Abstract
Hypoparathyroidism is characterized by hypocalcemia, hyperphosphatemia, and absent or markedly reduced circulating concentrations of parathyroid hormone. The transcription factor GCMB is predominantly, if not exclusively, expressed in parathyroid cells and is critical for development of the parathyroid glands in mice. Thus, in the present study we examined the GCMB gene, mapped to 6p23–24, as a candidate for isolated hypoparathyroidism. We defined the boundaries of the five exons of the human GCMB gene and then identified a large intragenic mutation in the GCMB genes of the proband of an extensive kindred with isolated hypoparathyroidism. Her parents and several other unaffected relatives were heterozygous for the mutation. Despite an absence of any history of consanguinity, microsatellite analysis showed shared genotypes that flanked the GCMB gene over a span of 5 cM, suggesting that both of the proband’s GCMB alleles had been derived from a single common ancestor. Analysis of additional, unrelated cases did not disclose the same mutation. We conclude that homozygous loss of function of the GCMB gene impairs normal parathyroid gland embryology and is responsible for isolated hypoparathyroidism in a subset of patients with this disease.
Introduction
Normal mineral metabolism and skeletal development depend upon an intricate interplay of parathyroid, renal, and skeletal factors. Crucial in this respect is parathyroid hormone (PTH), which is synthesized and secreted from the parathyroid glands at a rate that is inversely proportional to the serum ionized calcium concentration. Hormone secretion is tightly regulated through the interaction of extracellular calcium (and to a lesser extent other divalent cations) with specific calcium-sensing receptors (1–3) that are present on the surface of the parathyroid cell. Hypoparathyroidism is an uncommon metabolic disorder characterized by hypocalcemia and hyperphosphatemia due to deficient or absent secretion of PTH. Hypoparathyroidism may occur in combination with other disorders, including autoimmune, e.g., autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) syndrome (4–7), or developmental defects, e.g., DiGeorge syndrome (DGS) (8–12), or the hypoparathyroidism, sensorineural deafness, renal anomaly (HDR) syndrome (13), or as an isolated endocrinopathy termed isolated hypoparathyroidism (14). Molecular genetic studies have indicated that isolated hypoparathyroidism may be caused by defects in a variety of genes (14). Mutations in the PTH gene are an unusual cause of hypoparathyroidism and have been associated with both autosomal-dominant (15) and recessive (16, 17) forms of familial isolated hypoparathyroidism. By contrast, gain-of-function mutations in the CASR gene encoding the calcium-sensing receptor have been identified in many subjects with a mild variant of hypoparathyroidism, termed autosomal dominant hypocalcemia, that is associated with low or low-normal levels of serum PTH and relative hypercalciuria (18, 19). However, most cases of isolated hypoparathyroidism remain unexplained, and thus other genes must be involved in the molecular pathogenesis. One candidate is the GCMB gene, which encodes a human orthologue of the Drosophila glial cells missing (gcm) gene, especially as this protein is predominantly, if not exclusively, expressed in developing (20) and mature parathyroid cells (21) and is critical for development of the parathyroid glands in mice (22). Here, we report that mutation of the human GCMB gene located at chromosome 6p23–24 (21, 23) is associated with autosomal recessive isolated hypoparathyroidism. These findings confirm the importance of this transcription factor as an embryological regulator of parathyroid cell development.
Methods
Case report.
The proband (subject IV-2 in Figure 1), a 32-month-old female, was the product of a full-term pregnancy complicated only by hyperemesis gravidarum, for which the mother was hospitalized twice for administration of intravenous fluids. The proband had a birth weight of 3.04 kg and a birth length of 48 cm. There was no neonatal distress and no dysmorphic features were noted on physical examination. At 5 weeks of age she was brought to medical attention because of generalized seizures. Evaluation revealed a serum calcium concentration of 5.1 mg/dl and a serum phosphorus concentration of 9.8 mg/dl. The serum concentration of magnesium was 1.6 mg/dl, the serum concentration of 25-hydroxyvitamin D was 7 ng/ml (normal range 5-42), and the serum concentration of 1,25-dihydroxyvitamin D was 34 pg/ml (normal range 8–72). The serum concentration of intact PTH (Nichols Intact PTH immunometric assay) was undetectable. FISH analysis for DGS did not reveal a diagnostic deletion on chromosome 22 and high-resolution karyotype analysis using the standard trypsin-Wright G-banding (GTW) method revealed a normal complement of chromosomes (46 XX) without macro deletions or translocations. The diagnosis was idiopathic hypoparathyroidism, presumably congenital, and the infant was treated with calcitriol plus supplemental calcium.
