Abstract
Objective
To investigate whether epigenetic mechanisms can regulate leptin's expression and affect its downstream targets as metalloproteinases 3,9,13 in osteoarthritic chondrocytes.
Methods
DNA methylation in leptin promoter was measured by DNA bisulfite sequencing, and mRNA expression levels were measured by real‐time quantitative PCR in osteoarthritic as well as in normal cartilage. Osteoarthritic articular cartilage samples were obtained from two distinct locations of the knee (n = 15); from the main defective area of maximum load (advanced osteoarthritis (OA)) and from adjacent macroscopically intact regions (minimal OA). Using small interference RNA, we tested if leptin downregulation would affect matrix metalloproteinase (MMP)‐13 activity. We also evaluated the effect of the demethylating agent, 5′‐Aza‐2‐deoxycytidine (AZA) and of the histone deacetylase inhibitor trichostatin A (TSA) on leptin expression in chondrocyte cultures. Furthermore, we performed chromatin immunoprecipitation in leptin's promoter area.
Results
We found a correlation between leptin expression and DNA methylation and also that leptin controls MMP‐13 activity in chondrocytes. Leptin's downregulation with small interference RNA inhibited MMP‐13 expression dramatically. After 5‐AZA application in normal chondrocytes, leptin's methylation was decreased, while its expression was upregulated, and MMP‐13 was activated. Furthermore, TSA application in normal chondrocyte cultures increased leptin's expression. Also, chromatin immunoprecipitation in leptin's promoter after TSA treatment revealed that histone H3 lysines 9 and 14 were acetylated.
Conclusion
We found that epigenetic mechanisms regulate leptin's expression in chondrocytes affecting its downstream target MMP‐13. Small interference RNA against leptin deactivated directly MMP‐13, which was upregulated after leptin's epigenetic reactivation, raising the issue of leptin's therapeutic potential for osteoarthritis.
Osteoarthritis (OA) is a complex disease with genetic, mechanical and environmental components leading to the destruction of the articular cartilage.1 As articular chondrocytes seem to be involved in the initiation and progression of osteoarthritis, a detailed study of the molecular changes that occur in chondrocytes during the development of osteoarthritis is of utmost importance.2
Recent studies have shown that several genes are up‐ or downregulated in osteoarthritic chondrocytes compared with normal chondrocytes,3,4 suggesting that osteoarthritic chondrocytes possess a modulated phenotype.5 However, the regulatory mechanisms responsible for these alterations have not yet been clarified. It is abundantly clear that gene expression is regulated by genetic alterations such as mutations and by epigenetic mechanisms such as alterations in the DNA methylation status, covalent modifications of core nucleosomal histones and rearrangement of histones.6,7 DNA hypermethylation and histone hypoacetylation are hallmarks of gene silencing, while DNA hypomethylation and acetylated histones, specifically in histone H3 at lysines 9 and 14, promote active gene transcription.8,9,10,11 Epigenetic gene deregulation has been studied extensively in cancer and leukemia,12,13,14,15 while there are very limited studies suggesting that epigenetic gene regulation contributes to the pathology of non‐neoplastic diseases as osteoarthritis.16,17 Since epigenetic alterations are reversible, the possibility of reversing epigenetic marks, in contrast to genetic code, may provide new molecular targets for emerging therapeutic intervention.17
Recently, we have shown that one of the genes that are upregulated in osteoarthritic chondrocytes is leptin (unpublished data). Leptin, a 16‐kDa non‐glycosylated protein product of the obese (ob) gene, is a cytokine‐like peptide hormone secreted mainly by white adipose tissue18 that has been found to act as a regulator of bone growth, inducing osteoblast proliferation, collagen synthesis, bone mineralisation and also stimulating endochondrial ossification.19,20,21,22,23 Recently, a key role for leptin has been demonstrated in osteoarthritis, as it has been shown that leptin exhibits, in concert with other pro‐inflammatory cytokines, a detrimental effect on articular cartilage by promoting nitric oxide synthesis in chondrocytes.24 Leptin's 3‐kb promoter region is embedded within a CpG island and contains many putative binding sites for known transcription factors, as Sp‐1 sites, cAMP response element, glucocorticoid response element and a functional CCAAT/enhancer binding protein (C/EBP‐α) site which contains a CG dinucleotide and is sufficient for tissue‐specific gene expression.25,26,27 It has been shown that leptin's promoter is subject to epigenetic programming, and leptin's expression can be modulated by DNA methylation, thus raising a number of biological and clinical questions.26,28
In the present study, we investigated for the first time whether epigenetic mechanisms can regulate leptin's expression levels in osteoarthritic and normal chondrocytes and how they affect its downstream targets. In addition, in order to better understand how epigenetic changes may relate to the pathogenesis of osteoarthritis we combined pharmacological inhibition of DNA methylation and histone acetylation in normal chondrocytes. Furthermore, using small interference RNA technology against leptin, we investigated its role on MMP‐13 regulation, thus testing its potential use as a molecular target for therapeutic intervention in osteoarthritis.
