Abstract
Objective
To test whether HLA‐DR alleles influence the production of particular autoantibodies in rheumatoid arthritis (RA) patients, we screened synovial proteins with sera of RA patients homozygous for different HLA‐DR alleles by using 2D blots. We found that sera of RA patients homozygous for HLA‐DRB1*0404 recognised a 100‐kDa synovial protein identified as calpastatin. We studied B and T cell epitopes on calpastatin and their association with HLA‐DRB1*0404.
Methods
The frequency of positive sera in patients expressing different RA‐associated HLA‐DR allele combinations was calculated by inhouse ELISA using purified synovial calpastatin or calpastatin peptides encompassing the entire calpastatin protein as immunosorbent. Interaction between calpastatin peptides and HLA‐DR alleles was tested by a direct binding assay. T cell responses to calpastatin were measured in RA patients and controls.
Results
We found that RA‐associated HLA‐DR alleles are associated with presence of autoantibodies to synovial calpastatin in RA patients' sera. HLA‐DRB1*0404 is strongly associated with antisynovial calpastatin in RA sera. One linear B cell epitope is preferentially associated with HLA‐DRB1*0404. Multiple peptides from calpastatin bind every tested HLA‐DR allele associated or not with RA. Peptides from domain 1 and 4 of calpastatin are the best HLA‐DR allele binders. The T cell response to calpastatin is frequent in RA patients and independent of the HLA‐DR background.
Conclusions
HLA‐DRB1*0404 is strongly associated with anticalpastatin antibodies in rheumatoid arthritis.
Rheumatoid arthritis (RA) is a chronic inflammatory joint disease with a prevalence of 0.5% worldwide.1 The aetiology of RA is unknown, but a genetic predisposition to RA is well established.2 Most patients with rheumatoid arthritis express particular HLA‐DR alleles, like HLA‐DRB1*0401, *0404, *0405, *0408, *0101, *0102, *1001 and *1402. RA‐associated HLA‐DR alleles share a highly conserved amino acid motif expressed in the third hypervariable region of their DRB1 chain. This motif is called the shared epitope (SE). A dose effect has been observed in SE positive HLA‐DRB1 genotypes. Indeed, HLA‐DR genotypes containing two RA susceptibility alleles (“double dose” genotypes) confer a higher risk than genotypes containing only one susceptibility allele (“single dose genotypes”) which confer a higher risk than DR genotypes containing no susceptibility allele. The maximal risk to develop RA is observed in individuals expressing both HLA‐DRB1*0401 and HLA‐DRB1*0404. How these HLA‐DRB1 alleles influence the development of RA is unknown.
To test whether HLA‐DR alleles influence the production of specific autoantibodies in RA patients, we screened synovial proteins with sera of RA patients homozygous for HLA‐DR alleles. We observed that sera from RA patients homozygous for HLA‐DRB1*0404 recognised a 100‐kDa synovial protein identified as calpastatin. Calpastatin is an endogenous calpain (calcium‐dependent cysteine protease) inhibitor, distributed in most mammalian tissues. It includes an N‐terminal L domain and four repetitive calpain inhibition domains.3 Autoantibodies against calpastatin have been previously described in rheumatoid arthritis, but their specificity remains controversial.4,5,6,7
To test the influence of different RA‐associated alleles on anticalpastatin production, we calculated the frequency of positive sera in patients expressing two, one or no RA‐associated HLA‐DR allele by inhouse ELISA using purified synovial calpastatin as immunosorbent. To identify B cell epitopes, we tested RA sera against peptides encompassing the entire calpastatin. Calpastatin comprises five domains of about 140 amino acids each. They are called domains L, 1, 2, 3 and 4. We used 94 overlapping 15 mer peptides encompassing the five domains of calpastatin to analyse RA sera reactivity. We then analysed the interaction between calpastatin peptides and HLA‐DR alleles by a direct binding assay. The 94 overlapping 15 mer peptides encompassing the five domains of calpastatin were tested for binding to purified HLA‐DRB1*0401, *0404, *0101 (RA‐associated alleles) and HLA‐DRB1*0402, *0701 (RA non‐associated alleles). Finally, we measured T cell proliferative responses to calpastatin in RA patients and controls.