Figure 1.
Pedigree of family described in text. The figure above shows the pedigree of the family, with the proband indicated by the arrow. Squares denote male family members, circles female family members, and solid symbols subjects with symptomatic hypocalcemia. Markers under each symbol indicate the genotype for the GCMB gene allele; WT denotes wild-type allele; M denotes ΔGCMB allele. NT, not tested.
The infant experienced no further seizures after treatment of hypocalcemia, and she has reached all cognitive and developmental milestones appropriately. Serum concentration of intact PTH at 8 months of age was below the detectable limit with corresponding serum calcium concentrations of 7.0 and 8.0 mg/dl. At 19 months of age the serum concentration of intact PTH was 10 pg/ml (normal range 10–65) with a corresponding serum calcium concentration of 9.6 mg/dl. Her physical examination is normal, and her growth has been parallel, but below, the second percentile for age and is consistent with a predicted adult height that reflects her parents’ short stature (father’s height 149.9 cm and mother’s height 157.5 cm). Her weight has remained along the fifth percentile, and her head circumference has continued along the 75th percentile.
The proband’s 6-year-old brother (Subject IV-1), 30-year-old mother (Subject III-2), and 31-year-old father (Subject III-1) have normal serum concentrations of calcium and intact PTH. A 22-year-old paternal uncle (Subject III-9) developed seizures at 2 months of age and is presumed to have hypoparathyroidism. He is institutionalized in a chronic care facility and cannot walk, talk, or attend to his daily needs. A 24-year-old female cousin (Subject III-1) was given a diagnosis of idiopathic hypoparathyroidism at 2 weeks of age when she experienced seizures and was found to have a serum calcium concentration of 5.8 mg/dl. The serum phosphorus concentration was 12.2 mg/dl, and serum magnesium concentration was 1.3 mg/dl. The serum concentration of immunoreactive PTH was 55 μl Eq/ml (normal range 25–90) when measured in an assay that is primarily reactive with midmolecule-carboxyl terminal PTH fragments with a corresponding serum calcium concentration of 6.6 mg/dl. She is treated with vitamin D and calcium and currently attends college.
The proband’s father and all paternal relatives are from Jalisco, Mexico, whereas the mother and her family are from northern Guatemala. There is no indication of parental consanguinity. The Joint Committee on Clinical Investigation of The Johns Hopkins University School of Medicine approved the study protocol, and written informed consent was obtained from all subjects or their parents.
DNA analysis.
DNA was extracted from peripheral-blood leukocytes by standard methods (24). Based on analysis of published sequences for the complete complementary and partial genomic DNAs for rodent Gcm2 (25–27) and human GCMB (6p23–24) (21, 23, 28) (plus GenBank accession number 8979543), we devised a series of oligonucleotide primers that enabled us to ascertain the exon-intron organization of the human gene by PCR. To analyze the GCMB gene for mutations, ten oligonucleotide primers were designed to amplify and sequence the five exons plus the exon-intron junctions from genomic DNA (Table 1). The PCR was performed with 2.5 mM magnesium chloride, 0.2 mM deoxynucleoside triphosphates, 0.5 μM of each primer, and 1 unit of Taq polymerase (Amplitaq; Perkin-Elmer Applied Biosystems, Foster City, California, USA) with the manufacturer’s buffer; information on the primers and PCR conditions is presented in Table 1. Amplified DNA was analyzed first by polyacrylamide gel electrophoresis to assess the size of the amplicons. Fragments were excised from gels and directly sequenced by the dideoxy chain termination method using the AmpliTaq Cycle Sequencing Kit (Perkin Elmer, Branchberg, New Jersey, USA).