Materials and methods
Patients and cartilage samples
Articular cartilage samples were obtained from femoral heads, femoral condyles and tibial plateaus of patients with primary OA undergoing hip or knee replacement surgery at the Orthopaedics Department of the University Hospital of Larissa. A total of 15 patients were included in the study (5 male, 10 female; mean age 62.33 (SD 13.9) years, range 40–82; mean body mass index 29.85 (3.26), range 23.88–35.56). Two specimens were taken from every patient. Each sample was categorised according to its gross morphology, as either advanced OA cartilage, which was taken from the main defective area of maximal load, or minimal OA taken from areas with no obvious surface defects. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the Local Ethical Committee of the University Hospital of Larissa.
DNA methylation studies
DNA was extracted from articular cartilage using a DNA extraction kit according to the manufacturer's instructions (Qiagen, Chatsworth, CA). DNA was treated with sodium bisulfite using the CpGenome DNA modification Kit (Invitrogen, Life Technologies, Paisley, UK) according to the manufacturer's protocol. PCRs were performed on modified DNA using primers on leptin promoter area previously described (A). PCR products have been purified by agarose gel electrophoresis and inserted for sequencing into a pGEM‐T Easy vector (Promega, Madison, WI). Plasmids were isolated using the Qiagen plasmid kit and custom‐sequenced using standard sequence primers.
Isolation of RNA from human cartilage
Fresh tissue, within 1 h from surgery, was dissected, and total cellular RNA was extracted using Trizol reagent (Invitrogen, Life Technologies). RNA was further purified using an RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturers' instructions. Preservation of 28S and 18S rRNA species was used to assess RNA integrity. All the samples included in the study were with prominent 28S and 18S rRNA components. The yield was quantified spectrophotometrically.
Real‐time PCR of leptin and MMP‐13 mRNA
Transcription of 0.1 μg RNA to cDNA was performed using the AMV Kit (Roche, Indianapolis, IN). Retinoic acid receptor alpha cDNA sequences (RARα) were amplified in separate reactions as positive cDNA controls. LightCycler‐FastStart DNA master SYBR Green, which contains Taq DNA polymerase, dNTP mix, SYBR Green 1 dye and MgCl2 (Roche), was used as a reaction mix for PCR. LightCycler Primer set (Roche, Indianapolis, IN) was used for the primers for porphobilinogen deaminase (PBGD) as a housekeeping gene. Standard PBGD cDNAs with known copy numbers were included in the LightCycler Primer set. Quantification was performed by real‐time PCR (Light Cycler Instrument, Roche Molecular Systems, Alameda, CA) in 0.2 μl of cDNA for each analysed sample using the LightCycler FastStart DNA Master HybProbe Kit (Roche, Penzberg, Germany) according to the manufacturers' instructions. The oligonucleotide primers used for leptin were 5′‐TTCTTGTGGCTTTGGCCCTA‐3′ (forward, 81–100 in exon 2) and 5′‐GGAGACTGACTGCGTGTGTGTGAA‐3′ (reverse, 191–212 in exon 2), and for MMP‐13 forward, 5′‐CGC CAG AAG AAT CTG TCT TTA AA‐3′, and reverse, 5′‐CCA AAT TAT GGA GGA GAT GC‐3′. All samples were analysed in duplicate, and the average value of the duplicates was used for quantification. The variation in the two measurements for each sample was generally in a range of 0.1–10%. If the variation of a sample exceeded 10%, a novel triplicate assay was carried out for this sample. The data were expressed as the ratio of the levels of the target gene mRNA on that of the housekeeping gene PBGD (leptin or MMP‐13 mRNA copies/PBGD copies), which was used as an internal control.