Patients and methods
RA patients and controls
A total of 155 RA patients were chosen from the Rheumatology Ward at Hospital La Conception, Marseille, France. These patients fulfilled the 1987 American College of Rheumatology criteria for RA. Eighty‐two volunteers from the laboratory staff and the Marseille Blood Transfusion Center staff served as normal controls. For every patient and control, HLA‐DR oligotyping was performed. We studied 49 patients expressing two RA susceptibility HLA‐DR alleles (the most common were HLA‐DRB1*0101, DRB1*0404 and DRB1*0401), 71 patients expressing one RA susceptibility HLA‐DR allele (the most common were HLA‐DRB1*0101, DRB1*0404 and DRB1*0401) and 35 patients without any RA susceptibility HLA‐DR allele. Among the 82 controls, 28 expressed one RA susceptibility HLA‐DR allele. All participants had given informed consent.
Two‐dimensional gel electrophoresis and immunoblotting
Proteins were extracted from synovial tissue using the ReadyPrep sequential Extraction Kit (Bio‐Rad, France). Briefly, proteins were suspended in 8 M urea, 4% CHAPS, 10 mM DTT, 40 mM Tris and 0.2% Bio‐Lyte 3/10 ampholyte. First dimension separation was by isoelectric focusing using IPG ready strip pH4 to pH7. Second dimension separation was on 10% SDS PAGE gels. Proteins were then transferred onto PVDF membranes. Blots were revealed by sera of RA patients homozygous for HLA‐DR followed by peroxidase‐conjugated antihuman IgG. Blots were revealed by chemiluminescence (Roche diagnostics, Meylan, France).
Synovial calpastatin purification
Synovial tissue was lysed in 10 mM Tris pH8, 10 mM NaCl, 10 mM MgCl2, 1% Triton ×100, 0.05 mg/ml Dnase and protease inhibitors. Total protein extracts were immunoprecipitated by anticalpastatin C19 antibody covalently coupled on cyanogen bromide‐activated sepharose 4B (Sigma, St. Quentin, France). After washing, synovial calpastatin was eluted in PBS pH 2, neutralised and quantified.
Detection of anticalpastatin antibodies by ELISA
Plates were coated overnight with 0.1 µg/well synovial calpastatin or with 10 µg/well calpastatin peptides diluted in phosphate buffer saline (PBS), pH 7.4. Plates were blocked with PBS containing 5% milk. Sera diluted to 1:100 in PBS were incubated for 2 h. After washing with 0.1% Tween 20, peroxidase conjugated antihuman IgG, (Sigma, France) was added. The optical density was read at 405 nm. Background OD was obtained by adding each serum to a well without protein. Positive sera were defined as an OD value more than twice the background OD.
Synthetic peptides
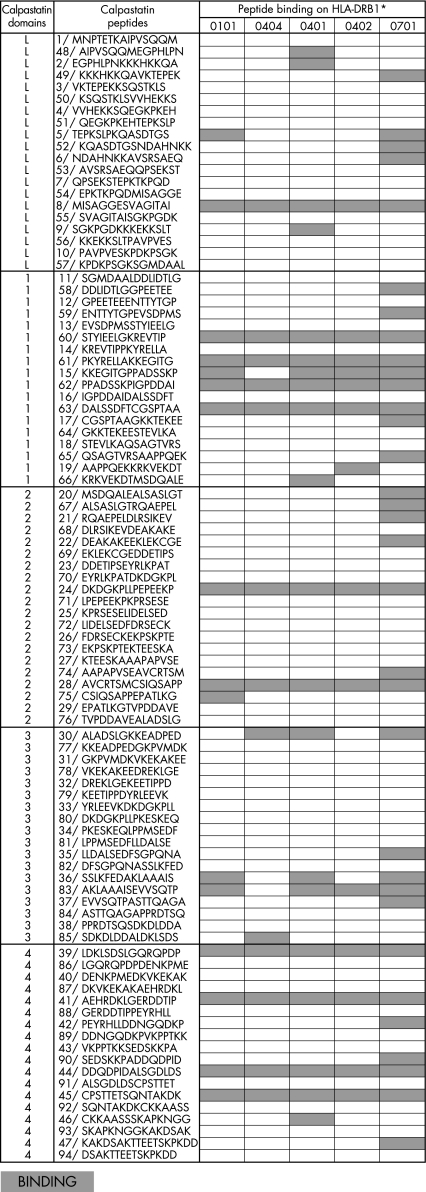
Peptides (Neosystem, Strasbourg, France) were synthesised using the solid‐phase system and purified (>60%). Ninety‐four 15‐amino‐acid peptides overlapping on 7 amino acids derived from calpastatin isoform A (locus NP_001741), residues 1–708. The amino acid sequence of the peptides from calpastatin is shown in Figure 2.