Table 1.
PCR conditions for GCMB gene analysis
For restriction endonuclease analysis, 10-μg aliquots of genomic DNA were digested with EcoRI (New England Biolabs Inc., Beverly, Massachusetts, USA) according to the manufacturer’s recommendations. Products were separated on 1% agarose gels (SeaKem; FMC Corp., Rockland, Maine, USA), transferred to nylon membranes, and hybridized to radiolabeled GCMB genomic probes by techniques described previously (29).
Microsatellite marker analysis.
Simple sequence repeat polymorphisms were analyzed using markers D6S289 and D6S422 (from the Perkin-Elmer Applied Biosystems mapping set), D6S1006 (from Weber v.6 mapping set), and D6S470, D6S1721, and D6S1653 (from the map of chromosome 6 at http://cedar.genetics.soton.ac.uk/pub/chrom6/map) with GCTTCT added to the 5′ end of each reverse primer to help eliminate problems associated with dinucleotide repeats. For D6S470, D6S1721, D6S1653, three distinct alleles were resolved in the proband, parents, and three normal controls. For D6S1006, three alleles were observed and for D6S289 four alleles were observed.
Results
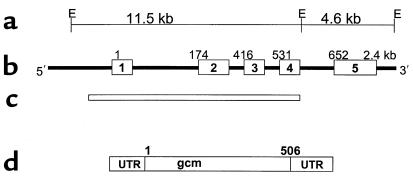
To analyze the GCMB gene for possible mutations, we first determined the intron-exon organization of the gene. The gene spans approximately 15 kb and comprises 5 exons based on a comparison with the human cDNA sequence (21, 23) (Figure 2). By comparison, the murine Gcm2 gene consists of only four exons (22, 25). The transcription start site and the translational-initiator codon are present in exon 1, and the termination codon is present in exon 5. The open reading frame of the gene agrees with that of the GCMB cDNA sequence and encodes a predicted peptide of 506 amino acids.
Figure 2.
GCMB gene. (a) An EcoRI restriction map of the human GCMB gene, with approximate genomic distances between restriction sites noted. (b) The genomic organization of the human GCMB gene, with exons 1–5 indicated by the boxes and introns indicated by the connecting lines. The numbers above each exon indicate the corresponding nucleotide number of the cloned cDNA. (c) The area of the gene deleted in the ΔGCMB allele. (d) The cDNA, with the 5′ and 3′ untranslated regions (UTR), and the open reading frame that predicts a protein of 506 amino acids.
In our preliminary analyses, we used oligonucleotide primers corresponding to exons 1 and 2 of the GCMB gene to amplify genomic DNA from the propositus and three other patients with isolated hypoparathyroidism (Figure 3). Figure 3 shows that products of the expected sizes (and appropriate sequences) were generated for patients 1, 3, and 4, but no specific product was generated for patient 2 (the propositus, subject IV-2 in Figure 1). By contrast, PCR of genomic DNA from the parents of subject 2 resulted in normal products for both exon 1 and 2 (not shown). PCR analysis of exons 3 through 5 of the GCMB gene showed amplification of normal products for both of the parents, but only an exon 5 product for the proband (Figure 4), suggesting that exons 1–4 were deleted. To exclude the possibility that contaminating substances were inhibiting amplification of exons 1 through 4 we performed mixing experiments in which PCR was performed on samples containing equal amounts of genomic DNA from the proband and a normal control subject. PCR of these hybrid samples resulted in successful amplification of fragments corresponding to exons 1–4 (data not shown). Moreover, PCR of genomic DNA from the proband resulted in successful amplification of multiple exons from other complex genes (e.g., GNAS1 and PTH), providing further evidence that a large, homozygous deletion of a portion of the GCMB gene (ΔGCMB), and not artifact, accounted for the inability to amplify exons 1 through 4 of GCMB.