Primary cultures of human articular chondrocytes, normal and osteoarthritic
Articular cartilage was transported from the surgical room in HBSS (Hanks Balanced Salt Solution) medium, immediately dissected and subjected to sequential digestion with 1 mg/ml pronase (Roche Applied Science, Germany) for 90 min and 1 mg/ml collagenase P (Roche Applied Science, Germany) for 3 h at 37°C. Chondrocytes were counted and checked for viability using Trypan Blue staining. More than 95% of the cells were viable after isolation. Chondrocytes were then seeded in 6‐well plates with Dulbecco's Modified Eagles Medium/Ham's F‐12 (DMEM/F‐12) (GIBCO BRL, Paisley, UK) plus 5% fetal bovine serum (FBS) and 100 U/ml penicillin‐streptomycin, and were incubated at 37°C under a humidified 5% CO2 atmosphere. After reaching confluence, cells were cultured in serum‐deprived DMEM/F‐12 medium for 24 h and were subsequently stimulated with leptin.
Protein expression on chondrocytes
Osteoarthritic and normal cells were trypsinised, collected and centrifuged for 10 min at 1700 rpm. The cell pellet was washed with PBS and then centrifuged as previously. The cell pellet was lysed using Nonidet P‐40 lysis buffer containing 30 mM Tris (pH 7.5), 150 mM NaCl, 10% glycerol, 1% Nonidet P‐40 and a cocktail of protease inhibitors for 30 min on ice, followed by centrifugation for 15 min at 12 000 rpm. The supernatant was transferred in another Eppendorf and stored at −20°C. Protein concentration was quantified using the Bio‐Rad Bradford protein assay (Bio‐Rad, Hercules, CA) with bovine serum albumin as standard.
Western blot analysis
Cell lysates from normal and OA chondrocytes were electrophoresed and separated on a 4–20% Tris‐HCl gel (Bio‐Rad) and transferred to a Hybond‐ECL nitrocellulose membrane (Amersan Biosciences, Piscataway, NJ). The membrane was probed with antileptin (1:2000 dilution) (Sigma, St Louis, MO), and signals were detected using antirabbit immunoglobulin IgG conjugated with horseradish peroxidase (1:5000 dilution). The nitrocellulose membranes were then exposed to photographic film, which was scanned, and the intensities of the protein bands, which were expressed as arbitrary units (a.u.), were determined by computerised densitometry.
siRNA expreriments
Chondrocytes were seeded in 6‐well plates and transfected with 50 nM siRNA against leptin (Ambion, Austin, TX) using SiPORT transfection agent. SiPORT is a lipid transfection agent consisting of a mixture of lipids that spontaneously complex small interference RNA and facilitate its transfer to the chondrocytes. Transfection with 50 nM scrambled siRNA was used as a control. No cell toxicity was detected due to the transfection agent. Protein was extracted 24 and 48 h after siRNA transfection, and a western blot analysis for leptin was performed as described above.
In addition, at the same time points, mRNA was extracted, and a real‐time PCR analysis was performed for MMP‐13 expression. In both cases, glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) levels were used as loading control.
DNA methyltransferase and histone deacetylase inhibitors treatment of chondrocytes
Cells were seeded in 100‐mm plates and were treated with 1, 2, 5 and 10 μM of the demethylating agent, 5‐Aza‐2‐deoxycytidine (Sigma) for 2–5 days and/or the histone deacetylase inhibitor, trichostatin A (Sigma) for 1 day (1 μM concentration). DNA and RNA were extracted, and DNA methylation and mRNA expression studies were performed as described above.
Chromatin immunoprecipitation analysis
Cells were seeded in 100‐mm plates and were treated with different doses (0.2, 0.5 and 1 μM) of trichostatin A. Chromatin immunoprecipitation was performed using the Acetul‐Histone H3 Immunoprecipitation (ChIP) Assay Kit (UpState Biotechnology, New York) according to the manufacturer's protocol. PCR was performed in leptin's promoter, and the PCR products were analysed in an agarose gel. Values were normalised according to the input.
Statistical analysis
All calculations were performed on a Microsoft computer, using the SPSS software (version 11.0). Data were analysed by unpaired t test, paired t test as well as analysis of variance (ANOVA). Correlation coefficients were calculated by Pearson rank correlation (r) and Spearman rank correlation where applicable.