Figure 2 Binding of calpastatin peptides to HLA‐DR alleles. ELISA plates were coated with 10 µg of calpastatin peptide per well. One microgram of purified HLA‐DR molecule was then added to plates. After washing, bound HLA‐DR was detected by biotinylated anti‐HLA‐DR antibody followed by peroxidase conjugated avidin. Optical density was read at 405 nm. Positive binding was defined as an OD value higher than twice the OD for HLA‐DR without peptide.
Lymphoblastoid cell lines
The HLA homozygous lymphoblastoid cell lines JESTHOM (HLA‐DRB1*0101), SAVC (HLA‐DRB1*0401), YAR (HLA‐DRB1*0402), PEYSSON (HLA‐DRB1*0404) and MOU (HLA‐DRB1*0701) were cultured in RPMI 1640 with 10% fetal calf serum.
Antibodies
Anti‐HLA‐DR monoclonal antibodies were LB3.1 (gift from M F Del Guercio, Cytel, San Diego, CA) and biotinylated B8122 (Immunotech, Marseille, France). Peroxidase‐conjugated antihuman IgG and peroxidase‐conjugated avidin was supplied by Sigma (St. Quentin, France). Anticalpastatin polyclonal antibodies were C19 and H300 (Tebu Bio, Paris, France).
Purification of HLA‐DR molecules
HLA‐DRB1 homozygous cells (2×109) were lysed in 10 mM Tris pH 8, 10 mM NaCl, 10 mM MgCl2, 1% Triton ×100, 0.05 mg/ml Dnase and protease inhibitors. Total protein extracts were immunoprecipitated by antiHLA‐DR LB3.1 antibody covalently coupled on cyanogen bromide‐activated sepharose 4B (Sigma, St. Quentin, France). After washing, HLA‐DR molecules were eluted in PBS pH 2 with 0.5% n‐octylglucoside, neutralised in 1 M Tris and quantified.8
Peptide‐binding assay
ELISA plates were coated with 10 µg of peptide/well and blocked in 1% bovine serum albumin. 1 µg of purified HLA‐DR molecule was added to plates. After washing, bound HLA‐DR was detected by biotinylated antiHLA‐DR antibody followed by peroxidase‐conjugated avidin. The optical density was read at 405 nm. Positive binding was defined as an OD value more than twice the OD for HLA‐DR without peptide. To confirm positive binding, we used a known positive peptide from flu haemagglutinin in each test.8
Statistical analysis
The p values were calculated using the χ2 test.
Results
HLA‐DRB1*0404 homozygous RA sera recognise synovial calpastatin
To identify new autoantigens in RA patients homozygous for HLA‐DR alleles, we screened synovial proteins in two‐dimensional SDS PAGE gel with sera from RA patients homozygous for HLA‐DR alleles.