Figure 3.
Amplification of exons 1 and 2 by PCR. Genomic DNA from four unrelated subjects with familial isolated hypoparathyroidism was amplified using oligonucleotide primers flanking exon 1 (left panel) or exon 2 (right panel); the proband shown in Figure 1 corresponds to subject 2. The positions of molecular-weight markers are indicated on the left. DNA from subjects 1, 3, and 4 showed amplification of appropriate-sized fragments, whereas DNA from subject 2 failed to amplify a fragment corresponding to either exon 1 or exon 2.
Figure 4.
Amplification of exons 3, 4, and 5 by PCR. Genomic DNA from the proband (Pt), her father (F) and mother (M), and two unrelated normal subjects (NL) was amplified using oligonucleotide primers flanking exon 3 (upper panel), exon 4 (middle panel), or exon 5 (bottom panel). Arrows indicate the position of the appropriately sized amplified product. DNA from the proband (Pt) failed to amplify fragments corresponding to exons 3 and 4 but did yield an appropriate PCR product corresponding to exon 5. Genomic DNA from the parents and two normal subjects yielded appropriate-sized PCR products for all three exons.
To verify the deletion, we performed restriction endonuclease analysis of genomic DNA from the proband, her parents, and normal control subjects. Hybridization of a radiolabeled probe corresponding to exon 3 of GCMB to genomic DNA that had been digested with EcoRI revealed a single predicted band of approximately 10–11 kb in normal subjects and the propositus’ parents but not in the propositus (Figure 5, left panel), confirming homozygous deletion of this portion of the gene (Figure 2). To further refine the ΔGCMB allele, we hybridized a radiolabeled DNA probe spanning exons 3 to 5 to equivalent amounts of EcoRI-digested DNA from these individuals. DNA from the normal subjects revealed two hybridizing fragments of expected sizes (11.5 kb and 4.6 kb spanning exons 1-4 and exon 5, respectively; Figure 2) that were also present but at reduced (∼50%) signal intensity in DNA from the propositus’ parents (Figure 5, right panel). By contrast, DNA from the proband hybridized with only a single fragment of abnormal size (∼8.0 kb), which was also present at reduced signal intensity in her two parents (Figure 5, right panel). These results indicated that the proband was homozygous for a partial deletion of the GCMB gene that includes the EcoRI site present at the exon 4-intron 4 boundary. To delineate the deletion more precisely, we designed a series of sense oligonucleotide primers that hybridized to sequences in the 5′ region of the GCMB gene and tested the ability of each to amplify genomic DNA from the proband using an antisense primer located in intron 4. These experiments demonstrated that that the 5′ boundary of the deletion is located 1901 bp upstream from the initiator codon, and the 3′ boundary is located at the exon 4-intron 4 border, with loss of the EcoRI site. The calculated size of the deletion is 7,792 bp, which agrees well with the size (approximately 8 kb) predicted from the results of the restriction endonuclease analyses (Figure 5). In addition, our analyses indicated that each parent was heterozygous for the same gene deletion. Similar molecular analyses of ten additional, unrelated patients with isolated hypoparathyroidism failed to disclose an analogous mutation.
Figure 5.
Southern blot analysis of GCMB gene. Genomic DNA from the proband (Pt), her father (F) and mother (M), and two unrelated normal subjects (NL) was digested using EcoRI and hybridized to a radiolabeled genomic DNA corresponding to either exon 3 (left) or exons 3–5 (right). The positions of molecular-weight markers are indicated. Genomic DNA from the proband (Pt) failed to hybridize to the exon 3 probe, and a unique 8.4-kb EcoRI fragment hybridized to the exon 3–5 probe. The proband’s parents showed hybridization of the exon 3–5 probe with two restriction fragments of expected size and with the abnormal 8.4-kb fragment, all with reduced signal intensity (see text).
