Abstract
Introduction: Selective dissemination of information (SDI) services regularly alert users to new information on their chosen topics. This type of service can increase a user's ability to keep current and may have a positive impact on efficiency and productivity. Currently, there are many venues available where users can establish, store, and automatically run MEDLINE searches.
Purpose: To describe, evaluate, and compare SDI services for MEDLINE.
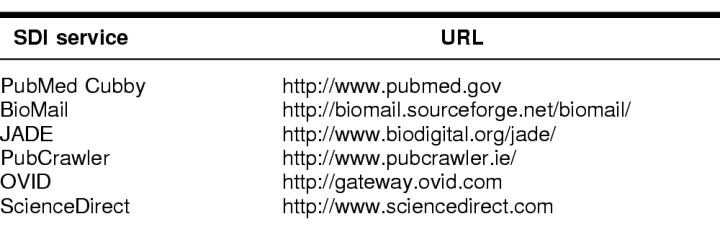
Resources: The following SDI services were selected for this study: PubMed Cubby, BioMail, JADE, PubCrawler, OVID, and ScienceDirect.
Methodology: Identical searches were established in four of the six selected SDI services and were run on a weekly basis over a period of two months. Eight search strategies were used in each system to test performance under various search conditions. The PubMed Cubby system was used as the baseline against which the other systems were compared. Other aspects were evaluated in all six services and include ease of use, frequency of results, ability to use MeSH, ability to access and edit existing search strategies, and ability to download to a bibliographic management program.
Results: Not all MEDLINE SDI services retrieve identical results, even when identical search strategies are used. This study also showed that the services vary in terms of features and functions offered.
INTRODUCTION
Selective dissemination of information (SDI) is defined as “a service provided by a library or other information agency whereby its users are periodically notified of new publications, report literature, or other sources of information in subjects in which they have specified an interest” [1]. SDI services offer an important option to users for keeping current with research. This has always been particularly valued in the health sciences, where new information can affect patient care.
While SDI is often equated with automated searching, it can be provided in a variety of ways, both manually and electronically. Librarians often provide this type of service as part of a daily reference function by pointing out new items (articles, books, news items) to patrons whom they know to be interested in a particular topic. SDI service, while perhaps not using that label, was available long before automation. Anderson even mused “a medieval monk who kept an eye out for special manuscripts for an ecclesiastical superior was practicing SDI” [2]. Ohta and Evans mentioned an SDI type of service that began in the 1940s at the Library of the Columbia University College of Physicians and Surgeons [3].
The literature points to Luhn as the pioneer of the more modern and automated SDI service in the 1950s [4–5]. The library and information science literature shows a steady growth of publications on SDI services, both manual and automated, during the 1960s and 1970s. The literature is replete with surveys, evaluations, and observations that note the benefits of such services to users, information professionals, and libraries.
Brandli reported that the College of Medicine and Dentistry of New Jersey began a current awareness service in 1964, which subsequently began using the State University of New York's automatic MEDLARS services and then moved to the National Library of Medicine's (NLM) SDILINE [6]. This institution's continuing current awareness services were later updated and reported on by Kapadia [7]. NLM began providing SDI services with the initiation of SDILINE in 1972. This fee-based service offered stored search requests that were run against Index Medicus on a monthly basis. The resulting citations were printed and mailed from NLM to the participating libraries.
Some libraries that had been performing manual SDI began incorporating this and other online services as part of a combined manual/automated function. Yunis reported on one such service in 1973, which provided regular manual SDI output from the printed Index Medicus, and its benefits to the user population [8]. Wood's study documents the move from a manual to an automated service and the resulting effects on SDI users, who preferred the automated search to the manual even when results were delayed [9]. While many of the manual services were replaced by their electronic counterparts, Strube reported on the benefits of a manual SDI service as recently as 1988 [10]. Strube also discussed benefits to the library, including the development of a professional rapport with patrons, positive public relations for the library, and the knowledge that the librarian gains by becoming more familiar with the subject areas of the users.