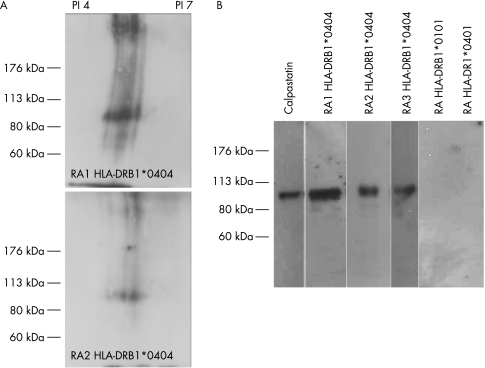
We observed that two sera from RA patients homozygous for HLA‐DRB1*0404 recognised a 100‐kDa synovial protein (fig 1A). No reactivity was observed with sera from RA patients homozygous for HLA‐DRB1*0401 or HLA‐DRB1*0101. The 100‐kDa synovial protein was identified as calpastatin, as it bound two antibodies specific for calpastatin.
Figure 1 HLA‐DRB1*0404 homozygous RA sera recognise the synovial calpastatin. (A) Synovial proteins were separated in two‐dimensional SDS PAGE gel and analysed with sera of RA patients homozygous for HLA‐DR alleles. (B) Synovial proteins were immunoprecipitated with anticalpastatin antibody and analysed by western blotting with sera of RA patients homozygous for HLA‐DR alleles.
This was confirmed by precipitation assay (fig 1B). Indeed, purified synovial calpastatin was recognised by sera of RA patients homozygous for HLA‐DRB1*0404.
Synovial calpastatin recognised by HLA‐DRB1*0404 patients is not citrullinated, as demonstrated by immunoprecipitation and analysis with anticitrulline antibody (data not shown/supplemental file: fig 1A).
HLA‐DRB1*0404 is most strongly associated with anticalpastatin antibodies in rheumatoid arthritis
The sera of 155 patients and 82 controls were tested for the presence of synovial calpastatin antibodies by ELISA using purified synovial calpastatin as immunosorbent. We observed that 42% of RA patients were positive to synovial calpastatin compared with 12% of controls (p<0.001). Among RA patients, the presence of the shared epitope was strongly associated with anticalpastatin positivity. Indeed, 73% of RA patients expressing two RA‐associated alleles (SE+/SE+) were positive to synovial calpastatin compared with 34% of RA patients expressing one RA‐associated allele (SE+/SE–). Only 14% of RA patients without RA‐associated allele (SE–) were positive (table 1).
Table 1 Distribution of SE and antisynovial calpastatin positivity.
| SE | n (%) | p |
|---|---|---|
| SE+/SE+ | 49 ()73 | p<0.001 |
| SE+/SE– | 71 (34) | p = 0.059 |
| SE– | 35 (14) |
p versus SE− group; n, number of patients.
To evaluate the influence of individual RA‐associated alleles on antisynovial calpastatin production, we compared the frequency of positive sera in patients expressing particular HLA‐DR allele combinations. We found that the RA‐associated HLA‐DRB1*0404 allele was very strongly associated with the presence of antisynovial calpastatin in RA sera (table 2). Eighty‐three percent of patients expressing both HLA‐DRB1*0404 and HLA‐DRB1*0401 were positive to synovial calpastatin. Seventy‐five percent of RA patients homozygous for HLA‐DRB1*0404 allele were positive to synovial calpastatin. Finally, 50% of RA patients expressing one HLA‐DRB1*0404 allele, but only 35% of RA patients expressing HLA‐DRB1*0401 and 25% of RA patients expressing HLA‐DRB1*0101, had autoantibodies to synovial calpastatin.
Table 2 HLA‐DRB1 genotypes and antisynovial calpastatin positivity.
| HLA‐DRB1* | n | F | % | p |
|---|---|---|---|---|
| 0401/0404 | 6 | 5 | 83 | 0.002 |
| 0404/0404 | 4 | 3 | 75 | 0.028 |
| 0401/0101 | 15 | 11 | 73 | <0.001 |
| 0401/0401 | 3 | 2 | 67 | 0.142 |
| 0101/0101 | 7 | 4 | 57 | 0.044 |
| 0404/SE– | 16 | 8 | 50 | 0.018 |
| 0401/SE– | 23 | 8 | 35 | 0.131 |
| 0101/SE– | 32 | 8 | 25 | 0.425 |
| SE– | 35 | 5 | 14 |
p versus SE− group; n, number of patients; F, positive frequency.