We performed a molecular analysis of the GCMB gene in all available relatives of the propositus by EcoRI restriction analysis of genomic DNA (Figure 1). In addition to her two parents, the propositus’ paternal grandfather was also heterozygous for the ΔGCMB allele. By contrast, her unaffected brother was homozygous for the wild-type GCMB allele. Three of her father’s six unaffected sibs were also heterozygous for the mutation. Regrettably, neither of the other two affected subjects, III-1 and III-9, agreed to participate in these studies.
Given the rarity of familial isolated hypoparathyroidism, an extended haplotype analysis using closely linked microsatellite markers was performed in the parents and proband (Table 2 and Figure 6), with genotype analyses performed for marker D6S470 on the proband’s paternal grandparents in order to establish phase assignments unambiguously. This analysis showed shared genotypes that flanked the GCMB gene over a span of 5 cM. Based on this limited data set, we believe that it is exceedingly unlikely that this mutation would have arisen independently twice on the same genetic background. Accordingly, a founder affect, with homozygosity by descent of the chromosomal segment containing the GCMB deletion, is the most probable explanation for our results (30) (Figure 6). This model is consistent with the observation of a rare allele occurring in the homozygous state and suggests that both of the proband’s ΔGCMB alleles are likely to have been derived from a single common ancestor.
Table 2.
Marker heterozygosity and allelic frequency
Figure 6.
Haplotype analysis of the GCMB locus. The figure above shows the haplotypes for the GCMB locus obtained using microsatellite polymorphisms. The proband is denoted by the solid circle, and her heterozygous father and mother are indicted by the half-filled square and circle, respectively. This figure demonstrates that the two parental mutant alleles (ΔGCMB) have identical haplotypes for a series of markers that span over 5 cM of genomic DNA on chromosome 6p23–24.
Discussion
We have identified mutation of the GCMB gene as a novel mechanism for isolated hypoparathyroidism. The genetic defects responsible for most cases of isolated hypoparathyroidism remain largely unknown. Two loci associated with monogenic early-onset forms of this disease have been identified, but in each case the molecular defect impairs synthesis (i.e., PTH gene defects) or secretion (i.e., CASR gene defects) of PTH rather than development of the parathyroid glands (15–19). Although candidate genes have recently been identified that account for parathyroid hypoplasia in patients with DGS (31, 32) or HDR (13), the expression of these genes in diverse tissues during embryogenesis suggests a more complex role in vertebrate development. By contrast, the GCMB gene is a likely a specific regulator of parathyroid gland development, as it is predominantly, if not exclusively, expressed in parathyroid cells, and Gcm2-deficient mice lack parathyroid tissue (22). Our data extend this role to humans and show that loss of GCMB is sufficient to produce the characteristic clinical and biochemical findings of severe early-onset hypoparathyroidism and that heterozygous mutation of the GCMB gene is associated with an asymptomatic carrier state. As expected for a rare mutation occurring in the homozygous state, both of the proband’s ΔGCMB alleles appear to have been derived from a single common ancestor. Despite the absence of a history of consanguinity, the restricted geographic area in which members of the extended family live and the lack of an obvious heterozygous phenotype likely account for the propagation of this allele and the occurrence of hypoparathyroidism in multiple members of this kindred.
The vertebrate glial cells missing (GCM) transcription factors are a small family of unique proteins that are orthologues of the gcm gene originally identified in Drosophila melanogaster (33). In the fly, the gcm gene is specifically and transiently expressed in the glial lineage and promotes choice between glial versus neuronal fate in a multipotential neural stem cell. Loss of function mutations in the gcm gene result in conversion of presumptive glial cells into neurons (33). Based on its nuclear localization and sequence-specific DNA-binding activity, GCM has been proposed to be a transcriptional activator of glial-specific genes. Two mammalian GCM homologues have been identified, encoded by the GCMA and GCMB genes. Both genes encode peptides of 504–506 amino acids and contain a number of motifs that are signatures of the GCM family of nuclear transcription factors, including an evolutionarily conserved DNA-binding domain in the amino-terminal region, a nuclear localization signal, and several potential PEST sequences, which are typical of proteins displaying a rapid turnover (20, 23, 25, 26, 33). Despite the significant homology of the GCMA and GCMB proteins to their Drosophila orthologue, the unique expression patterns of these two mammalian genes suggest that the roles of these proteins have evolved and diverged significantly from those in the fly. Although GCMB is detectable at low levels in embryonic neural tissues and kidney (21, 23, 25), transcripts for both GCMA and GCMB are found primarily in non-neural tissues. Whereas GCMA is most highly expressed in the placenta (25, 26), where it plays a critical role in trophoblast differentiation (34), the GCMB gene is expressed predominantly, if not exclusively, in the PTH-secreting cells of the developing and mature parathyroid gland (25).