Results
DNA methylation status of leptin promoter and expression levels in normal and osteoarthritic chondrocytes
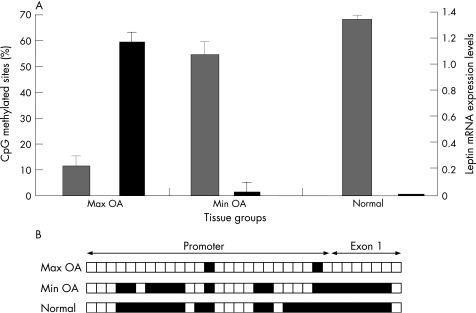
We observed that leptin was almost not expressed (as assessed by real‐time PCR) and was highly methylated (68%) in normal chondrocytes (fig 1). In Min OA chondrocytes, leptin was expressed in very low levels and was methylated (54%), while in Max OA chondrocytes, leptin was highly expressed and had very low levels (12%) of promoter methylation. Overall an association was found between leptin's DNA methylation and expression levels, suggesting leptin's epigenetic regulation in osteoarthritis.
Figure 1 Correlation between DNA methylation and leptin expression in normal, minimally affected (min OA) and max OA chondrocytes.
Leptin downregulation by siRNA affects MMP‐13 expression levels
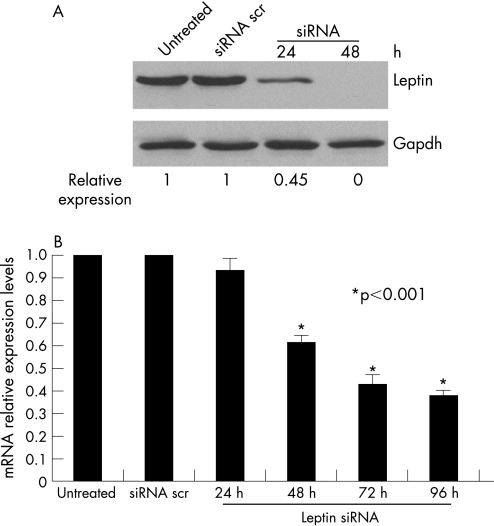
We transfected osteoarthritic chondrocytes using siRNA against leptin and monitored leptin's protein expression by western blot analysis. We found that leptin was around 65% downregulated 24 h after transfection and was completely shut off 48 h after siRNA transfection (fig 2A). We also observed that leptin's downregulation actually affected MMP‐13 expression levels (fig 2B). More specifically, 48 h after siRNA treatment against leptin, MMP‐13 expression was significantly reduced and continued to decrease even 96 h after treatment (p<0.001). We did not find any modulation of MMP‐3 or MMP‐9 expression levels after leptin downregulation (data not shown).
Figure 2 Leptin's downregulation in OA chondrocytes inhibits MMP‐13 activity. (A) Leptin's protein expression levels after siRNA treatment. Leptin was downregulated after 24 h of treatment and was completely suppressed after 48 h of 50 nM siRNA treatment. Treatment with siRNA scrambled was used as a control. GAPDH protein levels were used as a loading control. (B) MMP‐13 mRNA levels after leptin siRNA treatment. MMP‐13 levels were downregulated 48 h after leptin siRNA treatment and were highly downregulated after 72 h.
Epigenetic reactivation of leptin in chondrocytes
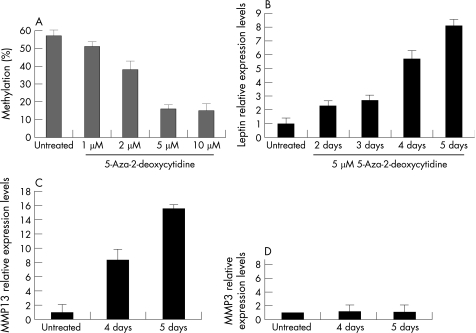
Normal chondrocytes were treated with different doses of 5‐Aza‐2‐deoxycytidine (fig 3A). We found that 1 and 2 μM of AZA were not able to alter the methylation status of leptin in normal chondrocytes. However, 5 and 10 μM of AZA were able to demethylate leptin's promoter drastically (fig 3A). We used the lower effective (5 μM) concentration of AZA, as it is known that in very high concentrations AZA has cytotoxic and not gene‐specific effects. AZA treatment of normal chondrocytes was time‐dependent (fig 3B). Two and 3 days of AZA treatment increased leptin mRNA expression 2–3‐fold, 4 days of treatment increased leptin expression 5‐fold, while 5 days of treatment had the highest effect, and leptin expression was found to be upregulated by 7‐fold. The downstream target MMP‐13 was found to be upregulated after leptin's epigenetic reactivation (fig 3C).