One linear B cell epitope in calpastatin is associated with HLA‐DRB1*0404 allele
To map B cell epitopes in calpastatin, we synthesised 94 overlapping 15 mer peptides encompassing the five domains of calpastatin. In a first step, we screened these peptides with the sera of 17 RA patients expressing different HLA‐DR alleles. Among the 94 overlapping peptides, only a few peptides are recognised by the sera of RA patients. However, we observed that HLA‐DRB1*0404 sera preferentially detected one calpastatin peptide, peptide 28 from domain 2 (data not shown/supplemental file: Figure B).
To confirm these reactivities, the sera of 126 patients and 82 controls were tested by ELISA using peptide 28 as immunosorbent. Autoantibodies against this peptide were found in more RA patients than controls (table 3). Forty‐four percent of RA patients were positive to peptide 28 compared with 14% of controls (p<0.001). No significant association was found between RA‐associated HLA‐DR alleles and antipeptide 28 positivity. There was only a trend toward association with HLA‐DRB1*0404. Indeed, 50% of HLA‐DRB1*0404 patients were positive to peptide 28.
Table 3 SE and anticalpastatin peptide positivity.
| Peptide 28 n (%) | |
|---|---|
| RA patients | 126 (44) |
| SE+/SE+ | 49 (59) |
| SE+/SE– | 51 (31) |
| HLA‐DRB1* | |
| 0404/SE– | 16 (50) |
| 0101/SE– | 18 (33) |
| 0401/SE– | 17 (12) |
| SE– | 26 (38) |
| Controls | 82 (14) |
n, number of patients.
Binding of calpastatin peptides to HLA‐DR alleles
The association between HLA‐DRB1*0404 and the production of anticalpastatin antibodies suggested that HLA‐DRB1*0404 could preferentially bind calpastatin peptides. The binding of purified HLA‐DRB1*0101, *0401, *0404 (RA‐associated alleles) and DRB1*0701 and *0402 (RA non‐associated alleles) was assayed on ELISA plates coated with 94 15 mer peptides encompassing the five domains of calpastatin.8 We used a known positive peptide from flu haemagglutinin to confirm positive binding.
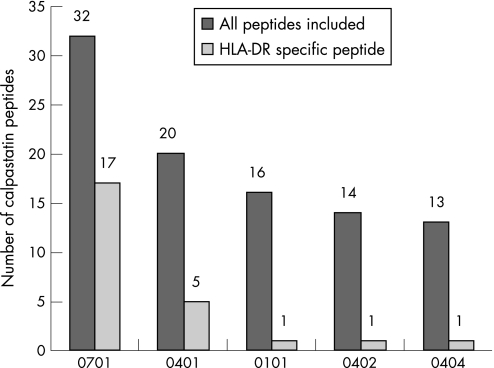
We found that every tested HLA‐DR allele was capable of binding peptides from calpastatin (fig 2). A total of 40 peptides bound to HLA‐DR alleles. The allele which bound the highest number of calpastatin peptides was non‐RA‐associated HLA‐DRB1*0701 (fig 3). The second best binder was HLA‐DRB1*0401. Surprisingly, HLA‐DRB1*0404 bound fewer calpastatin peptides than any other allele. Only one peptide from calpastatin specifically bound to HLA‐DRB1*0404. Peptides from domain 1 and 4 of calpastatin were the best HLA‐DR binders. Peptide 28, the B epitope associated with HLA‐DRB1*0404, bound every HLA‐DR allele tested (fig 2).
Figure 3 Binding of calpastatin peptides to HLA‐DR alleles. ELISA plates were coated with 10 µg of calpastatin peptide per well. One microgram of purified HLA‐DR molecule was then added to plates. After washing, bound HLA‐DR was detected by biotinylated anti‐HLA‐DR antibody followed by peroxidase conjugated avidin. The optical density was read at 405 nm. Positive binding was defined as an OD value higher than twice the OD for HLA‐DR without peptide. HLA‐DR specific peptide defined a peptide that binds only one HLA‐DR allele.
