and C-reactive protein in hyperlipidemic patients (Madej et al., 1998; Staels et al., 1998). In addition, numerous studies have clarified the role of PPARγ agonists on inflammatory responses. For example, members of antidiabetic thiazolidinediones, which are synthetic PPARγ ligands, have been shown to reduce inflammation in a mouse model of inflammatory bowel disease (Su et al., 1999) and in adjuvant-induced arthritis in rats (Kawahito et al., 2000). Rosiglitazone has also been reported to decrease plasma concentrations of C-reactive protein and matrix metalloproteinase-9 (Haffner et al., 2002), and inhibit the development of atherosclerosis in low-density lipoprotein receptor-deficient mice (Li et al., 2000). However, although there are a large number of studies reporting anti-inflammatory actions of PPAR ligands, some observations suggest that PPAR agonists may also have pro-inflammatory effects (Thieringer et al., 2000; Guyton et al., 2001; Moore et al., 2001b; Fu et al., 2002). Thus, PPARs have been shown to have in vivo relevance with inflammatory processes, and that is the reason why the mechanisms underlying these effects are highly interesting to clarify.
In the present study, PPARα agonists suppressed LPS-induced NO production and iNOS protein expression in a dose-dependent manner, but they had no effect on iNOS mRNA levels or on activation of NF-κB or STAT1, which are important transcription factors for iNOS. These results together suggest that the suppressive effects of PPARα agonists on iNOS expression and NO production are mediated through post-transcriptional mechanisms.
When we investigated the effects of PPARα agonists on iNOS protein expression at different time points, we found that PPARα agonists reduced LPS-induced iNOS protein expression significantly more when measured 24 h after addition of LPS than at the 8 h time point. These findings suggest that PPARα agonists GW7647 and WY14643 enhance the degradation of iNOS protein in macrophages and this is the mechanism for the inhibition of NO production by PPARα agonists. This idea is also supported by the fact that the suppressing effect of PPARα agonists was greater on iNOS protein than on NO levels at the equal time point. There is evidence showing that iNOS protein is degraded by the proteasome pathway (Felley-Bosco et al., 2000; Musial and Eissa, 2001). In the present study, we found that two proteasome inhibitors lactacystin and MG132 reversed the effects of PPARα agonists on iNOS protein expression. Therefore, we proposed that PPARα agonists reduced NO production through iNOS pathway by enhancing the degradation of iNOS protein by proteasomal enzymes. This assumption is supported by the recent data from mRNA microarrays showing that PPARα agonists enhance expression of proteasomal genes in cynomolgus monkey liver (Cariello et al., 2005) and in murine hepatocytes (Anderson et al., 2004).
The regulation of iNOS protein degradation is poorly known. However, there are data that support the importance of the proteasome pathway in this degradation process (Felley-Bosco et al., 2000; Musial and Eissa, 2001). In those reports, the proteasome inhibitor lactacystin was shown to enhance iNOS protein levels in murine RAW 264.7 macrophages and in human cell lines (Felley-Bosco et al., 2000; Musial and Eissa, 2001). The present findings support the earlier data by showing that two proteasome inhibitors, lactacystin and MG132, inhibited iNOS protein degradation in LPS-treated J774 macrophages supporting the significant role of proteasomes in the degradation of iNOS protein.
In the literature, only a few factors have been described to regulate iNOS protein stability. TGF-β, in addition to its effects on iNOS mRNA stability and translation, has been found to increase degradation of iNOS protein in macrophages (Vodovotz et al., 1993; Mitani et al., 2005) and in chondrocytes (Vuolteenaho et al., 2005). In addition, dexamethasone has been reported to decrease iNOS protein stability in IL-1-stimulated mesangial cells (Kunz et al., 1996).