Figure 3 5‐Aza‐2‐deoxycytidine (AZA) treatment of normal chondrocytes. (A) AZA treatment demethylated leptin promoter area. 1 μM was not sufficient, but 5 μM AZA almost completely demethylated leptin's promoter area. (B) Treatment with 5 μM AZA in different time points. 5 days' treatment with AZA upregulated leptin levels (seven fold) suggesting that DNA methylation controls leptin expression levels. (C) Epigenetic upregulation of leptin activates MMP‐13. (D) Epigenetic upregulation of leptin does not affect MMP‐3.
Histone modifications contribute to leptin gene regulation in chondrocytes
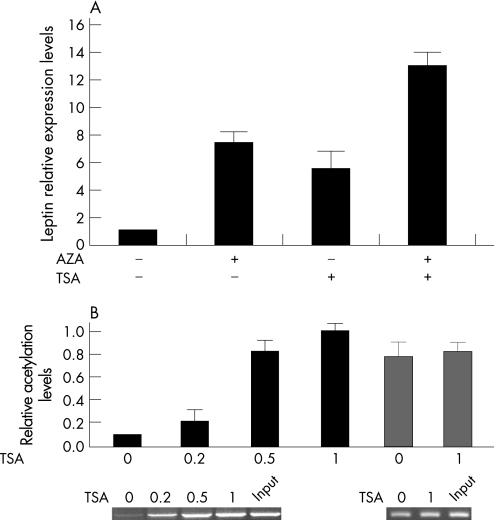
Treatment of normal chondrocytes with the histone deacetylase inhibitor trichostatin A increased leptin's expression 3‐fold. When normal chondrocytes were treated with 5 μM AZA for 5 days and 1 μM TSA for 1 day consecutively, we observed a synergistic effect resulting in a 13‐fold increase of leptin expression (fig 4A). We tested the acetylation levels of histone H3 and found that treatment with 0.5 and 1 μM of TSA increased H3 acetylation levels dramatically (fig 4B).
Figure 4 DNA methylation and histone acetylation regulate leptin expression in normal chondrocytes. (A) Treatment with 5 μM AZA for 5 days increased leptin expression seven fold, while 1 μM TSA for 1 day's treatment increased leptin expression six fold. AZA and TSA treatment had a synergistic effect (14‐fold) in leptin expression levels. (B) After TSA treatment, H3 K9/14 was found to be acetylated in the leptin promoter.
Discussion
In order to elucidate the signalling cascades in osteoarthritis, the regulatory mechanisms of genes altered in osteoarthritic chondrocytes need to be determined. Epigenetic mechanisms have been involved in gene regulation in carcinogenesis and in complex non‐Mendelian diseases as Alzheimer's, multiple sclerosis or atherosclerosis;29,30 however, data are scarce regarding specific genes that are epigenetically deregulated in the process of osteoarthritis.
As genomic methylation appears to be an important factor for tissue‐ and cell‐specific differentiation during chondroneogenesis,31 the methylation status of a gene appears to be important for its general activity in a defined tissue of cell type such as chondrocytes.31,32 In the present study, we showed, for the first time, that leptin can be regulated by epigenetic mechanisms in osteoarthritis. Leptin was found to be methylated in normal chondrocytes and umnethylated in mildy and severely affected osteoarthritic chondrocytes, and also its methylation status was correlated with its expression levels. In addition, using the chromatin immunoprecipitation approach, we found that leptin was also regulated by histone acetylation in its promoter region.
It is known that DNA methylation patterns change with increasing age, and age‐dependent hypomethylation may contribute to pathological processes.33 As osteoarthritis is an age‐related disorder, ageing may be one of the factors contributing to the loss of methylation in OA through altered expression or function of DNA methyltransferases and demethylases.34
In the present study, the importance of the observed heritable epigenetic alterations, namely DNA methylation and histone acetylation, regulating leptin's expression, lies in the fact that they are reversible and thus may have theraupeutic potential. DNA methylation can be inhibited by using DNA methyltransferase inhibitors, enzymes that inhibit the methyltransferases to methylate DNA. 5‐Aza‐2‐deoxycytidine is a potent DNA methyltransferase inhibitor which has been widely used in vitro, in animal models and recently in clinical trials in patients with melanoma, solid tumours and haematological malignancies.35,36,37 In addition, histone acetylation is also reversible using histone deacetylase inhibitors such as trichostatin A and SAHA which have also been used in clinical trials.38,39,40,41 We found that leptin's expression was highly upregulated in normal chondrocytes after AZA and TSA treatment. Previous studies have suggested that DNA methylation and histone modification collaborate in gene regulation.42 In the present study, we observed that the combined treatment of AZA and TSA in normal chondrocytes had a synergistic effect in increasing leptin's expression.