Recently, parathyroid agenesis was demonstrated in Gcm2-deficient mice that were generated by targeted deletion of the Gcm2 gene (22). Similar to the patient we describe in this report, Gcm2-deficient mice develop severe hypocalcemia and hyperphosphatemia shortly after birth. Serum levels of intact PTH were marked reduced or undetectable in our patient and her affected cousin, III-1, but circulating PTH in Gcm2-deficient mice was unexpectedly normal (although inappropriately low relative to the hypocalcemia) and shown to arise from the thymus, which shares a common embryological origin with the parathyroids (22). This discrepancy might reflect differences in the performance characteristics of the PTH assays used in these studies or could indicate that the thymus plays a less important role as an auxiliary source of PTH in humans than in mice, which have only two parathyroid glands. Although we cannot exclude the possibility that the low levels of PTH in the serum of the patients we describe here derive from residual parathyroid tissue, it is tempting to speculate that the human thymus does contain PTH-secreting cells. In that case, the measurable levels of circulating PTH detected in subjects III-1 and IV-2 might indicate that the thymus can provide an auxiliary source of PTH in humans as well as mice. Moreover, it would also suggest that the occurrence of parathyroid rests or adenomas in patients with hyperparathyroidism (35–37) may not always represent ectopic migration of an errant parathyroid gland (38).
In summary, we describe a syndrome of autosomal recessive isolated hypoparathyroidism due to absence of the GCMB transcription factor. Loss of the GCMB gene appears to cause a highly specific embryological defect that is limited to parathyroid cells. The restricted phenotype of this syndrome confirms that GCMB controls differentiation of cells in the third and fourth branchial pouches into parathyroid glands and indicates that further study of GCMB and its regulated genes will help to elucidate the embryological pathways for parathyroid development.
Acknowledgments
This work was supported in part by United States Public Health Service Grant DK34281 (to M.A. Levine) and by General Clinical Research Center Grant RR-0035 from the NIH. We are indebted to Laura Kasch for performing the haplotype analyses and to Aravinda Chakravarti for assistance in analyzing genotypes.
References
- 1.Hebert SC, Brown EM. The extracellular calcium receptor. Curr Opin Cell Biol. 1995;7:484–492. doi: 10.1016/0955-0674(95)80004-2. [DOI] [PubMed] [Google Scholar]
- 2.Brown EM, et al. Cloning and characterization of an extracellular Ca2+-sensing receptor from bovine parathyroid. Nature. 1993;366:575–580. doi: 10.1038/366575a0. [DOI] [PubMed] [Google Scholar]
- 3.Chattopadhyay N, Mithal A, Brown EM. The calcium-sensing receptor: a window into the physiology and pathophysiology of mineral ion metabolism. Endocr Rev. 1996;17:289–307. doi: 10.1210/edrv-17-4-289. [DOI] [PubMed] [Google Scholar]
- 4.Ahonen P. Autoimmune polyendocrinopathy-candidosis-ectodermal dystrophy (APECED): autosomal recessive inheritance. Clin Genet. 1985;27:535–542. doi: 10.1111/j.1399-0004.1985.tb02037.x. [DOI] [PubMed] [Google Scholar]
- 5.Ahonen P, Myllarniemi S, Sipila I, Perheentupa J. Clinical variation of autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) in a series of 68 patients. N Engl J Med. 1990;322:1829–1836. doi: 10.1056/NEJM199006283222601. [DOI] [PubMed] [Google Scholar]
- 6.Scott HS, et al. Common mutations in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy patients of different origins. Mol Endocrinol. 1998;12:1112–1119. doi: 10.1210/mend.12.8.0143. [DOI] [PubMed] [Google Scholar]