There are some data on the role of PPARs in the regulation of iNOS expression and NO production, but most of the interest has been focused on PPARγ. PPARγ agonists have been shown to decrease NO production and iNOS expression in macrophages (Ricote et al., 1998; Castrillo et al., 2000; Alleva et al., 2002; Chen et al., 2003), and this inhibitory effect seems to take place at a transcriptional level by inhibiting the action of transcription factors NF-κB and STAT1 (Ricote et al., 1998; Chen et al., 2003). There are, however, only two previous reports on the effects of PPARα agonists on iNOS expression and NO production in macrophages showing contradictory results (Colville-Nash et al., 1998; Cernuda-Morollón et al., 2002). Colville-Nash et al. (1998) found that a selective PPARα ligand WY14643 reduced interferon-γ and LPS-induced NO production in RAW 264.7 macrophages. Cernuda-Morollón et al. (2002) reported that WY14643 amplified LPS- or LPS and interferon-γ-stimulated iNOS protein expression in RAW 264.7 macrophages. The present results are in line with those reported by Colville-Nash et al. (1998) and they extend the earlier data by showing a cellular mechanism that could, at least in part, explain the inhibitory effect of PPARα agonists on LPS-induced iNOS protein expression and NO production in activated macrophages. As the proteasome pathway is involved in the degradation of several inflammatory proteins (Ben-Neriah, 2002; Colmegna et al., 2005), PPARα agonists may well regulate the levels of an array of inflammatory factors by the same mechanism.
In conclusion, the present data show that PPARα agonists GW7647 and WY14643 suppress LPS-induced iNOS protein expression and NO production in macrophages, and this effect is likely to be mediated by enhanced iNOS protein degradation through the proteasome pathway. These results offer an additional mechanism for the anti-inflammatory effects of PPARα agonists and point to the significance of proteasomes in the degradation of iNOS protein and as a target of anti-inflammatory drugs.
Acknowledgments
We thank Dr Daniela Ungureanu for her expert methodological advice, Mrs Niina Ikonen and Mrs Salla Hietakangas for their excellent technical assistance and Mrs Heli Määttä for her skilful secretarial help. This study was supported by grants from the Academy of Finland, from the National Technology Agency of Finland and from the Medical Research Fund of Tampere University Hospital.
Abbreviations
- 15d-PGJ2
15-deoxy-Δ12,14-prostaglandin J2
- CRP
C-reactive protein
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IFN-γ
interferon-γ
- IL-6
interleukin-6
- iNOS
inducible nitric oxide synthase
- LPS
lipopolysaccharide
- MMP-9
matrix metalloproteinase-9
- NF-κB
nuclear factor κB
- NO
nitric oxide
- PPAR
peroxisome proliferator-activated receptor
- STAT1
signal transducer and activator of transcription 1
- TNF-α
tumour necrosis factor-α
Conflict of interest
The authors state no conflict of interest.