Until now, there have been very limited data regarding gene epigenetic regulation in osteoarthritis. Poschl et al43 could not associate hypermethylation with silencing of aggregan expression in osteoarthritis, while Roach et al17 found an association between methylation status and MMP‐9 expression for specific CpG sites. High‐through analysis of the genes that are epigenetically regulated in osteoarthritis, taking into account not only DNA methylation but also histone modifications, could be of great importance.
Furthermore, we found that leptin's epigenetic regulation affected its downstream catabolic target MMP‐13. In addition, treatment with siRNA against leptin transfected into osteoarthritic chondrocytes together with liposomes, downregulated MMP‐13 expression and had no effect on the expression levels of other metalloproteinases. The specificity of the interaction between leptin and MMP‐13 points towards a possible therapeutic potential of siRNA treatment against leptin for osteoarthritis treatment (fig 5).
Figure 5 Model for epigenetic and siRNA modulation of expression in chondrocytes. A gene can be shutoff in normal chondrocytes due to DNA methylation (grey circles) or deacetylation of histones. When the histones around the promoter area become acetylated, chromatin opens, and gene expression is starting. In addition to histone acetylation, DNA hypomethylation (white circles) has a high gene expression as as result. High gene expression (such as leptin) in osteoarthritic chondrocytes induces signalling pathways and specifically catabolic activity. MMP‐13 levels were increased, leading to cartilage destruction. Inhibition of MMP‐13 expression by siRNA against leptin transferred by liposomes inhibits the destruction of the cartilage.
Taking all our findings together, we showed, for the first time, that leptin is epigenetically regulated in osteoarthritis and that it directly affects MMP‐13 expression levels. We also propose, for the first time, that epigenetic therapy or targeted gene therapy using small interference RNA transferred with liposomes in chondrocytes could possibly have therapeutic potential for osteoarthritis treatment especially in early stages. However, further studies are required to fully understand the molecular profile of osteoarthritis.
Abbreviations
AZA - 5′‐Aza‐2‐deoxycytidine
MMP - matrix metalloproteinase
OA - osteoarthritis
TSA - trichostatin A
Footnotes
Competing interests: None declared.
References
- 1.Mahr S, Burmester G R, Hilke D, Gobel U, Grutzkau A, Haupl T.et al Cis‐ and trans‐acting gene regulation is associated with osteoarthritis. Am J Hum Genet 200678793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas C M, Fuller C J, Whittles C E, Sharif M. Chondrocyte death by apoptosis is associated with cartilage matrix degradation. Osteo Cart 2006;19 Jul [Epub ahead of print] [DOI] [PubMed]
- 3.Eleswarapu S V, Leipzig N D, Athanasiou K A. Gene expression of single articular chondrocytes. Cell Tissue Res 2006;31 Aug [Epub ahead of print] [DOI] [PubMed]
- 4.Burrage P S, Mix K S, Brinckerhoff C E. Matrix metalloproteinases: role in arthritis. Front Biosci 200611529–543. [DOI] [PubMed] [Google Scholar]
- 5.Forsyth C B, Cole A, Murphy G, Bienias J L, Im H J, Loeser R F., Jr Increased matrix metalloproteinase‐13 production with aging by human articular chondrocytes in response to catabolic stimuli. J Gerontol A Biol Sci Med Sci 2005601118–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meng J, Ma X, Ma D, Xu C. Microarray analysis of differential gene expression in temporomandibular joint condylar cartilage after experimentally induced osteoarthritis. Osteo Cart 2005131115–1125. [DOI] [PubMed] [Google Scholar]