Discussion
Susceptibility to rheumatoid arthritis is carried by different HLA‐DRB1 alleles. RA‐associated HLA‐DRB1 chains share a highly conserved motif in their third hypervariable region. This motif called “shared epitope” is (according to the one letter amino acid code) QKRAA in HLA‐DRB1*0401, QRRAA in HLA‐DRB1*0101, *0102, *0404, *0405, *0408, *1402 and RRRAA in HLA‐DRB1*1001. Respective contribution of each allele to the pathogenesis of RA is not understood. In particular, individuals expressing both HLA‐DRB1*0401 and HLA‐DRB1*0404 are exposed to maximal risk to develop RA. In any case, RA‐associated HLA‐DR alleles could contribute to the development of RA by different mechanisms.
Previously, our group has observed unique functions of the QKRAA motif of HLA‐DRB1*0401:
It can constitute B and T cell epitopes on infectious agents.9,10
It can shape the T cell repertoire.11
It is over‐represented in protein databases.12
It is a binding motif for the bacterial 70‐kDa heat shock protein, dnaK.13
It is a binding motif for the human 70‐kDa heat shock protein hsp73, causing original trafficking of HLA‐DRB1*0401 in B cells. Binding of hsp73 to HLA‐DRB1*0401 may cause particular antigen processing and presentation in rheumatoid arthritis patients.14,15,16 The following mechanism can be suggested to explain how hsp73 contributes to rheumatoid arthritis in HLA‐DRB1*0401 patients. Hsp73 binds HLA‐DRB1*0401 and transports it directly from endoplasmic reticulum to lysosomes. Hsp73 binds autoantigens in lysosomes. Hsp73 may cause autoantigenic peptide loading on HLA‐DRB1*0401. T cell responses to autoantigenic peptides presented by HLA‐DRB1*0401 lead to autoimmunity.17 Evidence for this model was provided by Melchers, who demonstrated that processing of antigen is more efficient in cells that express HLA‐DRB1*0401. Efficient processing of antigen requires both hsp70 and HLA‐DRB1*0401. Abnormalities in the expression and distribution of hsp70 in rheumatoid arthritis patients have also been reported.18 Indeed, overexpression of hsp73 was observed in the synovial tissue of patients with rheumatoid arthritis and overexpression of hsp73 is known to enhance antigen processing.19 These data confirm that a special interaction between hsp73 and HLA‐DRB1*0401 can affect antigen processing or presentation in rheumatoid arthritis patients expressing HLA‐DRB1*0401 allele.
To identify new autoantigens in RA, we screened synovial proteins with sera of RA patients homozygous for HLA‐DR alleles. We found that sera of RA patients homozygous for HLA‐DRB1*0404 recognised a 100‐kDa synovial protein identified as calpastatin. RA patients' sera contain autoantibodies to citrullinated proteins (filaggrin, vimentin, fibrinogen and so on) which are specific for RA. Synovial calpastatin recognised by HLA‐DRB1*0404 patients is not citrullinated.
Calpastatin is a ubiquitous protein that specifically inhibits calpain, a calcium‐dependent cysteine protease involved in the development of inflammation. Calpain is overexpressed in rheumatoid synovial tissue, and calpain/calpastatin imbalance may be associated with cartilage destruction. Anticalpastatin antibodies have been identified in RA by several groups. However, epitope mapping in calpastatin remains controversial. Calpastatin contains an N‐terminal domain L and four calpain inhibition domains (domains 1, 2, 3 and 4). The N‐terminal fragment encompassing domain L and domain 1 (amino acids 1–188) is recognised by antibodies from RA patients and controls. The C‐terminal fragment (amino acids 682–708) of domain 4 is recognised by sera from patients with systemic rheumatic diseases.