These studies echo an earlier report by Lavendel on the specific benefits of SDI services to the patron, library, and librarian [11]. The patron benefits by receiving updated information and substantial time savings. The librarian benefits by developing professional relationships with patrons and developing increased professional expertise and subject knowledge. The library benefits from an enhanced reputation and increased usage. Lavendel also suggested that the librarian's role as SDI mediator should connect to the acquisitions department in terms of forecasting which subjects and materials users need [12]. Yunis, Brandli, and others all evaluated their SDI services through surveys and found that participants were happy with the services and that it added value to their work [13–18]. Mondschein performed an in-depth study on the effect of SDI services on productivity in a corporate research and development department, which showed a positive relationship between the use of an SDI service and an increase in productivity. Productivity was measured as the number of articles authored during the time the SDI service was offered [19].
By 1993, NLM began offering libraries the option of receiving SDILINE output via ftp (file transfer protocol) rather than waiting for mailed printouts. SDILINE was finally retired in February 2001, and the task was taken on by the new PubMed Cubby service, which allows users to create, store, and run their own SDI searches [20].
More recent literature offers comparisons of various SDI services [21, 22]. Eberle described six SDI services for the MEDLINE/PubMed system and compared their features and functions. Eberle's article was written prior to the introduction of PubMed's Cubby feature [23].
Multiple vendors provide access to the MEDLINE database for searching, for example, MD Consult, PubMed, OVID, ScienceDirect, OCLC FirstSearch. Many of these vendors also provide users with the option of saving searches to be run at a later date or having them automatically rerun on a regular schedule.
Given the known benefits of SDI and current awareness services and the history of SDI use for MEDLINE, this study compares the features and effectiveness of six vendors providing MEDLINE SDI services. The purpose of this study is to provide librarians with background information for selecting a service and training users.
METHODOLOGY
Six MEDLINE SDI services were selected to be evaluated and compared: PubMed, BioMail, JADE, PubCrawler, OVID, and ScienceDirect (Table 1). PubMed was selected because it is a widely used search interface supported by NLM, which produces the MEDLINE database. ScienceDirect and OVID were selected as examples of fee-based, licensed resources and are two highly used systems at the author's institution. BioMail, JADE, and PubCrawler were selected as examples of freely available, non-NLM SDI services to MEDLINE.
Table 1 Selective dissemination of information (SDI) services
Two distinct methods were used to evaluate the SDI services. One method compared the actual search results in four of the selected SDI services. Although searches were established in all six SDI services, only Cubby, PubCrawler, BioMail, and OVID MEDLINE remained in the study for this comparison. As a result of technical difficulties, no results were received from JADE or ScienceDirect during the test period, and, therefore, they were dropped from this part of the study. The second method compared the special functions and features of each of the six services.
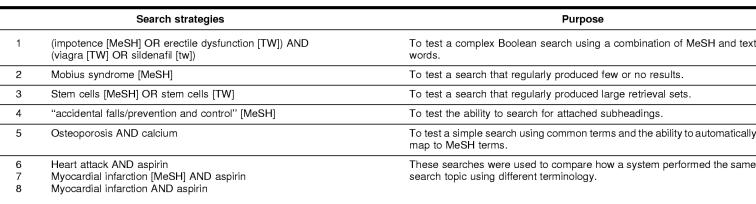
Identical search strategies were established in each of the selected SDI services and were executed on a weekly basis over a period of two months in the winter of 2002. Eight search strategies were used in each SDI system to test performance under various search conditions. The search strategies were based on representative search requests that we received while providing reference services in academic health science libraries. We selected these searches to reflect the diversity of approaches that searchers might use; and we included both simple and complex strategies. Table 2 lists each search strategy used and the related condition tested.
Table 2 Search strategies
JADE, PubCrawler, BioMail, and Cubby all searched the PubMed database to retrieve results for their SDI services. OVID and ScienceDirect each used versions of MEDLINE leased from NLM and mounted on their own servers. Since PubMed is produced by NLM, we viewed it as the official interface to MEDLINE and used the PubMed Cubby system as the baseline to evaluate the other systems. The results of the searches were analyzed to compare the efficiency and characteristics of each SDI provider.