- 7.Aigner T, Zien A, Hanisch D, Zimmer R. Gene expression in chondrocytes assessed with use of microarrays. J Bone Joint Surg Am 200385A(Suppl 2)117–123. [DOI] [PubMed] [Google Scholar]
- 8.Serman A, Vlahovic M, Serman L, Bulic‐Jakus F. DNA methylation as a regulatory mechanism for gene expression in mammals. Coll Antropol 200630665–671. [PubMed] [Google Scholar]
- 9.Villar‐Garea A, Imhof A. The analysis of histone modifications. Biochim Biophys Acta. 2006;26 Aug [Epub ahead of print] [DOI] [PubMed]
- 10.Schob H, Grossniklaus U. The first high‐resolution DNA “methylome”. Cell 20061261025–1028. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda H, Sano N, Muto S, Horikoshi M. Simple histone acetylation plays a complex role in the regulation of gene expression. Brief Funct Genomic Proteomic 20065190–208. [DOI] [PubMed] [Google Scholar]
- 12.Lu O, Oiu X, Hu N, Wen H, Su Y, Richardson B C. Epigenetics, disease and therapeytic interventions. Ageing Res Rev 20065449–467. [DOI] [PubMed] [Google Scholar]
- 13.Bhalla Kn Epigenetic and chromatin modifiers as targeted therapy of hematologic malignancies. J Clin Oncol 2005233971–3993. [DOI] [PubMed] [Google Scholar]
- 14.Stebbing J, Bower M, Syed N, Smith P, Yu Y, Crook T. Epigenetics:an emerging technology in the diagnosis and treatment of cancer. Pharmacogenomics 20067747–757. [DOI] [PubMed] [Google Scholar]
- 15.Galm O, Herman J G, Baylin S B. The fundamental role of epigenetics in hematologic malignancies. Blood Rev 2006201–13. [DOI] [PubMed] [Google Scholar]
- 16.Roach M I, AIgner T. DNA methylation in osteoarthritic chondrocytes: a new molecular target. Osteo Cart 2006;12 Aug [Epub ahead of print] [DOI] [PubMed]
- 17.Roach M I, Yamanda N, Cheung K S, Tilley S, Clarke N M, Oreffo R O.et al Association between the abnormal expression of matrix degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthr Rheum 2005523110–3124. [DOI] [PubMed] [Google Scholar]
- 18.Santos‐Reboucas C B, Pimentel M M. Implication of abnormal epigenetic patterns for human diseases. Eur J Hum Genet 2006;18 Oct [Epub ahead of print] [DOI] [PubMed]
- 19.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman J M. Positional cloning of the mouse obese gene and its human homologue. Nature 199472425–432. [DOI] [PubMed] [Google Scholar]
- 20.Steppan C M, Crawford D T, Chidsey‐Frink K L, Ke H, Swick A G. Leptin is a potent stimulator of bone growth in ob/ob mice. Regul Pept 20009273–78. [DOI] [PubMed] [Google Scholar]
- 21.Kume K, Satomura K, Nishisho S, Kitaoka E, Yamanouchi K, Tobiume S.et al Potential role of leptin on endochondral ossification. J Histochem Cytochem 200250159–169. [DOI] [PubMed] [Google Scholar]
- 22.Elefteriou F, Ahn Jong Deok, Takeda S, Starbuck M, Yang X, Liu X.et al Leptin regulation of bone resorption by the sympathetic nervous system and CART. Nature 2005434514–520. [DOI] [PubMed] [Google Scholar]
- 23.Elmquist J K, Strewler G J. Do neural signals remodel bone? Nature 2005434447–448. [DOI] [PubMed] [Google Scholar]
- 24.Gordeladge J O, Drevon C A, Syversen U, Reseland J E. Leptin stimulates human osteoblastic cell proliferation, de novo collagen synthesis, and mineralization: impact on differentiation markers, apoptosis, and osteoclastic signaling. J Cell Biochem 200285825–836. [DOI] [PubMed] [Google Scholar]
- 25.Gong D W, Bi S, Pratley R E, Weintraub B D. Genomic structure and promoter analysis of the humane obese gene. J Biol Chem 19962713971–3974. [DOI] [PubMed] [Google Scholar]
- 26.Stoger R. In vivo methylation patterns of the leptin promoter in human and mouse. Epigenetics 20061155–162. [DOI] [PubMed] [Google Scholar]