- 7.Obermayer-Straub P, Manns MP. Autoimmune polyglandular syndromes. Baillieres Clin Gastroenterol. 1998;12:293–315. doi: 10.1016/s0950-3528(98)90136-1. [DOI] [PubMed] [Google Scholar]
- 8.Greig F, Paul E, DiMartino-Nardi J, Saenger P. Transient congenital hypoparathyroidism: resolution and recurrence in chromosome 22q11 deletion. J Pediatr. 1996;128:563–567. doi: 10.1016/s0022-3476(96)70372-4. [DOI] [PubMed] [Google Scholar]
- 9.Hur H, Kim YJ, Noh CI, Seo JW, Kim MH. Molecular genetic analysis of the DiGeorge syndrome among Korean patients with congenital heart disease. Mol Cells. 1999;9:72–77. [PubMed] [Google Scholar]
- 10.Monaco G, et al. DiGeorge anomaly associated with 10p deletion. Am J Med Genet. 1991;19:215–216. doi: 10.1002/ajmg.1320390220. [DOI] [PubMed] [Google Scholar]
- 11.Daw SC, et al. A common region of 10p deleted in DiGeorge and velocardiofacial syndromes. Nat Genet. 1996;13:458–460. doi: 10.1038/ng0896-458. [DOI] [PubMed] [Google Scholar]
- 12.Yamagishi H, Garg V, Matsuoka R, Thomas T, Srivastava D. A molecular pathway revealing a genetic basis for human cardiac and craniofacial defects. Science. 1999;283:1158–1161. doi: 10.1126/science.283.5405.1158. [DOI] [PubMed] [Google Scholar]
- 13.Van Esch H, et al. GATA3 haplo-insufficiency causes human HDR syndrome. Nature. 2000;406:419–422. doi: 10.1038/35019088. [DOI] [PubMed] [Google Scholar]
- 14.Ahn TG, Antonarakis SE, Kronenberg HM, Igarashi T, Levine MA. Familial isolated hypoparathyroidism: a molecular genetic analysis of 8 families with 23 affected persons. Medicine. 1986;65:73–81. [PubMed] [Google Scholar]
- 15.Arnold A, et al. Mutation of the signal peptide-encoding region of the preproparathyroid hormone gene in familial isolated hypoparathyroidism. J Clin Invest. 1990;86:1084–1087. doi: 10.1172/JCI114811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkinson DB, Thakker RV. A donor splice site mutation in the parathyroid hormone gene is associated with autosomal recessive hypoparathyroidism. Nat Genet. 1992;1:149–152. doi: 10.1038/ng0592-149. [DOI] [PubMed] [Google Scholar]
- 17.Sunthornthepvarakul T, Churesigaew S, Ngowngarmratana S. A novel mutation of the signal peptide of the preproparathyroid hormone gene associated with autosomal recessive familial isolated hypoparathyroidism. J Clin Endocrinol Metab. 1999;84:3792–3796. doi: 10.1210/jcem.84.10.6070. [DOI] [PubMed] [Google Scholar]
- 18.Pearce SH, et al. A familial syndrome of hypocalcemia with hypercalciuria due to mutations in the calcium-sensing receptor. N Engl J Med. 1996;335:1115–1122. doi: 10.1056/NEJM199610103351505. [DOI] [PubMed] [Google Scholar]
- 19.Bai M, et al. Expression and characterization of inactivating and activating mutations in the human Ca2+o-sensing receptor. J Biol Chem. 1996;271:19537–19545. doi: 10.1074/jbc.271.32.19537. [DOI] [PubMed] [Google Scholar]
- 20.Jones BW, Fetter RD, Tear G, Goodman CS. Glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell. 1995;82:1013–1023. doi: 10.1016/0092-8674(95)90280-5. [DOI] [PubMed] [Google Scholar]
- 21.Kanemura Y, et al. Isolation and expression analysis of a novel human homologue of the Drosophila glial cells missing (gcm) gene. FEBS Lett. 1999;442:151–156. doi: 10.1016/s0014-5793(98)01650-0. [DOI] [PubMed] [Google Scholar]
- 22.Gunther T, et al. Genetic ablation of parathyroid glands reveals another source of parathyroid hormone. Nature. 2000;406:199–203. doi: 10.1038/35018111. [DOI] [PubMed] [Google Scholar]
- 23.Kammerer M, Pirola B, Giglio S, Giangrande A. GCMB, a second human homolog of the fly glide/gcm gene. Cytogenet Cell Genet. 1999;84:43–47. doi: 10.1159/000015210. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press. Plainview, New York. 280–281.