In this study, we tested whether the HLA‐DR background may influence autoantibody production in RA patients. We found that RA‐associated HLA‐DR alleles are associated with the presence of autoantibodies to synovial calpastatin in RA patients' sera. The allele most strongly associated with antisynovial calpastatin production is HLA‐DRB1*0404. A peptide‐based ELISA with peptides encompassing the complete sequence of calpastatin was used to identify B cell epitopes. We detected one linear B cell epitope among patients expressing HLA‐DRB1*0404. In order to find T cell epitopes, we used a direct binding assay between purified HLA‐DR alleles and calpastatin peptides. We found that every tested HLA‐DR allele was capable of binding calpastatin peptides. No particular binding was detected with HLA‐DRB1*0404. We observed that peptides from domain 1 and 4 of calpastatin are the best HLA‐DR binders. Finally, when we measured T cell proliferative responses to calpastatin in 17 RA patients and 13 controls, we found that T cell proliferative responses to calpastatin are very frequent in RA patients. Seventy‐five percent of RA patients and 30% of controls have T cell responses to calpastatin, and HLA‐DR does not influence these proliferative response (data not shown). This results are consistent with the fact that either RA‐associated or RA non‐associated HLA‐DR alleles bind multiple peptides from calpastatin.
Our most striking finding is that HLA‐DRB1*0404 is strongly associated with anticalpastatin antibodies in rheumatoid arthritis. Fifty percent of RA patients expressing HLA‐DRB1*0404 have autoantibodies to synovial calpastatin. We recently observed that HLA‐DRB1*0404 is also associated with anticitrullinated fibrinogen in RA sera. Indeed, 83% of RA patients expressing HLA‐DRB1*0404 have autoantibodies to citrullinated fibrinogen.8 Moreover, we also observed that HLA‐DRB1*0404 is associated with anticyclic citrullinated peptide (anti‐CCP) antibodies in our RA patients (unpublished data). These data support the idea that HLA‐DRB1*0404 is associated with many autoantibody responses.
In the case of antibody response to citrullinated fibrinogen, we found that HLA‐DRB1*0404 binds more fibrinogen peptides than any other allele. Thus, we suggested that HLA‐DRB1*0404 is associated with anticitrullinated fibrinogen because, by binding more peptides, it can activate more helper T cells. However, this is quite different in the case of antibody response to synovial calpastatin. Indeed, HLA‐DRB1*0404 is not the allele that binds the highest number of peptides from calpastatin. This finding should lead us to reconsider the simple explanation, more peptide bound, more T cell help, which may be irrelevant to the association of HLA‐DRB1*0404 with a good antibody response.
In summary, HLA‐DRB1*0401 and HLA‐DRB1*0404 could act differently in the development of RA. HLA‐DRB1*0401, because it binds HSP70s, could be involved in particular autoantigen processing. HLA‐DRB1*0404 is associated with autoantibody production in RA patients, and this fact is not explained. These original functions carried by HLA‐DRB1*0401 and HLA‐DRB1*0404 may explain why the maximal risk and the severity to develop RA are present in RA patients expressing both HLA‐DRB1*0401 and HLA‐DRB1*0404.
Acknowledgements
Supported by grants from INSERM, Association pour la Recherche contre la Polyarthrite, Société Française de Rhumatologie.
Abbreviations
OD - optical density
RA - rheumatoid arthritis
SE - shared epitope
Footnotes
Competing interests: None declared.