References
- Abramson SB, Amin AR, Clancy RM, Attur M. The role of nitric oxide in tissue destruction. Best Pract Res Clin Rheumatol. 2001;15:831–845. doi: 10.1053/berh.2001.0196. [DOI] [PubMed] [Google Scholar]
- Aktan F. iNOS-mediated nitric oxide production and its regulation. Life Sci. 2004;75:639–653. doi: 10.1016/j.lfs.2003.10.042. [DOI] [PubMed] [Google Scholar]
- Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: structure, function and inhibition. Biochem J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alleva DG, Johnson EB, Lio FM, Boehme SA, Conlon PJ, Crowe PD. Regulation of murine macrophage proinflammatory and anti-inflammatory cytokines by ligands for peroxisome proliferator-activated receptor-γ: counter-regulatory activity by IFN-γ. J Leukoc Biol. 2002;71:677–685. [PubMed] [Google Scholar]
- Anderson SP, Howroyd P, Liu J, Qian X, Bahnemann R, Swanson C, et al. The transcriptional response to a peroxisome proliferator-activated receptor α agonist includes increased expression of proteome maintenance genes. J Biol Chem. 2004;279:52390–52398. doi: 10.1074/jbc.M409347200. [DOI] [PubMed] [Google Scholar]
- Ben-Neriah Y. Regulatory functions of ubiquitination in the immune system. Nat Immunol. 2002;3:20–26. doi: 10.1038/ni0102-20. [DOI] [PubMed] [Google Scholar]
- Berger J, Moller DE. The mechanisms of action of PPARs. Annu Rev Med. 2002;53:409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cabrero A, Laguna JC, Vázquez M. Peroxisome proliferator-activated receptors and the control of inflammation. Curr Drug Targets Inflamm Allergy. 2002;1:243–248. doi: 10.2174/1568010023344616. [DOI] [PubMed] [Google Scholar]
- Cariello NF, Romach EH, Colton HM, Ni H, Yoon L, Falls JG, et al. Gene expression profiling of the PPAR-alpha agonist ciprofibrate in the cynomolgus monkey liver. Toxicol Sci. 2005;88:250–264. doi: 10.1093/toxsci/kfi273. [DOI] [PubMed] [Google Scholar]
- Castrillo A, Díaz-Guerra MJ, Hortelano S, Martín-Sanz P, Boscá L. Inhibition of IκB kinase and IκB phosphorylation by 15-deoxy-Δ12,14-prostaglandin J2 in activated murine macrophages. Mol Cell Biol. 2000;20:1692–1698. doi: 10.1128/mcb.20.5.1692-1698.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernuda-Morollón E, Rodríguez-Pascual F, Klatt P, Lamas S, Pérez-Sala D. PPAR agonists amplify iNOS expression while inhibiting NF-κB: implications for mesangial cell activation by cytokines. J Am Soc Nephrol. 2002;13:2223–2231. doi: 10.1097/01.asn.0000025786.87646.b1. [DOI] [PubMed] [Google Scholar]
- Chen CW, Chang YH, Tsi CJ, Lin WW. Inhibition of IFN-γ-mediated inducible nitric oxide synthase induction by the peroxisome proliferator-activated receptor γ agonist, 15-deoxy-Δ12,14-prostaglandin J2, involves inhibition of the upstream janus kinase/STAT1 signaling pathway. J Immunol. 2003;171:979–988. doi: 10.4049/jimmunol.171.2.979. [DOI] [PubMed] [Google Scholar]
- Clark RB. The role of PPARs in inflammation and immunity. J Leukoc Biol. 2002;71:388–400. [PubMed] [Google Scholar]
- Colmegna I, Sainz B, Jr, Garry RF, Espinoza LR. The proteasome and its implications in rheumatology. J Rheumatol. 2005;32:1192–1198. [PubMed] [Google Scholar]
- Colville-Nash PR, Qureshi SS, Willis D, Willoughby DA. Inhibition of inducible nitric oxide synthase by peroxisome proliferator-activated receptor agonists: correlation with induction of heme oxygenase 1. J Immunol. 1998;161:978–984. [PubMed] [Google Scholar]
- Delerive P, Fruchart JC, Staels B. Peroxisome proliferator-activated receptors in inflammation control. J Endocrinol. 2001;169:453–459. doi: 10.1677/joe.0.1690453. [DOI] [PubMed] [Google Scholar]