The authors set up identical searches in Cubby using our personal accounts. Because the other selected SDI services were scheduled to automatically run at various times of the week, we each tested different systems, using the Cubby search results as the baseline. The PubMed Cubby system does not automatically run searches, but rather users must log in and select the search to be run. We ran the Cubby searches as close as possible to the automated time of the other search systems. For example, the BioMail SDI searches are always automatically executed on Fridays, so for an accurate comparison it was necessary for the Cubby searches to be executed on Fridays. Each week we ran the searches, saved the results of each search, and recorded the total number of citations retrieved from each search strategy in each system. The results from each of the searches were compared against the results retrieved from the Cubby searches, and the number of citations that duplicated those found in Cubby were recorded. These data were used to provide evidence of any difference in the average number of citations produced weekly by the selected interfaces and to determine the amount of overlap with the baseline system (Cubby).
Each system was also observed and evaluated in terms of features and functions offered to users. The features evaluated were selected based on a previous study by Eberle [24] and also included items that we considered useful based on our professional experiences. The features evaluated included ease of use; options for frequency of results; ability to use MeSH; ability to access and edit existing search strategies; ability to download to a bibliographic management program; fields displayed in results; and maximum number of search strategies allowed.
Determining the ease of use of each of the SDI services was subjective. We used our own experience (11 cumulative years in medical librarianship) with online systems to determine how easily and quickly we were able to find help screens, input searches successfully, and learn how to perform other functions in the system.
RESULTS
Comparison of SDI search results.
As mentioned previously, searches were established in all six SDI services, but no results were received from JADE or ScienceDirect during the test period, and, therefore, they were dropped from this part of the study. We attempted to contact these SDI service providers to determine the nature of the problem and request resolution. ScienceDirect responded to emails and made efforts to resolve the problem. No replies to requests for assistance were received from JADE. The testing was completed for the remaining services: PubMed Cubby, BioMail, OVID, and PubCrawler.
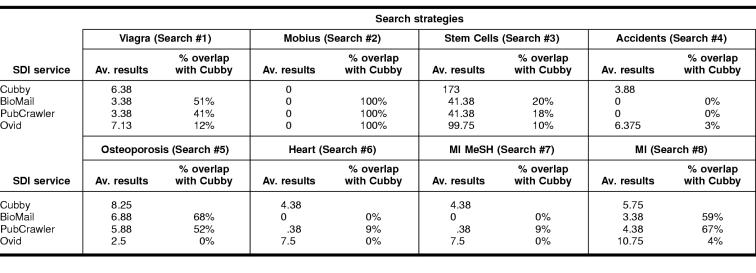
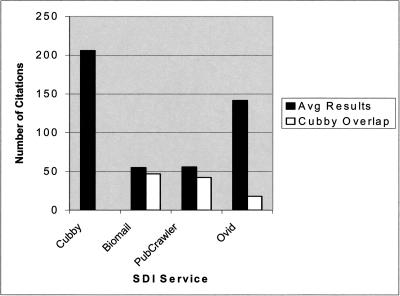
The data collection portion of the study showed that different MEDLINE SDI services retrieve varying numbers of results, even with identical search strategies. Table 3 shows the average weekly results over an eight-week period for each search strategy and the percentage of results in BioMail, PubCrawler, and OVID that overlapped with the results retrieved from Cubby. Figure 1 presents the total average results retrieved from all the searches established in each of the SDI services.
Table 3 Average weekly SDI results over an eight-week period
Figure 1.
Total average results of all SDI searches
Although OVID generally received more weekly citations than either BioMail or PubCrawler, both BioMail and PubCrawler had greater overall overlap with the citations retrieved through Cubby. It was observed in this study that there is a lag time between when citations appeared in PubMed and when they appeared in OVID MEDLINE. This is likely because OVID uses a licensed copy of MEDLINE rather than using PubMed directly, as the other SDI systems do. This lag time of one to three months would explain the small amount of overlap between OVID MEDLINE and Cubby.