References
- 1.Symmons D P M. What is rheumatoid arthritis? Br Med Bull 199551243–248. [DOI] [PubMed] [Google Scholar]
- 2.Gregersen P K, Silver J, Winchester R J. The shared epitope hypothesis: an approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum 1987301205–1213. [DOI] [PubMed] [Google Scholar]
- 3.Menard H A, El‐Amine M. The calpain–calpastatin system in rheumatoid arthritis. Immunol Today 199617545–547. [DOI] [PubMed] [Google Scholar]
- 4.Lackner K J, Schlosser U, Lang B, Schmitz G. Autoantibodies against human calpastatin in rheumatoid arthritis: epitope mapping and analysis of patient sera. Br J Rheumatol 1998371164–1171. [DOI] [PubMed] [Google Scholar]
- 5.Vittecoq O, Salle V, Jouen‐Beades F, Krzanowska K, Menard J F, Gayet A.et al Autoantibodies to the 27 C‐terminal amino acids of calpastatin are detected in a restricted set of connective tissue diseases and may be useful for diagnosis of rheumatoid arthritis in community cases of very early arthritis. Rheumatology (Oxford) 2001401126–1134. [DOI] [PubMed] [Google Scholar]
- 6.Saulot V, Vittecoq O, Salle V, Drouot L, Legoedec J, Le Loet X.et al Autoantibodies directed against the amino‐terminal domain I of human calpastatin (ACAST‐DI Ab) in connective tissue diseases. High levels of ACAST‐DI Ab are associated with vasculitis in lupus. J Autoimmun 20021955–61. [DOI] [PubMed] [Google Scholar]
- 7.Iwaki‐Egawa S, Matsuno H, Yudoh K, Nakazawa F, Miyazaki K, Ochiai A.et al High diagnostic value of anticalpastatin autoantibodies in rheumatoid arthritis detected by ELISA using human erythrocyte calpastatin as antigen. J Rheumatol 20043117–22. [PubMed] [Google Scholar]
- 8.Auger I, Sebbag M, Vincent C, Balandraud N, Guis S, Nogueira L.et al Influence of HLA‐DR genes on the production of rheumatoid arthritis‐specific autoantibodies to citrullinated fibrinogen. Arthritis Rheum 2005523424–3432. [DOI] [PubMed] [Google Scholar]
- 9.Albani S, Tuckwell J E, Esparza L, Carson D A, Roudier J. The susceptibility sequence to rheumatoid arthritis is a cross‐reactive B cell epitope shared by Escherichia coli heat shock protein dnaJ and Histocompatibility leukocyte antigen DRB1*0401 molecule. J Clin Invest 199289327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roudier J, Petersen J, Rhodes G, Luka J, Carson D A. Susceptibility to rheumatoid arthritis maps to a T cell epitope shared by the HLA‐Dw4DRB1 chain and the Epstein barr virus glycoprotein gp110. Proc Natl. Acad Sci USA 1989865104–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salvat S, Auger I, Rochelle L, Begovich A, Geburher L, Sette A.et al Tolerance to a self‐peptide from the third hypervariable region of HLA‐DRB1*0401 in rheumatoid arthritis patients and normal subjects. J Immunol 19941535321–5329. [PubMed] [Google Scholar]
- 12.Roudier C, Auger I, Roudier J. Molecular mimicry reflected through database screening: serendipity or survival strategy. Immunol Today 199617357–358. [DOI] [PubMed] [Google Scholar]
- 13.Auger I, Roudier J. A function for the QKRAA amino acid motif: mediating binding of dnaJ to dnaK. Implications for the association of rheumatoid arthritis with HLA‐DR4. J Clin Invest 1997991818–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auger I, Escola J M, Gorvel J P, Roudier J. HLA‐DR4 and HLA‐DR10 motifs that carry susceptibility to rheumatoid arthritis bind 70 kD heat shock proteins. Nat Med 19963306–310. [DOI] [PubMed] [Google Scholar]
- 15.Auger I, Roudier J. Heat shock proteins, HLA‐DR and rheumatoid arthritis. Nat Med 199841210–1211. [DOI] [PubMed] [Google Scholar]
- 16.Auger I, Lepecuchel L, Roudier J. Interaction between HSP73 and HLA‐DRB1 alleles associated or not with rheumatoid arthritis. Arthritis Rheum 200246929–933. [DOI] [PubMed] [Google Scholar]
- 17.Auger I, Roudier J. Interaction between HSP73 and HLA‐DRB1*0401: implications for the development of rheumatoid arthritis. Immunol Res 200531261–266. [DOI] [PubMed] [Google Scholar]
- 18.Roth S, Willcox N, Rzepka N, Mayer M P, Melchers I. Major differences in antigen‐processing correlate with a single Arg 71<‐>Lys in HLA‐DR predisposing to rheumatoid arthritis and their selective interactions with 70 kD heat shock protein chaperones. J Immunol 20021693015–3020. [DOI] [PubMed] [Google Scholar]
- 19.Schick C, Arbogast M, Lowka K, Rzepka R, Melchers I. Continuous enhanced expression of hsc70 but not hsp70 in rheumatoid arthritis synovial tissue. Arthritis Rheum 20045088–93. [DOI] [PubMed] [Google Scholar]