- 25.Kim J, et al. Isolation and characterization of mammalian homologs of the Drosophila gene glial cells missing. Proc Natl Acad Sci USA. 1998;95:12364–12369. doi: 10.1073/pnas.95.21.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altshuller Y, Copeland NG, Gilbert DJ, Jenkins NA, Frohman MA. Gcm1, a mammalian homolog of Drosophila glial cells missing. FEBS Lett. 1996;393:201–204. doi: 10.1016/0014-5793(96)00890-3. [DOI] [PubMed] [Google Scholar]
- 27.Akiyama Y, Hosoya T, Poole AM, Hotta Y. The gcm-motif: a novel DNA-binding motif conserved in Drosophila and mammals. Proc Natl Acad Sci USA. 1996;93:14912–14916. doi: 10.1073/pnas.93.25.14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akiyama-Oda Y, Hosoya T, Hotta Y. Alteration of cell fate by ectopic expression of Drosophila glial cells missing in non-neural cells. Dev Genes Evol. 1998;208:578–585. doi: 10.1007/s004270050217. [DOI] [PubMed] [Google Scholar]
- 29.Patten JL, et al. Mutation in the gene encoding the stimulatory G protein of adenylate cyclase in Albright’s hereditary osteodystrophy. N Engl J Med. 1990;322:1412–1419. doi: 10.1056/NEJM199005173222002. [DOI] [PubMed] [Google Scholar]
- 30.Clark AG. The size distribution of homozygous segments in the human genome. Am J Hum Genet. 1999;65:1489–1492. doi: 10.1086/302668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindsay EA, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 32.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 33.Hosoya T, Takizawa K, Nitta K, Hotta Y. Glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell. 1995;82:1025–1036. doi: 10.1016/0092-8674(95)90281-3. [DOI] [PubMed] [Google Scholar]
- 34.Schreiber J, et al. Placental failure in mice lacking the mammalian homolog of glial cells missing, GCMa. Mol Cell Biol. 2000;20:2466–2474. doi: 10.1128/mcb.20.7.2466-2474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Numano M, et al. Surgical significance of supernumerary parathyroid glands in renal hyperparathyroidism. World J Surg. 1998;22:1098–1102. doi: 10.1007/s002689900524. [DOI] [PubMed] [Google Scholar]
- 36.Wei JP, Tippins RB, Rao RN, Burke GJ, Mansberger AR., Jr Nonadenomatous thymic unencapsulated parathyroid tissue as a cause of persistent primary hyperparathyroidism. South Med J. 1994;87:1264–1268. doi: 10.1097/00007611-199412000-00011. [DOI] [PubMed] [Google Scholar]
- 37.Palmer JA, Sutton FR. Importance of a fifth parathyroid gland in the surgical treatment of hyperparathyroidism. Can J Surg. 1978;21:350–351. [PubMed] [Google Scholar]
- 38.Rizzoli R, Pache JC, Didierjean L, Burger A, Bonjour JP. A thymoma as a cause of true ectopic hyperparathyroidism. J Clin Endocrinol Metab. 1994;79:912–915. doi: 10.1210/jcem.79.3.8077382. [DOI] [PubMed] [Google Scholar]