Identical search strategies in BioMail and PubCrawler, which search the same PubMed interface, rarely resulted in an equal number of citations retrieved. A small percentage of the variance in citations retrieved could be attributed to differences in the time of day when the SDI services automatically ran their searches and the time of day we ran ours. However, if differences in time were the only explanation, the average results over the eight-week period would be closer in number.
The Accidental Falls [MeSH] “prevention and control” [subheading] search had been established to test the ability of the SDI services to use subheadings. Although this search strategy yielded results for Cubby and OVID, there were no results from BioMail or PubCrawler. We initially assumed that BioMail and PubCrawler were unable to handle subheadings. Eventually, we determined that BioMail and PubCrawler both had difficulty with subheadings that contained “and”. “Prevention and control” was translated by both systems to “prevention AND control,” thus resulting in zero citations. We tested both systems with other subheadings and found that the services could probably use simple subheadings, although further study is warranted.
Features and functions of SDI services.
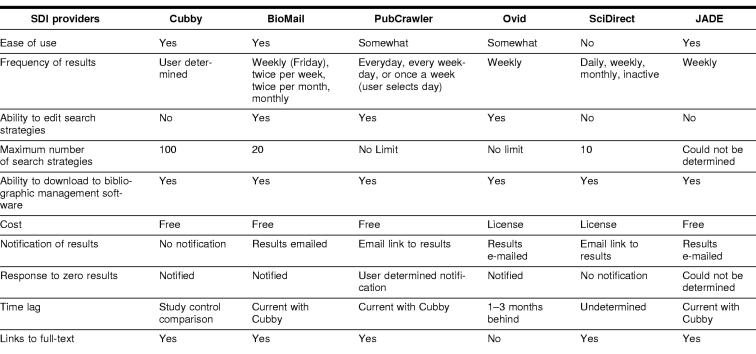
The features of the SDI services compared in this study are summarized in Table 4. All services allowed the results to be saved and imported into bibliographic software. Other features such as ease of use, help screens, fields displayed, and search set-up screens varied across the SDI services.
Table 4 Comparison of SDI provider features
Cubby
Users familiar with PubMed will have little difficulty establishing SDI searches in Cubby. Searches saved in PubMed's Cubby feature use the same interface and manner as usual searches run in PubMed. The only restriction to saving searches in Cubby is that these searches cannot be developed using PubMed's “History” feature. To save searches in Cubby, users need to establish a user name and password. Once a search strategy has been executed in PubMed, users click on “Cubby” from the sidebar menu. The search automatically appears in a “Cubby Search Name Box” with the actual search displayed above. Users can edit the name of the search and click “Store in Cubby.” Up to 100 searches can be saved in Cubby. Although search strategies cannot be directly edited, users can click on the existing search strategies, edit as needed, and save as a new search strategy.
PubMed allows the use of Boolean logic, MeSH, MeSH subheadings, keyword searching, author searching, title word searching, journal name searching, and the ability to use command language. As the citations retrieved through Cubby are displayed in the PubMed system, the features for displaying results remain the same as in a typical PubMed results screen. Citations may be displayed 5, 20, 50, 100, 200, or 500 at one time, Results may also be displayed in summary, brief, abstract, citation, and MEDLINE formats [25]. Links to the full text of articles are also available in the citations retrieved through Cubby, depending on a user's subscription status.
Cubby was the only SDI service examined in this study in which automatic emails were not sent notifying users of new citations. To retrieve the latest citations added to PubMed since the last time a saved Cubby search was run, a user must log in to Cubby. When check marks are placed next to the searches to be updated and “What's New For Selected” is clicked, Cubby will display each selected search along with the number of new citations since the last update for each search. When the number of new items is clicked, those citations are displayed, and the stored search is updated with the new date and time. If a user wants to keep a record of the citations retrieved, then the citations must be saved at this point, or they will be lost. If there are no new items, Cubby displays “0 new.” If zero hits were retrieved, the date will not change.
Being required to log in to PubMed in order to view the latest updates has advantages and disadvantages. Users may prefer to go into PubMed and get an update on the latest results in their field at a convenient time. This feature allows the user to choose what day of the week, or even the month, they would like to see the latest citations from their stored searches. On the other hand, some users may prefer a weekly reminder to look at the latest articles or prefer a weekly update just to browse through the latest results.