- Devchand PR, Keller H, Peters JM, Vazquez M, Gonzalez FJ, Wahli W. The PPARα-leukotriene B4 pathway to inflammation control. Nature. 1996;384:39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- Felley-Bosco E, Bender FC, Courjault-Gautier F, Bron C, Quest AF. Caveolin-1 down-regulates inducible nitric oxide synthase via the proteasome pathway in human colon carcinoma cells. Proc Natl Acad Sci USA. 2000;97:14334–14339. doi: 10.1073/pnas.250406797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Luo N, Lopes-Virella MF. Upregulation of interleukin-8 expression by prostaglandin D2 metabolite 15-deoxy-delta 12, 14 prostaglandin J2 (15d-PGJ2) in human THP-1 macrophages. Atherosclerosis. 2002;160:11–20. doi: 10.1016/s0021-9150(01)00541-x. [DOI] [PubMed] [Google Scholar]
- Genolet R, Wahli W, Michalik L. PPARs as drug targets to modulate inflammatory responses? Curr Drug Targets Inflamm Allergy. 2004;3:361–375. doi: 10.2174/1568010042634578. [DOI] [PubMed] [Google Scholar]
- Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, Tannenbaum SR. Analysis of nitrate, nitrite, and [15n]nitrate in biological fluids. Anal Biochem. 1982;126:131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- Guyton K, Bond R, Reilly C, Gilkeson G, Halushka P, Cook J. Differential effects of 15-deoxy-Δ12,14-prostaglandin J2 and a peroxisome proliferator-activated receptor γ agonist on macrophage activation. J Leukoc Biol. 2001;69:631–638. [PubMed] [Google Scholar]
- Haffner SM, Greenberg AS, Weston WM, Chen H, Williams K, Freed MI. Effect of rosiglitazone treatment on nontraditional markers of cardiovascular disease in patients with type 2 diabetes mellitus. Circulation. 2002;106:679–684. doi: 10.1161/01.cir.0000025403.20953.23. [DOI] [PubMed] [Google Scholar]
- Kawahito Y, Kondo M, Tsubouchi Y, Hashiramoto A, Bishop-Bailey D, Inoue K, et al. 15-deoxy-Δ12,14-PGJ2 induces synoviocyte apoptosis and suppresses adjuvant-induced arthritis in rats. J Clin Invest. 2000;106:189–197. doi: 10.1172/JCI9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinert H, Schwarz PM, Förstermann U. Regulation of the expression of inducible nitric oxide synthase. Biol Chem. 2003;384:1343–1364. doi: 10.1515/BC.2003.152. [DOI] [PubMed] [Google Scholar]
- Korhonen R, Lahti A, Kankaanranta H, Moilanen E. Nitric oxide production and signalling in inflammation. Curr Drug Targets Inflamm Allergy. 2005;4:471–479. doi: 10.2174/1568010054526359. [DOI] [PubMed] [Google Scholar]
- Kota BP, Huang TH, Roufogalis BD. An overview on biological mechanisms of PPARs. Pharmacol Res. 2005;51:85–94. doi: 10.1016/j.phrs.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Kunz D, Walker G, Eberhardt W, Pfeilschifter J. Molecular mechanisms of dexamethasone inhibition of nitric oxide synthase expression in interleukin 1β-stimulated mesangial cells: evidence for the involvement of transcriptional and posttranscriptional regulation. Proc Natl Acad Sci USA. 1996;93:255–259. doi: 10.1073/pnas.93.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti A, Jalonen U, Kankaanranta H, Moilanen E. c-Jun NH2-terminal kinase inhibitor anthra(1,9-cd)pyrazol-6(2 H)-one reduces inducible nitric-oxide synthase expression by destabilizing mRNA in activated macrophages. Mol Pharmacol. 2003;64:308–315. doi: 10.1124/mol.64.2.308. [DOI] [PubMed] [Google Scholar]