- 27.Miller S G, De Vos P, Guerre‐Millo M, Wong K, Herman T, Staels B.et al The adipocyte specific transcription factor C/EBPalpa modulates human ob gene expression. Proc Natl Acad Sci USA 1996935507–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melzner I, Scott V, Dorsch K, Fischer P, Wabitsch M, Bruderlein S.et al Leptin gene expression in human preadipocytes is switched on by maturation‐induced demethylation of distinct CpGs in its proximal promoter. J Biol Chem 200227745420–45427. [DOI] [PubMed] [Google Scholar]
- 29.Baylin S B, Esteller M, Rountree M R, Bachmann K E, Schuebel K, Herman J G. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet 200110687–692. [DOI] [PubMed] [Google Scholar]
- 30.Hiltunen M O, Yia‐Hertuala S. DNA methylation, smooth muscle cells and atherogenesis. Arterioscler Thromb Vasc Biol 2003231750–1753. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez M P, Young M F, Sobel M E. Methylation of type II and type I collagen genes in differentiated and dedifferentiated chondrocytes. J Biol Chem 19852602374–2378. [PubMed] [Google Scholar]
- 32.Aoyama T, Okamoto T, Nagayama S, Nishijo K, Ishibe T, Yasura K.et al Methylation in the core‐promoter region of the chondromodulin‐I gene determines the cell‐specific expression by regulating the binding of transcriptional activator Sp3. J Biol Chem 200427928789–28797. [DOI] [PubMed] [Google Scholar]
- 33.Richardson B. Impact of aging on DNA methylation. Ageing Res Rev 20032245–261. [DOI] [PubMed] [Google Scholar]
- 34.Richardson B C. Role of DNA methylation in the regulation of cell function: autoimmunity, aging and cancer. J Nutr 20021322401–2405. [DOI] [PubMed] [Google Scholar]
- 35.Gollob J A, Sciambi C J, Peterson B L, Richmond T, Thoreson M, Moran K.et al Phase I trial of sequential low dose 5′‐aza‐2′deoxycytidine plus high dose intravenous bolus interleukin‐2 in patients with melanoma or renal cell carcinoma. Clin Cancer Res 2006124619–4627. [DOI] [PubMed] [Google Scholar]
- 36.Schwartsmann G, Schunemann H, Gorini C N, Filho A F, Garbino C, Sabini G.et al A phase I trial of cisplatin plus decitabine, a new DNA hypomethylating agent in patients with advanced solid tumors and a follow‐up early phase II evaluation in patients with inoperable non‐small cell lung cancer. Invest New Drugs 2000183–91. [DOI] [PubMed] [Google Scholar]
- 37.Issa J P, Garcia‐Manero G, Giles F J, Mannari R, Thomas D, Faderl S.et al Phase I study of low dose prolonged exposure schedules of the hypomethylating agent 5‐aza‐2′deoxycitidine(decitabine) in hemotopoietic malignancies. Blood 20041031635–1640. [DOI] [PubMed] [Google Scholar]
- 38.Peixoto P, Lansiaux A. Histone deacetylases inhibitors:from TSA to SAHA. Bull Cancer 20069327–36. [PubMed] [Google Scholar]
- 39.Secrist J P, Zhou X, Richon V M. HDAC inhibitors for the treatment of cancer. Curr Opin Invest Drugs 200341422–1427. [PubMed] [Google Scholar]
- 40.Mei S, Ho A D, Mahlknecht U. Role of histone deacetylase inhibitors in the treatment of cancer. Int J Oncol 2004251509–1519. [PubMed] [Google Scholar]
- 41.Zhong S, Fields C R, Su N, Pan Y X, Robertson K D. Pharmacologic inhibition of epigenetic modifications, coupled with gene expression profiling, reveals novel targets of aberrant DNA methylation and histone deacetylation in lung cancer. Oncogene. 2006;Oct 9[Epub ahead of print] [DOI] [PubMed]
- 42.Cameron E E, Bachman K E, Myohanen S, Herman J G, Baylin S B. Synergy of demethylation and histone deacetylase inhibition in the re‐expression of genes silenced in cancer. Nature Genet 199921103–107. [DOI] [PubMed] [Google Scholar]
- 43.Poschl E, Fidler A, Schmidt B, Kallipolitou A, Schmidt E, Aignew T. DNA methylation is not likely to be responsible for aggregan down regulation in aged or osteoarthritic cartilage. Ann Rheum Dis 200564477–480. [DOI] [PMC free article] [PubMed] [Google Scholar]