BioMail
BioMail is free and produced by the Medical Informatics Department at the State University of New York at Stony Brook. BioMail searches the PubMed server for new citations to provide users with the most up-to-date additions to PubMed. To use BioMail, users must register with a username and password. In order to create or edit searches, users must be logged in to the system.
Up to 20 searches can be stored and run regularly in BioMail. A text box is provided in BioMail in which users can write their search strategy. The search input screen does not provide a great deal of help to users. Therefore, users need to be familiar with MeSH terms, command language, and Boolean logic in order to establish adequate searches in BioMail. As long as users are aware of the command language that can be used for all the fileds in PubMed, there does not appear to be a limit to the types of fields that can be searched in BioMail. Citations retrieved through BioMail will not be lost until the user deletes the SDI email alert containing them.
Only the basic citation information is available in the BioMail results unless the html option is used. By using the html option, the abstracts can be viewed after users have received the citations by clicking on a hyperlink within the citation. Users may also select desired citations and click on “see abstracts” to view the abstracts. Full-text links provided through PubMed are available to BioMail users in the abstract display based on institutional access. Users may click on the “MEDLINE Format” button to import selected citations into bibliographic software.
The search input screen allows users to select whether they want to retrieve citations weekly (Friday only), twice per week, twice per month, or monthly. Users may also select the maximum number of citations they wish to retrieve for each search. In addition, users may provide an email subject heading within the search input screen. BioMail also offers users a configuration page where they may select to have the results emailed in text format, html, or as an attachment. Individual emails are sent for each individual search established, although users may select an option to send all searches in one combined email. Users may request that no email be sent when zero results have been retrieved. BioMail also provides an option in which users may suspend the service while they are away on vacation.
PubCrawler
PubCrawler is another free SDI service. EMBnet supported the development of PubCrawler. EMBnet is a science-based group of collaborative nodes throughout Europe. To use PubCrawler, users need to establish a username and password. PubCrawler allows registered users to set up profiles, which includes establishing search queries that will search PubMed for the latest update. PubCrawler also allows users to select the frequency in which the SDIs are run and emailed to the user. Possible frequencies include every day, every weekday, or any day of the week selected by the user. Users may also select the specific time of day that PubCrawler will run the query. PubCrawler provides up to five text boxes where users can add search terms. Each text box has a pull-down menu for selecting the search field. Another pull-down menu allows users to select the Boolean operators AND or OR. Alternatively, users may establish an entire search within one text box using Boolean logic. PubMed command language may also be used in the text box. Users may request that no email be sent when zero results have been retrieved.
When PubCrawler is updated, a single email is sent to the user with notification of the latest update. The user is required to click on a URL and log in to access the latest update. PubCrawler provides the results of all searches, grouped by search query, in html format on one Web page in the PubMed interface. This allows users to browse all results at one time. Only basic citation information is displayed for initially viewing PubCrawler's results. Users must click on the individual citation to view the abstracts. Alternatively, as in PubMed, users may also select specific citations and view these results in summary, brief, abstract, citation, or MEDLINE format. The full-text links available through PubMed are also available in PubCrawler based on institutional access.
Displaying all results on a single Web page requires the searcher to use care. If users wish to save the results of a search, for example, in bibliographic software, they will need to select all citations from a particular search, download those citations, and then deselect the citations before moving to the next group of citations to save.
Because PubCrawler displays results through a Web page, citations retrieved on the latest update will replace citations from the previous update. This means users need to read or save their latest update prior to the next update being run.
JADE
JADE was the final SDI service evaluated that was both free and searched the PubMed database for the latest citations. JADE is provided by the National Center for Emergency Medicine Informatics. In order to establish SDI searches within JADE, a user simply enters their email address on the JADE home page. Once on the JADE home page, a user is asked for their email address and is then sent to the search input screen. A single text box states, “type new search here.” The search input screen provides very little assistance to help users establish searches. Therefore, users need to be familiar with MeSH terms, command language, and Boolean logic in order to establish adequate searches in JADE. Searches are run on a weekly basis in JADE.