- Li AC, Brown KK, Silvestre MJ, Willson TM, Palinski W, Glass CK. Peroxisome proliferator-activated receptor γ ligands inhibit development of atherosclerosis in LDL receptor-deficient mice. J Clin Invest. 2000;106:523–531. doi: 10.1172/JCI10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- Madej A, Okopien B, Kowalski J, Zielinski M, Wysocki J, Szygula B, et al. Effects of fenofibrate on plasma cytokine concentrations in patients with atherosclerosis and hyperlipoproteinemia IIb. Int J Clin Pharmacol Ther. 1998;36:345–349. [PubMed] [Google Scholar]
- Mitani T, Terashima M, Yoshimura H, Nariai Y, Tanigawa Y. TGF-β1 enhances degradation of IFN-γ-induced iNOS protein via proteasomes in RAW 264.7 cells. Nitric Oxide. 2005;13:78–87. doi: 10.1016/j.niox.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Moilanen E, Whittle BJR, Moncada S.Nitric oxide as a factor in inflammation Inflammation: Basic principles and Clinical Correlates 1999Lippincott, Williams & Wilkins: Philadelphia; 787–801.In: Gallin JI, Snyderman R (eds). [Google Scholar]
- Moore KJ, Fitzgerald ML, Freeman MW. Peroxisome proliferator-activated receptors in macrophage biology: friend or foe? Curr Opin Lipidol. 2001a;12:519–527. doi: 10.1097/00041433-200110000-00007. [DOI] [PubMed] [Google Scholar]
- Moore KJ, Rosen ED, Fitzgerald ML, Randow F, Andersson LP, Altshuler D, et al. The role of PPAR-γ in macrophage differentiation and cholesterol uptake. Nature Med. 2001b;7:41–47. doi: 10.1038/83328. [DOI] [PubMed] [Google Scholar]
- Musial A, Eissa NT. Inducible nitric-oxide synthase is regulated by the proteasome degradation pathway. J Biol Chem. 2001;276:24268–24273. doi: 10.1074/jbc.M100725200. [DOI] [PubMed] [Google Scholar]
- Petrova TV, Akama KT, Van Eldik LJ. Cyclopentenone prostaglandins suppress activation of microglia: down-regulation of inducible nitric-oxide synthase by 15-deoxy-Δ12,14-prostaglandin J2. Proc Natl Acad Sci USA. 1999;96:4668–4673. doi: 10.1073/pnas.96.8.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature. 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- Staels B, Koenig W, Habib A, Merval R, Lebret M, Torra IP, et al. Activation of human aortic smooth-muscle cells is inhibited by PPARα but not by PPARγ activators. Nature. 1998;393:790–793. doi: 10.1038/31701. [DOI] [PubMed] [Google Scholar]
- Su CG, Wen X, Bailey ST, Jiang W, Rangwala SM, Keilbaugh SA, et al. A novel therapy for colitis utilizing PPAR-γ ligands to inhibit the epithelial inflammatory response. J Clin Invest. 1999;104:383–389. doi: 10.1172/JCI7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieringer R, Fenyk-Melody JE, Le Grand CB, Shelton BA, Detmers PA, Somers EP, et al. Activation of peroxisome proliferator-activated receptor γ does not inhibit IL-6 or TNF-α responses of macrophages to lipopolysaccharide in vitro or in vivo. J Immunol. 2000;164:1046–1054. doi: 10.4049/jimmunol.164.2.1046. [DOI] [PubMed] [Google Scholar]
- Vallance P, Leiper J. Blocking NO synthesis: how, where and why? Nat Rev Drug Discov. 2002;1:939–950. doi: 10.1038/nrd960. [DOI] [PubMed] [Google Scholar]
- Vodovotz BY, Bogdan C, Paik J, Xie QW, Nathan C. Mechanisms of suppression of macrophage nitric oxide release by transforming growth factor β. J Exp Med. 1993;178:605–613. doi: 10.1084/jem.178.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuolteenaho K, Moilanen T, Jalonen U, Lahti A, Nieminen R, van Beuningen HM, et al. TGFβ inhibits IL-1–induced iNOS expression and NO production in immortalized chondrocytes. Inflamm Res. 2005;54:420–427. doi: 10.1007/s00011-005-1373-6. [DOI] [PubMed] [Google Scholar]
- Zhang X, Young HA. PPAR and immune system — what do we know? Int Immunopharmacol. 2002;2:1029–1044. doi: 10.1016/s1567-5769(02)00057-7. [DOI] [PMC free article] [PubMed] [Google Scholar]