Results in JADE are displayed with minimal citation information. On the main display page in JADE, there are links to “view abstract” and gain access to “full text” when available through PubMed. If users wish to save citations, they must click on the desired citations and next click “Store Selected Articles” to place them in the “File Cabinet.” Results are displayed either in citation or MEDLINE format in the File Cabinet. However, there appeared to be no way to remove citations that have been added to the “File Cabinet,” so there is no option to save citations temporarily. If users are determined to save citations in bibliographic software, users could click on “view abstract” to view individual citations in PubMed. The display format may at this time be changed to the “MEDLINE” display format, where it could then be saved to bibliographic software. Citations sent through this service will not be lost until the email message containing them is deleted.
ScienceDirect
ScienceDirect was one of two SDI services evaluated in this study that required a fee-based subscription to access the database that offered SDI service. ScienceDirect searches its own locally mounted MEDLINE database, which is licensed from NLM. A searcher must create a user name and password in ScienceDirect if they wish to establish SDI alert services. Once a user name and password have been established, users may log in to create their searches.
In order to create SDI searches in MEDLINE through ScienceDirect, users must limit their search to “Abstract Databases” and select MEDLINE. Pull-down menus next to the two text boxes allow users to select: abstract, title, keywords; authors; journal name; title; keywords; abstract; affiliate; all fields; MeSH terms; Major MeSH terms; and subheadings. Users may also limit searches by publication type, gender and population (age group). No help is offered when entering MeSH terms. In other words, if a non-MeSH term is entered and a MeSH field selected, the system will not try to map to a MeSH term, but instead will retrieve zero results. This is unlike PubMed or OVID MEDLINE where the interfaces are designed to assist the user in locating appropriate MeSH terms. In ScienceDirect, users must know MeSH terminology if they wish to search in that manner.
If a search run in ScienceDirect is too large, the search will be interrupted and a request will be given for the user to limit the search further. This means that the user may not save the strategy as an SDI search. This is problematic if a user wants to establish a general SDI search on a topic. For example the sample search, “Stem Cells”, could not be established as an SDI search because the initial search resulted in too many citations. The user cannot keep up-to-date on the much smaller number of citations that will be added to the database when updated on a weekly, daily, or monthly basis as the overall search was too large to be allowed by the system. One option to circumvent this situation is to limit the initial search to the current year. Users must then remember to re-create their search strategy each year. Citations retrieved through ScienceDirect can be exported into bibliographic software. It did not appear possible to edit search strategies once saved in ScienceDirect.
OVID
OVID MEDLINE was the other SDI service evaluated in the study that required a fee-based subscription to access the database where the SDI service was available. OVID MEDLINE searches it own locally mounted version of MEDLINE, which is licensed from NLM. The SDI service is an additional feature that must be enabled by the institution subscribing to the service. Some institutions may have the SDI service set up so that users must log in to OVID in order to add or edit SDI search strategies. For institutions that do not require a login, the strategies are accessible to all users. In this mode, a user has the ability to view, edit, or even delete another user's SDI search.
To establish SDI queries in OVID MEDLINE, users create their search query on the standard OVID search screen. Once a search strategy has been executed in OVID MEDLINE, the user needs to click on “save search.” The user may then select to have the search saved as an SDI query. Searches saved as SDI queries have “latest update” added to the search strategy so that only new citations are sent to the user. When the saved search is executed, it is the last line of the strategy that is limited to “latest update” to produce the citations to be emailed for the alert. When creating multiple saved searches, users should clear their search history prior to developing another search query to be saved as an SDI.
When the “save search” option is selected, users are prompted with a save search screen where they may select to save a search for 24 hours, permanently, or as an AutoAlert (SDI). Users are asked for search names and if they elect the SDI service, they must also enter an email address and indicate the format in which they wish SDIs to be sent. Users may select fields (citation; citation and abstract; citation, abstract and subject headings; complete reference) and citation format (OVID, BRS/Tagged, Reprint/Medlars).
The phrase “[Use Link to view the full text]” appears in the OVID email citations, however, in the two Web-based email programs used, there were no associated hypertext links to the full-text articles. Citations will not be lost until they are deleted with the email alert. OVID's email alerts appeared to take up much more memory than the email results sent by the other SDI services.
DISCUSSION
Of the features reviewed, only the ability to download to bibliographic software was common to all services tested. The remaining features varied across systems. For example, some services offered users a choice in frequency of results and others did not. Some of the systems required that the user have background knowledge about MeSH and available fields that can be searched, so they would not be appropriate for novice users. Some systems sent notification even when no results were retrieved. In those systems that did not send notification when the results were zero, users may be concerned that a problem with the system had occurred and have no way to know that zero results had been retrieved.
It was observed throughout the course of the study that the Cubby service retrieved the largest sets and was the most current, retrieving recent citations that did not appear in the other systems. As shown in the results section, the retrieval sets of the various interfaces differed even with identical search strategies. For example, when looking at the average number of results retrieved for the “Stem Cells” search, it can be noted that the results varied widely, with the Cubby system retrieving an average of 173 citations per week while BioMail and PubCrawler averaged 41.
PubMed Cubby was used as the baseline for this study as it is the PubMed/MEDLINE interface offered by NLM. The weekly results of each search were compared against the baseline and the overlap (identical citations) with Cubby was noted to evaluate the effectiveness of each system. Looking to the previous example of “Stem Cells,” OVID averaged 100 citations per week for this search, but because of the time delay, the overlap with Cubby was much smaller.
Although we attempted to run the Cubby searches as close to the times the other SDI searches were run, this was not always possible. PubCrawler, for example, defaulted to run its searches at 6:00 a.m. Irish time, while we did not run the search until approximately 9:00 a.m. CST. It is possible citations were added to the PubMed database during the time delay.
We also observed at times that the other SDI services retrieved one or two unique citations that were not retrieved that week from Cubby. The unique citations of the other services were not closely examined during this study. Further research is warranted to determine why these results were retrieved. Future studies should also include search strategies to test other subheadings, author searches, journal title searches, title word searches, and major MeSH terms.
CONCLUSION
This study has shown that not all MEDLINE SDI services retrieve identical results even when identical search strategies are used. The observational portion of this study also showed that the services vary in terms of features and functions offered. This information will be useful to information professionals when selecting SDI services and when assisting users.
Earlier studies showed the value of librarians in assisting with the development of finely tuned search strategies for SDI. As with most online and electronic systems, there has been a movement away from librarian-mediated search support for SDIs to user-initiated and controlled services. Librarians still have an important role to play by promoting SDI and current awareness services, providing training to users, assisting patrons in deciding which service to use, explaining the differences between the systems, and in the development and refinement of complex search strategies.
Acknowledgments
The authors would like to give special thanks to the following individuals for their assistance and encouragement throughout the course of this project: Victoria Pifalo and Ann Weller, Library of the Health Sciences, University of Illinois at Chicago.
Footnotes
* This study was originally presented as a poster presentation at the 102nd annual meeting of the Medical Library Association, Dallas, Texas, May 17–23, 2002.
Contributor Information
Mary Shultz, Email: shultz@uic.edu.
Sandra L. De Groote, Email: sgroote@uic.edu.
REFERENCES
- Young H. The ALA glossary of library and information science. Chicago: American Library Association, 1983. [Google Scholar]
- Anderson CR. The Exchange: proactive reference. Ref User Serv Q. 1998 Winter; 38(2):139–40. [Google Scholar]
- Ohta M, Evans GT. Mechanization of library procedures in the medium-sized medical library: XII. An information retrieval system: a combination of a manual selective dissemination of information, and a personal file indexing system by computer. Bull Med Libr Assoc. 1970 April; 58(2):112–9. [PMC free article] [PubMed] [Google Scholar]
- Luhn HP. A business intelligence system. IBM J Res Dev. 1958 Oct; 2(4):314–319. [Google Scholar]
- Luhn HP. Selective dissemination of new scientific information with the aid of electronic processing equipment. Yorktown Heights, NY: International Business Machines Corporation, 1959. [Google Scholar]
- Brandli MJ. Current awareness services—observations of the past and present, and implications for the future. Spec Libr. 1976 Jan; 67(1):40–4. [Google Scholar]
- Kapadia S, Rettino J. SDI Service at the Library of the College of Medicine and Dentistry of New Jersey: development and future trends. Sci Tech Libr. 1981 Fall; 2(1):17–29. [Google Scholar]
- Yunis SS. The implementation, evaluation, and refinement of a manual SDI service. Bull Med Libr Assoc. 1973 Jan; 61(1):4–14. [PMC free article] [PubMed] [Google Scholar]
- Wood MS, Seeds RS. Development of SDI services from a manual current awareness service to SDILINE. Bull Med Libr Assoc. 1974 Oct; 62(4):374–84. [PMC free article] [PubMed] [Google Scholar]
- Strube K, Antoniewicz CM. Manual selective dissemination of information from journal holdings of an academic medical library. Med Ref Serv Q. 1988 Spring; 7(1):1–8. [Google Scholar]
- Lavendel GA. SDI in scientific and technical libraries: an overview of the options. Sci Tech Libr. 1981 Fall; 2(1):3–16. [Google Scholar]
- Lavendel GA. SDI in scientific and technical libraries: an overview of the options. Sci Tech Libr. 1981 Fall; 2(1):3–16. [Google Scholar]
- Yunis SS. The implementation, evaluation, and refinement of a manual SDI service. Bull Med Libr Assoc. 1973 Jan; 61(1):9. [PMC free article] [PubMed] [Google Scholar]
- Brandli MJ. Current awareness services—observations of the past and present, and implications for the future. Spec Libr. 1976 Jan; 67(1):43. [Google Scholar]
- Richards BG, Roysdon CM, and Siegler SM. Manual SDI services in an academic library: case studies of sci-tech topics. Sci Tech Libr. 1981 Fall; 2(1):31–42. [Google Scholar]
- Ohta M, Evans GT. Mechanization of library procedures in the medium-sized medical library: XII. An information retrieval system: a combination of a manual selective dissemination of information, and a personal file indexing system by computer. Bull Med Libr Assoc. 1970 April; 58(2):119. [PMC free article] [PubMed] [Google Scholar]
- Deardorff TC, Garrison AO. Developing an automated current awareness program using microcomputers and electronic mail. Tech Serv Q 1997;14(4):1–12. [Google Scholar]
- Wood MS, Seeds RS. Development of SDI services from a manual current awareness service to SDILINE. Bull Med Libr Assoc. 1974 Oct; 62(4):376. [PMC free article] [PubMed] [Google Scholar]
- Mondschein LG. SDI use and productivity in the corporate research environment. Spec Libr. 1990 Fall; 81(4):265–79. [Google Scholar]
- Knecht L, Shooshan S. Internet Grateful Med to be retired; reminder of NLM Gateway availability. NLM Tech Bull 2001 Jan/Feb;318(e3). [Google Scholar]
- Bandemer J, Tannery NH. A Comparison of four current awareness services. Med Ref Serv Q. 1998 Summer; 17(2):29–36. [DOI] [PubMed] [Google Scholar]
- Eberle M. Current awareness using PubMed: current Web services and possibilities for local solutions. Internet Ref Serv Q 2000;5(2):21–9. [Google Scholar]
- Eberle M. Current awareness using PubMed: current Web services and possibilities for local solutions. Internet Ref Serv Q 2000;5(2):21–9. [Google Scholar]
- Eberle M. Current awareness using PubMed: current Web services and possibilities for local solutions. Internet Ref Serv Q 2000;5(2):21–9. [Google Scholar]
- National Center for Biotechnology Information. National Library of Medicine. PubMed Help. [Web document]. [cited 14 Jul 2003]. v.2002: NCBI, NLM, 2002. <http://www.ncbi.nlm.nih.gov/entrez/query/static/help/pmhelp.html>. [Google Scholar]